Abstract
The T2Rs belong to a multi-gene family of G-protein-coupled receptors responsible for the detection of ingested bitter-tasting compounds. The T2Rs are conserved among mammals with the human and mouse gene families consisting of about 25 members. In the present study we address the signalling properties of human and mouse T2Rs using an in vitro reconstitution system in which both the ligands and G-proteins being assayed can be manipulated independently and quantitatively assessed. We confirm that the mT2R5, hT2R43 and hT2R47 receptors respond selectively to micromolar concentrations of cycloheximide, aristolochic acid and denatonium respectively. We also demonstrate that hT2R14 is a receptor for aristolochic acid and report the first characterization of the ligand specificities of hT2R7, which is a broadly tuned receptor responding to strychnine, quinacrine, chloroquine and papaverine. Using these defined ligand–receptor interactions, we assayed the ability of the ligand-activated T2Rs to catalyse GTP binding on divergent members of the Gα family including three members of the Gαi subfamily (transducin, Gαi1 and Gαo) as well as Gαs and Gαq. The T2Rs coupled with each of the three Gαi members tested. However, none of the T2Rs coupled to either Gαs or Gαq, suggesting the T2Rs signal primarily through Gαi-mediated signal transduction pathways. Furthermore, we observed different G-protein selectivities among the T2Rs with respect to both Gαi subunits and Gβγ dimers, suggesting that bitter taste is transduced by multiple G-proteins that may differ among the T2Rs.
Keywords: bitter taste, G-protein-coupled receptor, in situ reconstitution, Gαi subunit, signal transduction, taste receptor cell
Abbreviations: AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride; GPCR, G-protein-coupled receptor; GTP[S], guanosine 5′-[γ-thio]triphosphate (GTPγS); PLC, phospholipase C; TRC, taste receptor cell
INTRODUCTION
Taste perception is mediated by specialized sensory cells, the TRCs (taste receptor cells) that are clustered in groups of 50–100 within taste buds present on the tongue and palate. Each TRC has a microvillar dendritic process that extends through a taste pore and is exposed to the environment. Ingested tastants interact with either ion channels or GPCRs (G-protein-coupled receptors) present on the microvilli of TRCs to initiate downstream signalling pathways (reviewed in [1]). Salty and sour tastes are evoked by ions, including sodium ions, and protons that directly interact with ion channels. In contrast, sweet and bitter-tasting compounds, as well as glutamate and other amino acids, activate GPCRs.
It is estimated from surveys of mouse and human genomic sequences that each species has about 25 different bitter receptor genes that constitute a multi-gene family referred to as the T2Rs, members of which display 21–90% amino acid identity [2–7]. Cell-based assays utilizing Gα16 to re-direct signalling to PLC (phospholipase C) and mobilization of intracellular calcium as well as in vitro reconstitution studies have been used to examine the ligand specificities of members of the T2R family [2,8–12]. To date, activating bitter ligands have been identified for two mouse receptors and seven human receptors.
In addition to bitter taste receptors, several G-proteins have been implicated in bitter taste transduction. G-proteins function as heterotrimers, which consist of a Gα subunit and a Gβγ dimer. Upon interaction with an activated receptor, an exchange of GDP for GTP occurs on the Gα subunit and dissociation of the heterotrimer ensues. The dissociated Gα subunit and Gβγ dimer are then competent to act independently to initiate downstream signalling pathways. There are several different Gα, Gβ and Gγ subunits, and, dependent on which G-proteins a given receptor couples to, different Gα-mediated and Gβγ-mediated downstream effectors and pathways can be activated. Therefore determining the G-protein selectivities of a given receptor is key to understanding its cellular transduction pathway. Gustducin is a taste-receptor-cell-specific Gα subunit that is highly homologous to the visual Gα subunit, transducin [13]. In vivo experiments with gustducin knockout mice [14] and in vitro biochemical experiments with mT2R5 and purified gustducin [9] have demonstrated that gustducin couples to the T2Rs and plays a key role in the transduction of bitter-tasting compounds. However, gustducin knockout mice retain some sensitivity to bitter substances, suggesting that bitter signal transduction may involve additional Gα subunits [14]. With respect to the Gβγ dimeric subunits involved in taste transduction, two possible candidates include Gβ1γ13 and Gβ3γ13, which are co-expressed with gustducin and the T2Rs in TRCs [15,16]. However, the ability of the T2Rs to couple to these Gβγ dimers has not been demonstrated.
In order to address the molecular basis for bitter taste transduction, we have used a quantitative in vitro assay of T2R–G-protein interactions [17,18]. Unlike cell-based assays, the in situ reconstitution assay is independent of the identities and actions of downstream effectors as it measures the first biochemical step in the GPCR signalling pathway, which is receptor-catalysed exchange of GDP for GTP on the Gα subunit. We examined the receptor–ligand interactions of four previously de-orphanized T2Rs, mT2R5, hT2R14, hT2R43 and hT2R47. In addition we define the first ligands for the hT2R7 receptor and demonstrate that it is a broadly tuned receptor activated by papaverine, chloroquine, strychnine and quinacrine. Each of these receptors is activated in vitro by micromolar concentrations of their respective ligands. Furthermore, we demonstrate that individual bitter receptors display different affinities for Gα and Gβγ subunits and different selectivities among divergent members of the Gαi family.
EXPERIMENTAL
Generation of rhodopsin T2R chimeric receptors
Chimeric receptors consisting of the first 39 amino acids of bovine rhodopsin fused in frame to the N-terminus of each of the receptors were generated using a bridge-overlap-extension PCR method [19]. Addition of an N-terminal rhodopsin signal sequence to other family 1 GPCRs, including mT2R5, has been demonstrated previously [9,20] to facilitate surface expression of these receptors without affecting their ability to respond to appropriate ligands. Briefly, using the bridge-overlap-extension PCR method, sequences to be spliced first are amplified independently using primers that contain complementary sequences encompassing the junction site. The two products are then mixed and amplified with primers from the desired 5′- and 3′-ends of the clone. Each chimeric PCR fragment was cloned into the baculoviral transfer vector, pVL1393 (BD Biosciences), and the production and use of the baculoviruses and the culturing of insect cells were performed according to standard manufacturer's protocols (BD Biosciences and Orbigen).
Membrane preparations
Log-phase Sf9 at a density of 106 cells/ml were infected with receptor-expressing baculoviruses at a multiplicity of infection of 2. Post-infection (60 h), P2 membranes from the infected cells were prepared as previously described [18]. Briefly, cells were harvested by centrifugation (500 g for 5 min at 25 °C), washed two times with PBS containing 100 μM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], and resuspended in a hypotonic homogenization solution [10 mM Mops (pH 7.4), 1 mM EGTA and 100 μM AEBSF] using 40 ml of solution per litre of infected cells. The cells were allowed to swell at 4 °C for 15 min then harvested and homogenized in a Dounce homogenizer (tight-fitting, 20 strokes). Cell debris and nuclei were removed by centrifugation (750 g for 10 min at 4 °C), and the P2 post-nuclear membranes collected from the supernatant by centrifugation at 48384 g for 30 min at 4 °C. Membranes enriched with the β2-adrenergic receptor were prepared using the same protocol from Sf9 cells infected for 48 h at a multiplicity of infection of 30 with a baculovirus encoding the β2-adrenergic receptor (gift of Dr Brian Kobilka, Department of Molecular and Cellular Physiology, Stanford University, Stanford, CA, U.S.A.).
Urea extraction of P2 membranes
Endogenous, membrane-associated GTP-binding proteins were removed from the P2 membranes by extraction with 7 M urea [21]. Briefly, the P2 membrane pellets were resuspended in homogenization solution plus 7 M urea. Following incubation on ice for 30 min, the membrane suspension was diluted with homogenization solution to lower the urea concentration to less than 4 M. Membranes were collected by centrifugation (142000 g for 30 min at 4 °C), washed with homogenization solution, and sedimented again (142000 g for 30 min at 4 °C). The washed membrane pellets were resuspended in homogenization solution plus 12% (w/v) sucrose and snap frozen and stored at −80 °C.
Western blot analysis of receptor expression
Membranes were prepared as described above. Total protein [3 μg, as determined by the BCA protein assay (Pierce Biotechnology)] from each sample was loaded per lane. Following separation by SDS/PAGE (4–20% gel), the samples were transferred onto a 0.2 μm nitrocellulose membrane by semi-dry transfer at 10 V for 1 h in transfer buffer [25 mM Tris/HCl (pH 8.5), 192 mM glycine and 10% methanol]. Immunoblotting was performed using a monoclonal antibody that recognizes the N-terminus of bovine rhodopsin [B6-30N, kindly provided by Dr Paul A. Hargrave (Department of Ophthalmology, University of Florida, Gainesville, FL, U.S.A.)]. Detection was performed by ECL (enhanced chemiluminescence; Pierce Biotechnology), and high-resolution chemiluminescent images were captured and analysed using a UVP bioimaging system and Labworks 4.0 software (UVP).
Purification of G-protein subunits
Bovine Gα transducin, bovine retinal Gβ1γ1 and squid Gαq were purified using previously published protocols [17,22–25]. Rat Gαs-s was expressed and purified from Escherichia coli as described by Graziano et al. [26]. Homogeneously myristoylated Gαi1 and Gαo were produced in E. coli strain JM109 co-transformed with a Gα-encoding plasmid and a plasmid-encoding yeast N-myristoyltransferase essentially as described previously [27]. The pQE6 plasmids containing rat Gαi1 and Gαo as well as the pBB131 plasmid with yeast NMT1 were a gift of Dr Maurine E. Linder (Department of Cell Biology and Physiology, Washington University School of Medicine, St Louis, MO, U.S.A.). Rhodopsin was isolated from retina of Sepia officinales essentially as described for Loligo forbesii [17].
GDP/GTP[S] (guanosine 5′-[γ-thio]triphosphate) exchange assay
The receptor-catalysed GDP/GTP[S] exchange on Gα was performed as described by Hartman and Northup [17] with slight modifications. Unless otherwise noted: urea-washed membranes containing ∼4–8 pmol of receptor were reconstituted on ice with bovine transducin and Gβ1γ1 subunits with or without test ligand in a total volume of 30 μl. Reaction solution (20 μl) was added to initiate the reactions. The combined solution contained 50 mM Mops (pH 7.5), 100 mM NaCl, 1 mM EDTA, 3 mM MgSO4, 1 mM DTT (dithiothrietol), 3 mg/ml BSA, 1 μM GTP[S] and 0.2–0.5 μCi [35S]GTP[S]. Reactions were allowed to proceed for 20 min at 30 °C to reflect initial rates of reaction and were terminated by the addition of 2 ml of ice-chilled solution B [20 mM Tris/HCl, 25 mM MgCl2 and 100 mM NaCl (pH 8.0)]. The terminated reaction was then filtered over nitrocellulose membranes on a vacuum manifold and washed four times with 2 ml each of ice-chilled solution B. Following drying, the radioactivity on the filters was determined using a liquid scintillation spectrometer. Due to their high spontaneous GTP-binding exchange rates, a competing nucleotide (1 μM GDP) was included in the reconstitution assays performed with Gαi1, Gαo and Gαs [28].
The following compounds were used in these studies: aristolochic acid, atropine, brucine, caffeic acid, caffeine, chloroquine, cycloheximide, denatonium, (−)-epicatechin, naringin, papavarine, PTC (phenyl thiocarbamide), quinacrine, salicin, strychnine, SOA (sucrose octaacetate) and yohimbine. All were obtained from Sigma–Aldrich.
RESULTS AND DISCUSSION
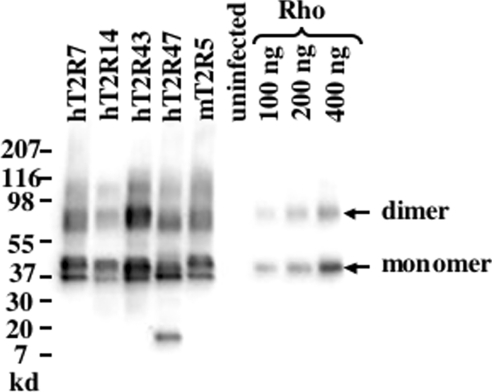
The five receptors presented in the present study, mT2R5, the mouse cycloheximide receptor, and four human bitter receptors, hT2R7, hT2R14, hT2R43 and hT2R47, were expressed as fusion proteins using a baculoviral expression system. To enhance surface expression and serve as an epitope tag, the N-terminal extracellular domain of bovine rhodopsin, consisting of 39 amino acids, was fused in frame to the N-terminus of each of the bitter receptors [20]. Baculovirus-infected Sf9 cells expressed the T2R constructs abundantly as determined by Western blot analysis of 7M urea-extracted P2 membranes (Figure 1). From the regression analysis of the bovine rhodopsin standards, we calculate that T2R expression, as estimated by rhodopsin-epitope tag immunoreactivity, ranged from 90 to 180 pmol/mg of total protein in the extracted P2 membrane fractions. Immunoreactivity of the extracellular rhodopsin epitope was readily detected in non-permeabilized Sf9 cells 60 h post-infection, indicating a plasma membrane localization of some portion of the T2R chimeric proteins (results not shown). No rhodopsin immunoreactivity was observed with P2 membranes isolated from uninfected Sf9 cells.
Figure 1. Western blot analysis of T2R expression.
Western blot analyses of urea-washed P2 membranes from uninfected Sf9 cells or cells infected with baculoviruses expressing the Rho-T2R chimeric receptors were performed to examine expression levels. Detection was performed with a monoclonal antibody (B6-30N) that recognizes the N-terminus of bovine rhodopsin. Total membrane protein (3 μg) or 30, 60 or 120 fmol of purified bovine rhodopsin (Rho) was loaded per lane. Electrophoresis and visualization was performed as described in the Experimental section. As is the case for most family 1 GPCRs, including rhodopsin and the expressed T2Rs, polymerization occurs in the presence of SDS resulting in the appearance of bands representing different states of polymerization. The integrated intensities of the monomer and dimer rhodopsin bands were used to establish a calibration curve, and the abundance of each T2R was determined from the regression line. The summated intensities of the various T2R oligomer bands were used to estimate the relative rhodopsin immunoreactivity and abundance of each T2R. Molecular mass markers are indicated.
To screen for agonist ligands, the urea-extracted, receptor-expressing membranes were reconstituted in vitro by the addition of purified transducin and Gβ1γ1 and examined for the ability of bitter-tasting compounds to stimulate receptor-catalysed exchange of GDP for [35S]GTP[S] on transducin. Transducin, which is highly related to gustducin, was used in these assays because gustducin is poorly expressed in heterologous systems, and its purification in quantities necessary for our ligand screening purposes was not feasible. In contrast, transducin as well as Gβ1γ1 can be purified in relatively large quantities from bovine retinal rod outer segment discs using established protocols [22–25]. Furthermore, in vitro findings demonstrate that transducin and gustducin are functionally identical with respect to their coupling properties to rhodopsin and Gβ1γ1, their ability to activate cyclic GMP-phosphodiesterase and their intrinsic GTPase activity [29]. Previous studies have also demonstrated that transducin effectively couples to the T2Rs [11].
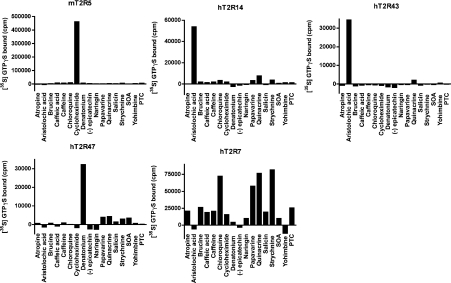
Sf9 membranes expressing mT2R5, hT2R14, hT2R43 or hT2R47, as well as the orphan receptor hT2R7, were reconstituted with transducin and Gβ1γ1, and the receptor-catalysed exchange of GDP for GTP[S] for each of the receptors was tested with a panel of 17 bitter-tasting compounds assayed at two concentrations, 100 μM and 1 mM. The responses of the receptor to the test compounds at 100 μM are shown in Figure 2. The values presented are the enhancement of GTP[S] binding to transducin in the presence of the indicated bitter-tasting compound. As previously reported and under our experimental conditions, mT2R5 specifically responded to cycloheximide, hT2R43 to aristolochic acid and hT2R47 to denatonium. In addition, hT2R14 specifically catalysed GDP/GTP[S] exchange on transducin in the presence of aristolochic acid, a previously unreported ligand for this receptor. To identify additional ligand–receptor interactions, we produced Sf9 membranes enriched in several ophan T2Rs and tested the ability of these same compounds to activate them. This allowed us to determine the ligand specificity of hT2R7. In contrast with the other responsive receptors, hT2R7 displayed a much broader response profile, displaying activation by at least four structurally unrelated compounds, chloroquine, papavarine, quinacrine and strychnine. Although hT2R7 appears to be particularly broadly tuned, it should be noted that additional ligands have been reported for the receptors tested here using different assay conditions and/or different panels of bitter compounds [8,10,11].
Figure 2. Ligand specificities of the T2Rs.
mT2R5-, hT2R7-, hT2R14-, hT2R43- or hT2R47-expressing membranes were reconstituted with 1 μM transducin and 1 μM Gβ1γ1 in the presence of 100 μM of the indicated test tastant, and GTP[S] (GTPγS) binding assays performed. [35S]GTP[S] binding determinations in the absence of test ligand were subtracted from previously reported values.
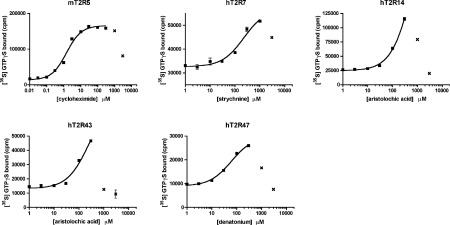
Representative concentration–response curves for the various receptor–ligand interactions are presented in Figure 3. Each of the receptors was half maximally activated by micromolar concentrations of its respective ligand. However, high concentrations of ligands profoundly inhibited the in vitro GTP[S] binding reactions. All three compounds also inhibited rhodopsin-catalysed GDP/GTP exchange on transducin indicating a non-competitive mechanism of inhibition that is independent of the receptor being tested. This observed inhibition raises the possibility that under our experimental conditions other bitter-tasting compounds may be inhibitory at concentrations well below their KD values, in which case receptor-activation would not be observable.
Figure 3. The T2Rs display micromolar sensitivities to bitter ligands.
Ligand saturations of mT2R5-, hT2R7-, hT2R14-, hT2R43- and hT2R47-catalysed [35S]GTP[S] (GTPγS) bindings are shown. Receptor-expressing membranes were reconstituted with 1 μM transducin and 1 μM Gβ1γ1 in the presence of the indicated concentrations of either cycloheximide (mT2R5), strychnine (hT2R7), aristolochic acid (hT2R14 and hT2R43) or denatonium (hT2R47). The data are averages of duplicate determinations from a representative experiment. The solid lines shown are the best fits for the data to single-site binding models. Data points (indicated by ×) at which ligand concentrations were inhibitory were excluded from the curve fitting analyses.
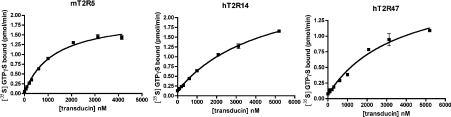
Despite this possible limitation, we were able to characterize five hT2R–ligand interactions using the in vitro system. A unique advantage of the in situ reconstitution system is the ability to manipulate each component independently. Therefore using these defined ligand–receptor interactions, we can quantitatively assess the G-protein-coupling properties of these receptors. To determine the affinities of the T2Rs for transducin, we chose to characterize mT2R5 and two representatives of the hT2R family. The binding affinities of these three T2Rs for transducin were determined by saturation analyses in the presence of saturating amounts of Gβ1γ1. These analyses yielded Km values of 1.2±0.1 μM, 3.6±1.3 μM and 2.6±1.4 μM for mT2R5, hT2R14 and hT2R47 respectively, indicating differences in the binding affinities for transducin among the T2R receptors (Figure 4 and Table 1).
Figure 4. The T2Rs display different binding affinities for transducin.
Transducin saturations of mT2R5-, hT2R14- and hT2R47-catalysed [35S]GTP[S] (GTPγS) binding are shown. The T2R-expressing membranes were reconstituted with the indicated concentrations of transducin in the presence of 2 μM Gβ1γ1 and activating ligands. The following concentrations of ligands were used: 30 μM cycloheximide for mT2R5, 200 μM aristolochic acid for hT2R14 and 200 μM denatonium for hT2R47. The data are averages of duplicate determinations from a representative experiment. The curves shown are the best fits for the data to single-site binding models.
Table 1. Summary of affinity data for T2Rs.
Values are averages±difference of values obtained from two experiments, each of which was performed with duplicate samples.
| Transducin saturation | Gβγ saturation K1/2 (nM) | ||||
|---|---|---|---|---|---|
| Receptor | Ligand | Km (μM) | Vmax (pmol bound/min) | Gβ1γ1 | Gβ1γ13 |
| mT2R5 | Cycloheximide | 1.2±0.1 | 1.9±0.1 | 208±89 | 815±11 |
| hT2R14 | Aristolochic acid | 3.6±1.3 | 3.5±0.9 | 642±12 | 710±45 |
| hT2R47 | Denatonium | 2.6±1.4 | 1.6±0.4 | 252±45 | 876±12 |
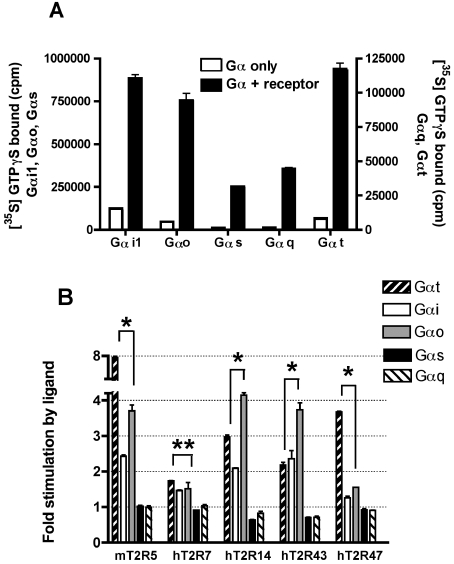
There are ∼20 different Gα subunits that fall into four major subfamilies, Gαs, Gα12, Gαi and Gαq. We examined the coupling properties of the T2Rs to members of three of these families. The Gα subunits examined included three members of the Gαi subfamily, namely transducin, Gαi1 and Gαo, as well as the more distantly related Gα subunits, Gαs and Gαq. The activities and concentrations of the purified Gα subunits were determined by reconstitution with Sepia rhodopsin, which couples effectively with transducin, Gαi1, Gαo and Gαq, or with β2-adrenergic receptor, which couples effectively with Gαs (Figure 5A). The reconstitutions of the T2Rs were performed with 200 nM of the indicated Gα subunit and saturating amounts of Gβ1γ1, in the presence or absence of activating ligand (Figure 5B). The T2Rs displayed ligand-dependent catalysis of GTP[S] binding to each of three Gαi subunits tested. For example, agonist-stimulated activity of mT2R5 was 2-, 4- and 8-fold on Gαi1, Gαo and transducin respectively, and activated hT2R14 catalysed the exchange of GDP for GTP 2-, 4- and 3-fold on Gαi1, Gαo and transducin respectively. hT2R7 coupled with approximately equal activity with all the Gαi subunits tested. However, for hT2R7 all four of the identified ligands activated the receptor with relatively low efficacy as compared with the activation of mT2R5 by cycloheximide. Overall, comparisons of the ratios of activities observed in the presence and absence of agonists for the other tested receptors indicate that mT2R5 and hT2R47 coupled most effectively with transducin, whereas hT2R14 and hT2R43 coupled most effectively with Gαo. These results demonstrate that members of the T2R family are capable of signalling through multiple Gαi subunits and that they differentially couple to the various Gαi subunits tested.
Figure 5. G-protein selectivity of T2Rs among members of the Gαi family.
(A) The activity of each purified Gα subunit was confirmed using either Sepia rhodopsin or, in the case of Gαs, Sf9-expressed β2-adrenergic receptor in the presence of 1 μM isoproterenol. Robust receptor-catalysed GDP/GTP exchange was observed for each Gα subunit tested. (B) Urea-washed membranes from mT2R5-, hT2R7-, hT2R14-, hT2R43- and hT2R47-expressing Sf9 cells containing approximately 8 pmol of rhodopsin epitope were incubated with 200 nM of the indicated Gα subunit and 1 μM Gβ1γ1 in the absence or presence of activating ligand (300 μM aristolochic acid for hT2R14 and hT2R43, 30 μM cycloheximide for mT2R5, 300 μM denatonium for hT2R47 and 300 μM strychnine for hT2R7). Receptor activation of Gαi1, Gαo and Gαs was measured using a competitive GDP reaction mixture for 4 min at 30 °C, while receptor activation for transducin (Gαt) and Gαq was measured with 1 μM GTP[S] (GTPγS) for 20 min at 30 °C. Data presented are fold-increases in GTP[S] binding comparing reactions which included ligand and those without ligand. *P<0.005; **P=0.2 (unpaired t test).
Although this result differs from the previously published report [9] of the exclusive activation of gustducin by mT2R5, the ability of the T2Rs to couple to multiple Gα subunits is consistent with studies of gustducin knockout mice [14,30]. Gustducin null mice retain a residual ability to respond to bitter-tasting compounds, and transgenic expression of transducin in TRCs only partially complements this deficiency, suggesting that in addition to gustducin and transducin the T2Rs couple in vivo to other Gα subunits. Furthermore, direct analyses of taste cells indicate that only half of the cells that responded to bitter compounds also expressed gustducin, strongly supporting the proposal for gustducin-independent bitter taste signalling pathways [31,32].
In contrast with the Gαi subunits, none of the receptors catalysed the GDP/GTP exchange on either Gαs or Gαq, suggesting that neither of these subunits play a role in the transduction of bitter taste. We note that the rat Gαs used in these studies is >99% identical and 100% similar at the amino acid level to the human Gαs, whereas the amino acid sequences of squid Gαq and human Gαq are 77% identical and 88% similar. Although unlikely in the case of Gαs, we cannot rule out the possibility that species differences may contribute to the lack of coupling observed between the hT2Rs and squid Gαq. However, thus far, of the 12 rodent and human GPCRs that we have tested which are known to signal through Gαq pathways, all but one have efficiently coupled in vitro with squid Gαq ([17,18,21,33,34], and J. Gutierrez, C. Chen, E. Hulme and J. K. Northup, unpublished work).
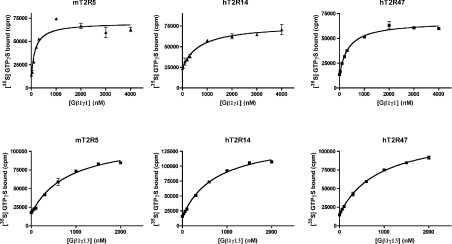
Our results suggest that in vivo the T2Rs selectively signal through Gαi subunits, activation of which leads to decreases in cyclic nucleotide levels by either activating phosphodiesterases (transducin/gustducin) or inhibiting adenyl cyclases (Gαi). Consistent with these findings, bitter compounds have been shown to elicit decreases in cyclic nucleotide levels when applied to murine taste tissue [35]. In addition to changes in cyclic nucleotide levels, application of bitter tastants to taste tissue also triggers the synthesis of InsP3 through the activation of a TRC-expressed PLC, PLCβ2 [15,35,36]. This pathway is critical as genetic ablation of PLCβ2 results in pronounced bitter taste deficits [37]. PLCβ2 is regulated by both a Gαq-mediated pathway and a Gβγ-mediated pathway. Our failure to observe coupling of the T2R to Gαq consequently supports a major role for Gβγ-mediated regulation of PLCβ2 in bitter taste signal transduction. In this respect, the Gγ13 subunit is of particular interest. Gγ13 has been implicated in bitter taste signal transduction as it is expressed in TRCs, co-localizes with gustducin, and physically interacts with Gβ1, which is also expressed in TRCs [15]. To directly assess the abilities of the T2Rs to couple to Gβγ dimers, we determined their affinities for both Gβ1γ1 and the TRC-specific dimer, Gβ1γ13 (Figure 6 and Table 1). Reconstitution analyses indicate that the affinities of mT2R5 and hT2R47 for Gβ1γ1 are approximately the same (K1/2=208±89 and 252±45 nM), whereas the affinity of hT2R14 for Gβ1γ1 is 2- to 3-fold lower (K1/2=642±12 nM). The T2Rs also coupled to Gβ1γ13 dimers, with hT2R14 (K1/2=710±45 nM) showing a slightly higher affinity for Gβ1γ13 than either mT2R5 (K1/2=815±11 nM) or hT2R47 (K1/2=876±12 nM). Similar to their coupling properties to Gα subunits, the T2Rs differentially couple to Gβγ dimers. For example, among the receptors tested, hT2R14 displayed the highest affinity for Gβ1γ13, yet the lowest affinity for Gβ1γ1.
Figure 6. The T2Rs couple to both Gβ1γ1 and the TRC-specific dimer, Gβ1γ13.
Gβγ saturations of mT2R5-, hT2R14- and hT2R47-catalysed [35S]GTP[S] (GTPγS) binding are shown. The T2R-expressing membranes were reconstituted with the indicated concentrations of either Gβ1γ1 (upper panels) or Gβ1γ13 (lower panels) in the presence of 1 μM transducin and activating ligands. The activating ligands used were 30 μM cycloheximide, 200 μM aristolochic acid and 200 μM denatonium for mT2R5, hT2R14 and hT2R47 respectively. The data are averages of duplicate determinations from a representative experiment. The curves shown are the best fits for the data to single-site binding models.
In summary, our in vitro reconstitution data demonstrate that the T2Rs couple to multiple Gαi/o subunits, including transducin, Gαi1 and Gαo, yet fail to couple to Gαs and Gαq. Our results provide further evidence that the Gα-mediated arm of the bitter taste transduction pathway works via modulation of intracellular cyclic nucleotides and that multiple Gαi subunits are likely to be involved. Furthermore, we directly demonstrate that the T2Rs couple to Gβ1γ13 lending support to the proposal that Gβ1γ13 plays a role in the bitter taste signal transduction pathway through the modulation of PLCβ2 and InsP3 levels. However, we note that the T2Rs displayed different affinities and selectivities among the G-proteins tested raising the possibility that, in vivo, different T2Rs may preferentially signal through different pathways. Furthermore, it appears that there are at least two classes of bitter receptors: those that are fairly specific, recognizing only a few structurally related compounds, and those, like hT2R7, that are broadly tuned [8]. Such a dichotomy may afford the advantage of having receptors specialized to recognizing bitter-tasting compounds of particular biological relevance, while also ensuring the ability to detect a wide spectrum of potentially poisonous compounds through the employment of a relatively small number of broadly tuned receptors.
Acknowledgments
We thank Dr Paul Randazzo [National Cancer Institute (NCI) National Institutes of Health (NIH)] and Dr Doris Wu [National Institute on Deafness and other Communication Disorders (NIDCD)/NIH] for helpful discussions and for reviewing the manuscript. This work was supported by the Divisions of Intramural Research of the NIDCD/NIH (Z01 DC000034-08 and Z01 DC000047-08) and the NINDS (National Institute of Neurological Disorders and Stroke)/NIH (Z01 NS003004-02).
References
- 1.Lindemann B. Receptors and transduction in taste. Nature. 2001;413:219–225. doi: 10.1038/35093032. [DOI] [PubMed] [Google Scholar]
- 2.Bufe B., Hofmann T., Krautwurst D., Raguse J. D., Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat. Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 3.Adler E., Hoon M. A., Mueller K. L., Chandrashekar J., Ryba N. J., Zuker C. S. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 4.Conte C., Ebeling M., Marcuz A., Nef P., Andres-Barquin P. J. Identification and characterization of human taste receptor genes belonging to the TAS2R family. Cytogenet. Genome Res. 2002;98:45–53. doi: 10.1159/000068546. [DOI] [PubMed] [Google Scholar]
- 5.Conte C., Ebeling M., Marcuz A., Nef P., Andres-Barquin P. J. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol. Genomics. 2003;14:73–82. doi: 10.1152/physiolgenomics.00060.2003. [DOI] [PubMed] [Google Scholar]
- 6.Matsunami H., Montmayeur J. P., Buck L. B. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 7.Shi P., Zhang J., Yang H., Zhang Y. P. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol. Biol. Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- 8.Behrens M., Brockhoff A., Kuhn C., Bufe B., Winnig M., Meyerhof W. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem. Biophys. Res. Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Chandrashekar J., Mueller K. L., Hoon M. A., Adler E., Feng L., Guo W., Zuker C. S., Ryba N. J. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn C., Bufe B., Winnig M., Hofmann T., Frank O., Behrens M., Lewtschenko T., Slack J. P., Ward C. D., Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J. Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pronin A. N., Tang H., Connor J., Keung W. Identification of ligands for two human bitter T2R receptors. Chem. Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- 12.Bufe B., Breslin P. A., Kuhn C., Reed D. R., Tharp C. D., Slack J. P., Kim U. K., Drayna D., Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaughlin S. K., McKinnon P. J., Margolskee R. F. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 1992;357:563–569. doi: 10.1038/357563a0. [DOI] [PubMed] [Google Scholar]
- 14.Wong G. T., Gannon K. S., Margolskee R. F. Transduction of bitter and sweet taste by gustducin. Nature. 1996;381:796–800. doi: 10.1038/381796a0. [DOI] [PubMed] [Google Scholar]
- 15.Huang L., Shanker Y. G., Dubauskaite J., Zheng J. Z., Yan W., Rosenzweig S., Spielman A. I., Max M., Margolskee R. F. Gγ13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat. Neurosci. 1999;2:1055–1062. doi: 10.1038/15981. [DOI] [PubMed] [Google Scholar]
- 16.Rossler P., Boekhoff I., Tareilus E., Beck S., Breer H., Freitag J. G protein βγ complexes in circumvallate taste cells involved in bitter transduction. Chem. Senses. 2000;25:413–421. doi: 10.1093/chemse/25.4.413. [DOI] [PubMed] [Google Scholar]
- 17.Hartman J. I., Northup J. K. Functional reconstitution in situ of 5-hydroxytryptamine2c (5HT2c) receptors with αq and inverse agonism of 5HT2c receptor antagonists. J. Biol. Chem. 1996;271:22591–22597. doi: 10.1074/jbc.271.37.22591. [DOI] [PubMed] [Google Scholar]
- 18.Hellmich M. R., Battey J. F., Northup J. K. Selective reconstitution of gastrin-releasing peptide receptor with Gαq. Proc. Natl. Acad. Sci. U.S.A. 1997;94:751–756. doi: 10.1073/pnas.94.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta R. K., Singh J. Bridge-overlap-extension PCR method for constructing chimeric genes. Biotechniques. 1999;26:1082–1086. doi: 10.2144/99266bm17. [DOI] [PubMed] [Google Scholar]
- 20.Krautwurst D., Yau K. W., Reed R. R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 21.Jian X., Sainz E., Clark W. A., Jensen R. T., Battey J. F., Northup J. K. The bombesin receptor subtypes have distinct G protein specificities. J. Biol. Chem. 1999;274:11573–11581. doi: 10.1074/jbc.274.17.11573. [DOI] [PubMed] [Google Scholar]
- 22.Papermaster D. S., Dreyer W. J. Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki A., Tatsumi M., Bitensky M. W. Purification of rod outer segment GTP-binding protein subunits and cGMP phosphodiesterase by single-step column chromatography. Methods Enzymol. 1988;159:702–710. doi: 10.1016/0076-6879(88)59065-1. [DOI] [PubMed] [Google Scholar]
- 24.Fawzi A. B., Fay D. S., Murphy E. A., Tamir H., Erdos J. J., Northup J. K. Rhodopsin and the retinal G-protein distinguish among G-protein βγ subunit forms. J. Biol. Chem. 1991;266:12194–12200. [PubMed] [Google Scholar]
- 25.Kuhn H. Light- and GTP-regulated interaction of GTPase and other proteins with bovine photoreceptor membranes. Nature. 1980;283:587–589. doi: 10.1038/283587a0. [DOI] [PubMed] [Google Scholar]
- 26.Graziano M. P., Freissmuth M., Gilman A. G. Purification of recombinant Gs α. Methods Enzymol. 1991;195:192–202. doi: 10.1016/0076-6879(91)95166-h. [DOI] [PubMed] [Google Scholar]
- 27.Mumby S. M., Linder M. E. Myristoylation of G-protein α subunits. Methods Enzymol. 1994;237:254–268. doi: 10.1016/s0076-6879(94)37067-2. [DOI] [PubMed] [Google Scholar]
- 28.Glass M., Northup J. K. Agonist selective regulation of G proteins by cannabinoid CB(1) and CB(2) receptors. Mol. Pharmacol. 1999;56:1362–1369. doi: 10.1124/mol.56.6.1362. [DOI] [PubMed] [Google Scholar]
- 29.Hoon M. A., Northup J. K., Margolskee R. F., Ryba N. J. Functional expression of the taste specific G-protein, α-gustducin. Biochem. J. 1995;309:629–636. doi: 10.1042/bj3090629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He W., Danilova V., Zou S., Hellekant G., Max M., Margolskee R. F., Damak S. Partial rescue of taste responses of α-gustducin null mice by transgenic expression of α-transducin. Chem. Senses. 2002;27:719–727. doi: 10.1093/chemse/27.8.719. [DOI] [PubMed] [Google Scholar]
- 31.Caicedo A., Pereira E., Margolskee R. F., Roper S. D. Role of the G-protein subunit α-gustducin in taste cell responses to bitter stimuli. J. Neurosci. 2003;23:9947–9952. doi: 10.1523/JNEUROSCI.23-30-09947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozeck M., Brust P., Xu H., Servant G. Receptors for bitter, sweet and umami taste couple to inhibitory G protein signaling pathways. Eur. J. Pharmacol. 2004;489:139–149. doi: 10.1016/j.ejphar.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Engel S., Neumann S., Kaur N., Monga V., Jain R., Northup J., Gershengorn M. C. Low affinity analogs of thyrotropin-releasing hormone are super-agonists. J. Biol. Chem. 2006;281:13103–13109. doi: 10.1074/jbc.M600440200. [DOI] [PubMed] [Google Scholar]
- 34.Okada M., Northup J. K., Ozaki N., Russell J. T., Linnoila M., Goldman D. Modification of human 5-HT(2C) receptor function by Cys23Ser, an abundant, naturally occurring amino-acid substitution. Mol. Psychiatry. 2004;9:55–64. doi: 10.1038/sj.mp.4001357. [DOI] [PubMed] [Google Scholar]
- 35.Yan W., Sunavala G., Rosenzweig S., Dasso M., Brand J. G., Spielman A. I. Bitter taste transduced by PLC-β(2)-dependent rise in IP(3) and α-gustducin-dependent fall in cyclic nucleotides. Am. J. Physiol. Cell Physiol. 2001;280:C742–C751. doi: 10.1152/ajpcell.2001.280.4.C742. [DOI] [PubMed] [Google Scholar]
- 36.Rossler P., Kroner C., Freitag J., Noe J., Breer H. Identification of a phospholipase C β subtype in rat taste cells. Eur. J. Cell Biol. 1998;77:253–261. doi: 10.1016/s0171-9335(98)80114-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Hoon M. A., Chandrashekar J., Mueller K. L., Cook B., Wu D., Zuker C. S., Ryba N. J. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]