Abstract
The IR (insulin receptor) and IGFR (type I insulin-like growth factor receptor) are found as homodimers, but the respective pro-receptors can also heterodimerize to form insulin–IGF hybrid receptors. There are conflicting data on the ligand affinity of hybrids, and especially on the influence of different IR isoforms. To investigate further the contribution of individual ligand binding epitopes to affinity and specificity in the IR/IGFR family, we generated hybrids incorporating both IR isoforms (A and B) and IR/IGFR domain-swap chimaeras, by ectopic co-expression of receptor constructs in Chinese hamster ovary cells, and studied ligand binding using both radioligand competition and bioluminescence resonance energy transfer assays. We found that IR-A–IGFR and IR-B–IGFR hybrids bound insulin with similar relatively low affinity, which was intermediate between that of homodimeric IR and homodimeric IGFR. However, both IR-A–IGFR and IR-B–IGFR hybrids bound IGF-I and IGF-II with high affinity, at a level comparable with homodimeric IGFR. Incorporation of a significant fraction of either IR-A or IR-B into hybrids resulted in abrogation of insulin- but not IGF-I-stimulated autophosphorylation. We conclude that the sequence of 12 amino acids encoded by exon 11 of the IR gene has little or no effect on ligand binding and activation of IR–IGFR hybrids, and that hybrid receptors bind IGFs but not insulin at physiological concentrations regardless of the IR isoform they contained. To reconstitute high affinity insulin binding within a hybrid receptor, chimaeras in which the IGFR L1 or L2 domains had been replaced by equivalent IR domains were co-expressed with full-length IR-A or IR-B. In the context of an IR-A–IGFR hybrid, replacement of IR residues 325–524 (containing the L2 domain and part of the first fibronectin domain) with the corresponding IGFR sequence increased the affinity for insulin by 20-fold. We conclude that the L2 and/or first fibronectin domains of IR contribute in trans with the L1 domain to create a high affinity insulin-binding site within a dimeric receptor.
Keywords: autophosphorylation, bioluminescence resonance energy transfer (BRET), fibronectin domain, hybrid receptor, insulin receptor (IR)
Abbreviations: BRET, bioluminescence resonance energy transfer; CHO, Chinese hamster ovary; FnIII, fibronectin type III; GFP, green fluorescent protein; IGF, insulin-like growth factor; IGFR, type I insulin-like growth factor receptor; IR, insulin receptor; R–luc, luciferase-tagged receptor; RLIC, radioligand immunocapture; YFP, yellow fluorescent protein; wt, wild-type
INTRODUCTION
The IR (insulin receptor) and IGFR (type I insulin-like growth factor receptor) are highly homologous members of subclass II of the tyrosine kinase receptor super-family [1,2]. Unlike other receptor tyrosine kinases that are activated by ligand-induced dimerization, IR and IGFR exist at the cell surface as pre-formed dimers composed of two αβ halves disulfide linked in a β–α–α–β structure. Dimerization of pro-receptors occurs post-translationally in the endoplasmic reticulum, prior to cleavage to generate α and β subunits [3]. When IR and IGFR are co-expressed, the pro-receptors can also heterodimerize to create insulin–IGF (insulin-like growth factor) hybrid receptors [4–6]. Heterodimerization occurs with similar efficiency to homodimerization, so that the proportion of hybrids is a function of the mole fractions of the individual receptors. The less abundant of the receptors (IR or IGFR) in any given cell type is thus found predominantly in hybrids rather than as classical homodimers [7,8].
The extracellular portions of IR and IGFR contain six structural domains, as initially revealed by molecular modelling and confirmed by crystallographic studies [9,10]. The N-terminal half consists of homologous L1 and L2 domains flanking a cysteine-rich domain, while the C-terminal half consists of three FnIII (fibronectin type III) domains. The central FnIII domain has an unstructured insert region, located within which is the site of cleavage between the α and β subunits. Ligand-binding determinants have been localized to the L1, cysteine-rich and L2 domains and to the C-terminal peptide sequence in the α subunit (reviewed in [11]). Surprisingly, in view of its symmetrical dimeric structure, the IR binds only a single molecule of insulin with high affinity, and binding displays negative co-operativity. These properties were accounted for in the models put forward by De Meyts [12] and Schäffer [13], who proposed that high affinity ligand-binding requires contacts with sites on both α-subunits, and that receptor activation is a consequence of cross-linking of α-subunits by ligand. In the absence of a crystal structure of the receptor in complex with the ligand, it remains unclear which of the several binding epitopes act in cis and which in trans, to achieve high affinity binding.
Alternative splicing gives rise to two isoforms of IR, IR-A (exon 11−) and IR-B (exon 11+), whereas there is only a single isoform of IGFR (effectively exon 11−). Exon 11 of the IR gene encodes 12 amino acids at the C-terminus of the α-subunit, immediately downstream of the C-terminal peptide that is essential for ligand binding [14,15]. The IR isoforms have very similar affinity for insulin [16,17], but IR-A has approx. 10-fold higher affinity for IGF-I and IGF-II than IR-B [18]. IR-A has been reported to bind IGF-II with affinity approaching that of insulin binding [19], and has been shown to mediate growth effects of IGF-II during embryonic development [20]. It has also been suggested that IGF-II acts through IR-A to elicit predominantly mitogenic rather than metabolic effects, and to act as a more potent mitogenic agonist than insulin [21]. Thus IR-A has been implicated along with IGFR in malignant transformation [22].
Early studies of IR–IGFR hybrids assembled from half-receptors in vitro suggested that hybrids bound both insulin and IGF-I with high affinity [6]. Subsequent studies of receptors purified from human placenta indicated that hybrids bound IGF-I with higher affinity than insulin, and had lower affinity for insulin than classical IR [23,24]. However, these studies did not take IR isoforms into account. Other previous work has suggested that IR-A–IGFR hybrids have significantly higher affinity for insulin, IGF-I and IGF-II than IR-B–IGFR hybrids, such that IR-A–IGFR hybrids might be responsive to all these ligands at near physiological concentrations [25]. Thus the influence of the IR exon 11 sequence on ligand binding may depend not only on which ligand is considered (insulin or IGF), but also on the receptor context (IR homodimer or hybrid). The fact that hybrid receptors (especially those incorporating the IR-B isoform) bind insulin with lower affinity than classical IR also suggests that interactions in trans from an IR half-receptor profoundly influence insulin binding. The aims of this study were to determine the influence of IR exon 11 on ligand binding, and to identify which IR domains are required in trans from the IR L1 domain to support high affinity insulin-binding, within the context of hybrid receptors.
EXPERIMENTAL
Materials
Bovine insulin was from Sigma, recombinant human IGF-I was a gift from Genentech and recombinant human IGF-II was from Calbiochem. 125I-insulin and 125I-IGF-I were from Amersham Biosciences. Anti-phosphotyrosine antibody 4G10 was from Upstate Biotechnology. Anti-IR monoclonal antibodies 83-7 and 83-14 and anti-IGFR monoclonal antibody 17-69 have been described previously [26,27]. Rabbit polyclonal anti-IR antibody used for blotting was raised in house against a 16-mer peptide corresponding to the C-terminal sequence of the human IR β-subunit. IR–GFP (green fluorescent protein) and IGF-1R–GFP constructs were a generous gift from Dr Rosemary O’Connor (Biosciences Institute, National University of Ireland, Cork, Ireland) and have been described previously [28]. CHO (Chinese hamster ovary) cells overexpressing IR-A [17] were a gift from Dr David Moller (Department of Metabolic Disorders, Merck Research Laboratories, Rahway, U.S.A.), CHO cells overexpressing IR-B [29] were a gift from Dr Leland Ellis (Institute of Bioscience and Technology, Texas A&M University, Houston, TX, U.S.A.) and NIH3T3 cells overexpressing IGFR [30] were a gift from Dr Axel Ullrich (Max Planck Institute of Biochemistry, Martinsried, Germany). Cell culture reagents (Dulbecco's modified Eagle's medium and foetal bovine serum) were from Gibco BRL. Antibiotics (G418 and puromycin) were from Sigma, and Coelenterazine was from Assay Designs.
Construction and expression of cDNA encoding tagged receptors
Full-length IGFR cDNA in which the stop codon had been mutated was excised from the plasmid pIGF-1R-GFP [28] using EcoRI and ligated in frame with the YFP (yellow fluorescent protein) in EcoRI digested pEYFP-N1 (Clontech) to generate the construct pIGFR-YFP. The coding sequence for Renilla luciferase was amplified by PCR using pRL-TK (Promega) as a template, and flanking BamHI and XbaI restriction sites were introduced, by using the primers LucFw: 5′-CACTATAGGCTGGATCCAATGACTTCG-3′, LucRev: 5′-GCGGCCGCTCTAGAATTATTGTTC-3′ restriction sites were introduced by the primers. The product was digested using BamHI and XbaI, and inserted into similarly digested pIGFR-YFP (to replace the YFP with the Renilla luciferase), and in frame with the stop-codon-mutated IGFR to generate pIGFR-Luc.
Full-length IR cDNA in which the stop and adjacent codons had been mutated into a HindIII site was inserted in frame into HindIII digested pEYFP-N1 to generate pIR-YFP. Finally, pIR-Luc was generated by excising the IGFR from pIGFR-Luc using EcoRI, and then religating the resultant Renilla luciferase plasmid and digesting this with HindIII prior to insertion of the full-length stop-mutated IR sequence. Restriction sites used to generate tagged receptors together with linkers from the expression plasmids added a sequence of 20 amino acids between IGFR and YFP in IGFR–YFP (GSSRILQSTVPRARDPPVAT), 16 amino acids between IGFR and luciferase in IGFR–Luc (GSSRILQSTVPRARDP), 19 amino acids between IR and YFP in IR–YFP (KLRILQSTVPRARDPPVAT) and 15 amino acids between IR and luciferase in IR–Luc (KLRILQSTVPRARDP).
Cell culture and transfection of tagged receptors
CHO cells were maintained in Hams F12 medium supplemented with 10% (v/v) newborn calf serum. To generate cells stably expressing IR-A–IGFR and IR-B–IGFR for analysis by RLIC (radioligand immunocapture) assays, CHO cells transfected previously with IR-A or IR-B [17,29] were seeded at 1.2×106 cells per 10–cm dish. Transfections were performed after 24–h, when the cells had reached 60–75% confluence, using Lipofectamine (Gibco BRL) with 8–μg of pCDNA3.1 vector encoding IGFR and a puromycin resistance gene. Following transfection, cells were grown in normal medium for 48–h before adding puromycin (15–μg/ml). After a further 5–days, cells were transferred into 75–cm2 flasks and the populations expanded. For transient expression of receptors for BRET (bioluminescence resonance energy transfer) and autophosphorylation assays, CHO cells were seeded at 5×105 cells per 10–cm dish and transfected 24–h later using Lipofectamine. For BRET experiments, cells were co-transfected with 0.5–μg of R–luc (luciferase-tagged receptor) cDNA (R being either IR or IGFR) and 0.5–μg of empty vector or with 0.5–μg of R–luc and 0.5–μg of R–YFP or R–GFP. For autophosphorylation experiments amounts of DNA used for transfection were as detailed in the legends to the respective Figures. Cells were harvested for receptor analysis 24–h after transfection.
RLIC binding assays
Ligand binding studies were conducted as described previously [15] using Immulon 4 HBX plates (Dynex Technologies) coated with monoclonal antibodies IR 83-7 or IGFR 17-69, which are highly specific for their respective receptors. The epitope of antibody 83-7 has been mapped to the cysteine-rich domain of the IR and this antibody has been shown to have no effect on insulin binding to solubilized receptors from placental membranes (M. A. Soos and K. Siddle, unpublished work). The epitope of antibody 17-69 has been mapped to the first FnIII domain of IGFR, and this antibody similarly has no effect on ligand binding [26].
Triton X-100 lysates from cells expressing IR-A, IR-B, IR-A plus IGFR or IR-B plus IGFR were diluted so that, following immunocapture, 15–20% of the added 125I-insulin or 125I-IGF-I tracer was bound in the absence of unlabelled ligand. Immunocaptured receptors were incubated with 50–pM 125Iinsulin or 25–pM 125I-IGF-I for 16–h at 4 °C, together with unlabelled insulin or IGF-I as specified for individual experiments. The plates were then washed with ice-cold PBS, and bound radioactivity was determined in a γ-counter. IC50 values were objectively determined using GraphPad Prism by curve-fitting with a one-site competition model.
BRET measurements
BRET measurements were made essentially as described previously [31,32]. Transfected CHO cells were cultured in Hams F12 medium containing 50–μg/ml streptomycin, 50–units/ml penicillin and 10% newborn calf serum, and lysed at 4 °C in 50–mM Hepes (pH 7.4), 150–mM NaCl, 10–mM EDTA, 30–mM NaF, 10–mM Na4P2O7, 1% (v/v) Triton X-100, 2.5–mM benzamidine and 1μg/ml each of pepstatin, antipain and leupeptin. Lysates (approx. 10–μg of protein) were pre-incubated with insulin or IGF in 96-well microplates for 45–min at 20 °C in a total volume of 60–μl containing 30–mM Mops and 1–mM Na3VO4. Coelenterazine (10μl, final concentration 15–μM) was added, and light emission acquisition at 410–nm (filter window 70–nm) and 530–nm (filter window 35–nm) was started immediately using a Fusion Microplate Analyser (Packard). The BRET ratio (BR) was defined in terms of emissions (E) at 530 and 410–nm as:
BR=[E530/E410 (R–luc+R–YFP)]−[E530′/E410′ (R–luc alone)]
Autophosphorylation of receptors
Receptor phosphorylation in intact cells was assessed by stimulation with insulin or IGF-I at 37 °C. Following stimulation, cells were placed on ice, lysed as described above, centrifuged at 15000–g for 15–min at 4 °C and the supernatants were immunoprecipitated then analysed by SDS/PAGE (6% gels) and immunoblotting.
RESULTS
Radioligand competition analysis of ligand binding to IR, IGFR and IR/IGFR hybrids
Before investigating ligand binding to hybrid receptors, we confirmed that we could replicate previously described characteristics of IR and IGFR homodimers using radioligand immunocapture assays with lysates of cells overexpressing IR-A, IR-B or IGFR (Figures 1 and 2). Concentrations of unlabelled ligand producing 50% inhibition of radioligand binding (IC50) were determined as a measure of affinity (Table 1). Both IR isoforms bound insulin with high affinity, approx. 2-fold higher for IR-A than IR-B. There was a more marked difference between the isoforms with respect to IGF binding, IR-A having approx. 10-fold higher affinity for IGF-I and 5-fold higher affinity for IGF-II than IR-B. Both IR isoforms had higher affinity for IGF-II than IGF-I, but even IR-A had approx. 10-fold lower affinity for IGF-II than for insulin. IGFR displayed high affinity for IGF-I but >1000-fold lower affinity for insulin. These results are broadly consistent with data published previously [18,19,25,33].
Figure 1. Ligand binding to IR-A and IR-B homodimers.
IR-A and IR-B expressed in CHO cells were immunocaptured using anti-IR antibody 83-7 and binding of 125I-insulin (50 pM) was measured in the presence of increasing concentrations of unlabelled insulin (A), unlabelled IGF-I (B), or unlabelled IGF-II (C). Binding to IR-A (○) and IR-B (●) is expressed as a percentage of the value in the absence of unlabelled ligand. Data points are the means±S.E.M. for three independent experiments in (A) and triplicate samples within a representative experiment in (B) and (C).
Figure 2. Ligand binding to IGFR homodimers.
IGF-I receptors expressed in NIH3T3 cells were immunocaptured using anti-IGFR antibody 17-69 and binding of 125I-IGF-I (25 pM) was measured in the presence of increasing concentrations of unlabelled insulin (○) or IGF-I (●). Binding is expressed as a percentage of the value in the absence of unlabelled ligand. Data points are the means±S.E.M. for triplicate samples within a representative experiment.
Table 1. IC50 values of IR, IGFR and hybrids as measured by ligand competition assays.
RLIC assays were performed using 125I-insulin or 125I-IGF-I as the tracer as described in the legends for Figures 1, 2 and 3. IC50 values were determined using GraphPad Prism by curvefitting with a one-site competition model. Values are the means±S.E.M. for three independent experiments performed with triplicate incubations.
| IC50 (nM) | ||||
|---|---|---|---|---|
| Receptor | Radioligand | Insulin | IGF-I | IGF-II |
| IR-A | 125I-insulin | 0.25±0.11 | 9.0±2.6 | 2.2±0.1 |
| IR-B | 125I-insulin | 0.51±0.12 | 90±6 | 10±1 |
| IR-A–IGFR | 125I-IGF-I | 70±12 | 0.5±0.2 | 0.7±0.1 |
| IR-B–IGFR | 125I-IGF-I | 76±12 | 0.3±0.1 | 0.3±0.1 |
| IGFR | 125I-IGF-I | >1μM | 0.5±0.1 | − |
Extracts of cells expressing IR-A plus IGFR or IR-B plus IGFR were used as a source of hybrid receptors. The IR-B–IGFR expressing cells expressed rather more IR than the IR-A–IGFR expressing cells, but expression of IGFR was similar in both cells and greater than that of IR (results not shown). Receptor concentrations in binding assays were adjusted to give similar binding of radioligand (approx. 20% of the total) in all cases. The percentage of hybrids was estimated as the fraction of 125I-IGF-I binding that was captured with the IR-specific antibody 83-7 compared with the IGFR-specific antibody, 17-69. Hybrid receptors accounted for approx. 20% of the 125I-IGF-I binding in IR-A–IGFR expressing cells, and 15% of the binding in IR-B–IGFR cells, consistent with the expression of the individual receptors. It would be expected that most IR would be incorporated into hybrids given that IGFR was expressed at a higher level than IR. The assay conditions used allowed ligand binding to hybrids to be specifically determined in crude cell lysates, as IGFR would not be captured by the IR-specific antibody, and the 125I-IGF-I tracer at 25 pM would not bind significantly to immunocaptured IR. These assumptions were verified by control experiments with extracts of cells expressing only IR or IGFR (results not shown). It was not possible to perform reciprocal assays using the IGF-specific capture antibody and the 125I-insulin tracer because of the low affinity of the hybrid receptors for insulin.
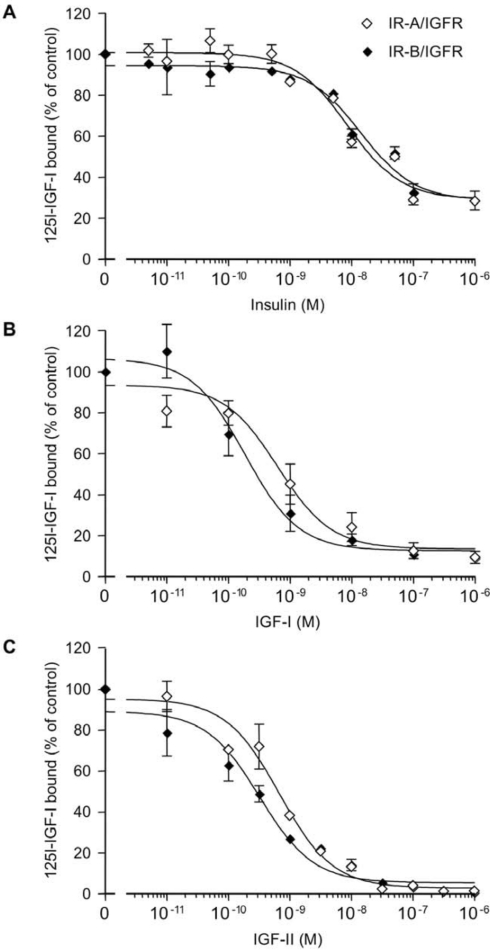
Competition curves using unlabelled insulin, IGF-I and IGF-II are shown in Figure 3, and the IC50 values are summarized in Table 1. IR-A–IGFR and IR-B–IGFR hybrids behaved similarly with all ligands. The affinity of hybrids for insulin was relatively low (IC50=70±12–nM for IR-A–IGFR and 76±12–nM for IR-B–IGFR), and intermediate between that of homodimeric IR (IC50=0.25–0.51–nM) and homodimeric IGFR (IC50>1μM). Hybrids bound both IGF-I and IGF-II with high affinity, similar with that of homodimeric IGFR, again regardless of the IR isoform. These findings differ from previous results [25], which suggested that IR-A–IGFR hybrids had significantly higher affinity than IR-B–IGFR hybrids for all ligands [IC50 values (nM) for Hybrid-RA and Hybrid-RB respectively of 3.7 and >100 for insulin; 0.3 and 2.5 for IGF-I; 0.6 and 15 for IGF-II].
Figure 3. Ligand binding to IR-A–IGFR and IR-B–IGFR hybrids.
Hybrid receptors IR-A–IGFR and IR-B–IGFR were immunocaptured using IR-specific antibody 83-7 and the binding of 125I-IGF-I (25 pM) was determined in the presence of unlabelled insulin (A), unlabelled IGF-I (B) or unlabelled IGF-II (C). Binding to IR-A–IGFR (○) and IR-B–IGFR (●) is expressed as a percentage of the value in the absence of unlabelled ligand. Data points are the means±S.E.M. for triplicate samples within a representative experiment in (A) and (C) and three independent experiments in (B).
BRET analysis of ligand binding to IR, IGFR and IR/IGFR hybrids
Because of the unexpected discordance between our own and previous data, we elected also to investigate ligand binding to hybrid receptors by BRET. This technique, which has been validated previously as a measure of ligand binding to homodimeric IR and IGFR [31,32], requires co-expression of receptor constructs tagged with luciferase (as donor) and fluorescent protein (as acceptor). Luciferase and YFP/GFP fused to the C-termini of the receptor β-subunits can engage in BRET, and the efficiency of energy transfer is influenced by changes in the conformation or proximity of the β subunits induced by ligand binding. A 3–4-fold increase in BRET is observed upon ligand binding to IR or IGFR [31,32], although the overall efficiency of BRET is low compared with what can be achieved by direct fusion of luciferase to GFP. It was previously established that energy transfer, and a resulting spectral shift, is only observed as an intramolecular event between two halves of the same receptor and not as an intermolecular event between dimeric receptors [32].
We created constructs in which IR and IGFR were tagged with luciferase, YFP or GFP and expressed the fusion proteins in CHO cells, which were subsequently stimulated with insulin and IGF-I. Western blotting of cell lysates with anti-IR, anti-IGFR or anti-phosphotyrosine antibodies (as appropriate) revealed proteins of apparent molecular masses of 125–135–kDa (results not shown) corresponding to the expected sizes of receptor β subunits fused to luciferase, YFP or GFP. It was concluded that the tagged receptors were efficiently expressed at the cell surface and retained their ability to undergo ligand-stimulated autophosphorylation in situ.
We next established that we could observe BRET from tagged IR expressed in CHO cells, and compared BRET signals from IR B homodimers and IR-A–IR-B heterodimers. IR-B–luc was co-expressed with either IR-B–YFP or IR-A–GFP and cell lysates were incubated with insulin. Although IR-B–luc/IR-A–GFP co-transfected cells will contain both IR-B–luc and IR-A–GFP homodimers, resonance energy transfer can only occur within IR-B–luc–IR-A–GFP heterodimers. BRET signals comparable in magnitude with those reported previously [31,32] were observed for both IR-B homodimers and IR-A/B heterodimers, and the BRET ratios were increased 3–4-fold by insulin (Figure 4). Under all conditions the BRET ratio was a small difference between two relatively large numbers (the 530/410 ratio for luciferase alone was approx 0.8) and the precision with which ratios could be determined was limited by levels of emission, which in turn were limited by measurement times and the amount of cell extract that could be contained in a well. Nevertheless, by objective curve-fitting for replicate independent experiments, the concentrations of ligand producing half-maximal increases in BRET ratio (EC50) could be determined with reasonable precision. EC50 values for insulin stimulation of IR-B homodimers and IR-A/B heterodimers were not significantly different (Table 2).
Figure 4. Effect of insulin on BRET signal of IR-B homodimers and IR-A/IR-B heterodimers.
Lysates of cells co-expressing IR-B–luc with either IR-B–YFP (A) or IR-A–GFP (B) were incubated with insulin and the BRET ratio was measured as described in the Experimental section. The results are shown as means±ranges for two independent experiments.
Table 2. EC50 values for IR, IGFR and IR–IGFR hybrids as measured by BRET assay.
BRET assays were performed as descirbed in the legends for Figures 4 and 5. EC50 values were determined using GraphPad Prism by curve-fitting with a one-site competition model. The values are means±range for two independent experiments each performed in duplicate.
| EC50 (nM) | |||
|---|---|---|---|
| Receptor | Tags | Insulin | IGF-I |
| IR-A | − | − | − |
| IR-B | IR–luc/IR–YFP | 0.1±0.1 | − |
| IR-A–IR-B | IR–luc/IR–GFP | 0.06±0.02 | − |
| IR-A–IGFR | IGFR–luc/IR–GFP | 60±10 | 4.0±0.5 |
| IR-B–IGFR | IGFR–luc/IR–YFP | 40±5 | 4.0±1.0 |
| IGFR | IGFR–luc/IGFR–YFP | 70±12 | 1.0±0.5 |
To investigate the activation of hybrid receptors, and to compare hybrids with IGFR homodimers, IGFR–luc was co-transfected with IR-A, IR-B or IGFR tagged with GFP or YFP. BRET ratios were again determined as a function of ligand concentration (Figure 5), and the EC50 values calculated (Table 2). IR-A–IGFR and IR-B–IGFR hybrids displayed very similar properties. Both had moderately high affinity for IGF-I and relatively low affinity for insulin. Affinity for IGF-I was slightly lower than that of homodimeric IGFR, whereas affinity for insulin was very much lower than that of homodimeric IR and comparable with homodimeric IGFR. The results of the BRET assays were therefore in broad agreement with those of RLIC assays, showing that hybrid receptors bound IGF-I with affinity comparable with homodimeric IGFR, but had low affinity for insulin compared with homodimeric IR, regardless of IR isoform.
Figure 5. Effects of insulin and IGF-I on BRET signal of IGFR and IR–IGFR hybrids.
Lysates of cells co-expressing IGFR–luc with IGFR–YFP, IR-A–GFP or IR-B–YFP were incubated with insulin (A) or IGF-I (B) and the BRET ratio was measured as described in the Experimental section. Results are means±ranges for two independent experiments each conducted in duplicate.
Ligand-induced autophosphorylation of IR and IR–IGFR hybrids
We next compared the properties of the hybrid receptors and homodimeric IR in intact cells, using autophosphorylation as a measure of activation. CHO cells were transiently transfected with IR-A or IR-B alone or with a 5-fold excess of IGFR, which is sufficient to drive most of the IR into hybrids. The concentrations of cDNA used were adjusted to give similar expression of total IR protein (as assessed by anti-IR immunoblotting) under all conditions. Following stimulation with 10–nM insulin or IGF-I (a concentration chosen to give near maximal activation of cognate homodimeric receptor with minimal activation of the heterologous receptor), cells were lysed and immunoprecipitated with either IR- or IGFR-specific antibodies. Hybrid receptors should be precipitated by both the anti-IR and anti-IGFR antibodies. The anti-IR immunoprecipitates will additionally contain homodimeric IR (although it is expected that approx. 80% of the IR will be in hybrids), whereas the anti-IGFR immunoprecipitates will also contain homodimeric IGFR (approx. 4-fold excess over hybrids). Immunoprecipitates were resolved by SDS/PAGE and the Western blots were probed with an anti-phosphotyrosine antibody. The blots were then stripped and reprobed with the anti-IR antibody. Under the conditions used the β-subunits of IR and IGFR were not resolved and the phospho-signal is therefore a composite value for both IR and IGFR protein. Assuming reciprocal trans-phosphorylation [34], it would be expected that both β-subunits would be phosphorylated equally within activated hybrids. Analyses of the IR-A–IGFR and IR-B–IGFR hybrids are shown in Figures 6 and 7 respectively.
Figure 6. Phosphorylation of IR-A in homodimers and in hybrids.
CHO cells were transfected with 40–ng of IR-A cDNA or 1–μg of IGFR plus 0.2–μg of IR-A cDNAs. The cells were stimulated with 10–nM insulin or IGF-I and lysates were immunoprecipitated (IP) with either anti-IR 83-14 (αIR) or anti-IGFR 17-69 (αIGFR) and resolved by SDS/PAGE. (A) Immunoblots (IB) were probed with anti-phosphotyrosine (pY) antibody (4G10). The membrane was then stripped and reprobed with anti-IR antibody (rabbit polyclonal recognizing the C-terminal domain). The blots shown are representative of three independent experiments. (B) Anti-phosphotyrosine blots [as in (A)] were scanned and quantified using MacBAS V2.2, and values were normalized for receptor expression as determined by scanning the anti-IR blots. The results are means±S.E.M. for three independent experiments. Insulin-stimulated phosphorylation was significantly less (*P<0.01) in presence of IGFR (lanes 5 and 6) compared with IR alone (lane 4) as assessed by two-tailed paired Student's t test.
Figure 7. Phosphorylation of IR-B in homodimers and in hybrids.
CHO cells were transfected with 80–ng of IR-B cDNA or 1–μg of IGFR plus 0.4–μg of IR-B cDNAs. The cells were stimulated with 10–nM insulin or IGF-I and lysates were immunoprecipitated with either anti-IR 83-14 (αIR) or anti-IGFR 17-69 (αIGFR) and resolved by SDS/PAGE. (A) Immunoblots were probed with anti-phosphotyrosine antibody (4G10). The membrane was then stripped and reprobed with an anti-IR antibody. The blots shown are representative of three independent experiments. (B) Anti-phosphotyrosine blots [as in (A)] were scanned and quantified using MacBAS V2.2, and values were normalized for receptor expression as determined by scanning the anti-IR blots. The results are means±S.E.M. for three independent experiments. Insulin-stimulated phosphorylation was significantly less (*P<0.02) in the presence of IGFR (lanes 5 and 6) compared with IR alone (lane 4) as assessed by two-tailed paired Student's t test.
Addition of 10–nM insulin to cells expressing IR alone markedly stimulated receptor phosphorylation (Figures 6 and 7, lanes 1 and 4). However, when the same level of IR protein was co-expressed with an excess of IGFR, and therefore the IR was mostly found in hybrids, phosphorylation in response to 10–nM insulin was reduced to near basal levels (Figures 6 and 7, lanes 5 and 6). The differences in phosphorylation of IR alone compared with co-expression with IGFR were highly significant (P<0.02) for both IR-A and IR-B. The slightly higher level of phophorylation in anti-IR compared with anti-IGFR immunoprecipitates (Figures 6 and 7, compare lanes 5 with lanes 6) is attributable to the presence of some IR homodimers in the former but not the latter. It is concluded that insulin induces little or no stimulation of either IR-A–IGFR or IR-B–IGFR hybrids at a concentration (10–nM) sufficient to maximally activate homodimeric IR. In contrast, 10–nM IGF-I stimulated greater phosphorylation of hybrid receptors than homodimeric IR (Figures 6 and 7, compare lanes 8 with lanes 7). Indeed, IGF-I-stimulated phosphorylation of hybrids approached that of insulin-stimulated IR homodimers (Figures 6 and 7, compare lanes 8 with lanes 4). There was no detectable phosphorylation of IR-B homodimers in response to IGF-I, but some phosphorylation of IR-A homodimers (compare Figure 6, lane 7 with Figure 7, lane 7), consistent with their 10-fold higher affinity for IGF-I. It is concluded that in intact cells both IR-A–IGFR and IR-B–IGFR hybrids respond well to IGF-I, but very poorly to insulin.
Reconstitution of high affinity insulin binding in a hybrid receptor
To investigate the domains required to support high affinity insulin-binding, IR was co-expressed with the previously described chimaeric IGFR constructs in which residues 1–137 (most of L1 domain) or 325–524 (L2 plus part of first FnIII domain) of the IR α-subunit had been replaced by equivalent residues from IGFR [35]. We confirmed by anti-IGFR immunoblotting that both chimaeras were processed and expressed with similar efficiency to wild-type IGFR when transfected into CHO cells (results not shown). The IGFR.IRL1 (IGF-1R/IR C1) and IGFR.IRL2/Fn (IGF-1R/IR C3′) chimaeras were then co-expressed with IR-A or IR-B so as to generate corresponding hybrid receptors, and binding properties were assessed using the RLIC assay. The IGFR cysteine-rich domain is a critical determinant of IGF-I binding [36] and as this domain is retained in both the chimaeras it was possible still to employ 125I-IGF-I as the tracer. The use of the IR-specific antibody 83-7 for immunocapture ensured that the observed binding was attributable to hybrid receptors rather than IGFR homodimers.
Binding data for the hybrid chimaeras are shown in Figure 8 and Table 3. Substitution of the IR-L2/Fn1 domains within the IGFR dramatically increased insulin binding affinity of hybrids with IR-A [IC50 values of 4–nM for IR-A–IGFR.IRL2/Fn compared with 70–nM for IR-A—IGFRwt (wild-type)], but had only a marginal effect on the affinity of hybrids with IR-B (IC50 values of 33–nM for IR-B–IGFR.IRL2/Fn compared with 76–nM for IR-B–IGFRwt). Replacement of the IGFR L1 domain with the equivalent IR domain produced only small increases in insulin affinity, within both IR-A–IGFR and IR-B–IGFR hybrids (approx. 1.4-fold for IR-A–IGFR and 2.3-fold for IR-B–IGFR), which were not statistically significant. It is concluded that the lack of the IR L2 and/or FnIII-1 domains in trans with other IR domains is in large part responsible for the low affinity of hybrid receptors for insulin, although the presence or absence of the region encoded by exon 11 of the IR gene modulates the contribution of an L2/Fn domain in trans.
Figure 8. Reconstitution of high affinity insulin-binding using chimaeric receptors.
(A) Schematic representation of the IR, IGFR and IGFR domain swap chimaeras (as previously described in [35]). Domains are indicated as L1, CR (cysteine-rich), L2, Fn1, Fn2, Fn3 (extracellular) and TK (intracellular), with IR (open bars) and IGFR (grey bars). (B) IR and chimaeric IGFR constructs were co-expressed in CHO cells as described in the Experimental section. Cells were lysed, hybrid receptors were immunocaptured using anti-IR antibody 83-7 and binding of 125I-IGF1 (25 pM) was measured in the presence of unlabelled insulin. Binding is expressed as percentage of the value in the absence of unlabelled ligand. Data points are the means±S.E.M. of triplicate samples within a representative experiment.
Table 3. IC50 values of the domain swap IR/IGFR chimaeric hybrids as measured by ligand competition assays.
Radioligand immunocapture assays were performed using 125I-IGF-I as the tracer, as described in the legend for Figure 8. IC50 values were determined using GraphPad Prism by curve-fitting with a one-site competition model. Values are the means±S.E.M. for three independent experiments performed with triplicate incubations.
| Receptor | Insulin IC50 (nM) |
|---|---|
| IR-A–IGFR | 70±12 |
| IR-B–IGFR | 76±12 |
| IR-A–IGFR.IRL1 | 50±15 |
| IR-A–IGFR.IRL2Fn | 4±2 |
| IR-B–IGFR.IRL1 | 33±18 |
| IR-B–IGFR.IRL2Fn | 33±13 |
DISCUSSION
It is still unclear whether insulin–IGF hybrid receptors have a specific physiological role, and uncertainties remain even with regard to their affinity for, and activation by, potential ligands. We have reported previously that hybrids bind insulin poorly compared with IR homodimers [24], although other studies have since produced conflicting data [25]. The first aim of this work was to re-evaluate the affinity of hybrid receptors for insulin and IGFs with particular regard to dependence on IR isoform. A second aim was to determine, within the context of hybrid receptors, which binding epitopes on individual α-subunits are required to create a high affinity insulin-binding site.
Radioligand competition binding assays are commonly used to study interactions between ligands and receptors, whereas autophosphorylation provides a convenient measure of insulin/IGF receptor activation in intact cells. We used both assays in conjunction with specific immunocapture or immunoprecipitation to characterize ligand interactions with hybrid receptors. We additionally employed a BRET technique that was developed previously to study activation of IR and IGFR [31,32] and very recently to study hybrid receptors [37]. An advantage of the BRET approach is that it allows activation of hybrids to be studied without purification or immunoprecipitation to remove homodimers.
A very clear conclusion from our studies was that heterodimeric hybrid receptors possess considerably higher affinity for IGF-I than for insulin. The affinity of hybrids for IGF-I and IGF-II is similar to (perhaps slightly lower than) that of homodimeric IGFR, whereas their affinity for insulin is very substantially less than that of homodimeric IR. This conclusion was supported by RLIC assays (Table 1), ligand-dependent BRET (Table 2) and autophosphorylation in intact cells (Figures 6 and 7), and is consistent with previous studies of ligand binding to immunoaffinity-purified hybrid receptors [23,24] and hybrid receptor activation in vascular endothelial or smooth muscle cells [38,39].
The last few years have seen renewed interest in IR isoforms generated by alternative splicing of exon 11, in particular in relation to the role of IR-A in mediating actions of IGFs [22], in malignant transformation [40] and in terms of isoform-specific signalling [41]. The sequence of 12 amino acids encoded by exon 11 lies immediately downstream of a sequence of 16 amino acids that is well conserved between IR and IGFR and makes a critical contribution to ligand binding in both receptors [14,42]. The exon 11-encoded sequence has an inhibitory influence on ligand binding. We found that IR-B (exon 11+) had approx. 2-fold lower affinity for insulin than IR-A (exon 11−) (Table 4), in agreement with most previous reports [16,17], although one study found IR-B to have higher affinity than IR-A [33]. We also found that IR-B had approx. 10-fold lower affinity for IGF-I and 5-fold lower affinity for IGF-II compared with IR-A, again broadly in line with previous work [18,25,33]. For both IR-A and IR-B, the rank order of affinities was insulin>IGF-II>IGF-I (Table 4).
Table 4. IC50 values for ligand binding to IR isoforms and hybrids.
Data are from the present work, Pandini et al. [25], Yamaguchi et al. [18] or Denley et al. [33]. See original publications for experimental details, but note that different studies used different conditions (intact cells or solubilized receptors, incubation at 4 °C or room temperature, 125I-insulin, Eu-labelled insulin or 125I-IGF-I as the tracer).
A previous study by Pandini and colleagues [25] reported that IR-A–IGFR hybrids have significantly higher affinity for insulin, IGF-I and IGF-II than IR-B–IGFR hybrids. They concluded that IR-B acts as a highly specific receptor for insulin, while IR-A expression up-regulates the IGF system both by increasing affinity of hybrids for IGFs and by allowing insulin to activate the IGFR in hybrids, and proposed that regulated expression of IR isoforms thus constitutes a molecular switch. While the present study was in preparation a report was published which questioned these findings [43]. We found no significant dependence of the properties of hybrids on IR isoform, whether assessed by RLIC, BRET or autophosphorylation. In RLIC assays, both IR-A and IR-B hybrids displayed low affinity for insulin, closer to that of IGFR than IR. We also found that both IR-A and IR-B hybrids had high affinity for IGFs, similar to that of IGFR. Our data therefore differ significantly from those of Pandini et al. [25] in several important respects (Table 4), and indicate that hybrids are responsive to IGFs and unresponsive to insulin regardless of the IR isoform they contain.
We can offer no explanation for these discordant findings. Both the present study and that of Pandini et al. [25] involved human receptors and RLIC assays with the same tracer (125I-IGF-I) and antibody (anti-IR 83-7). We extracted receptors from populations of transfected CHO cells and performed binding assays for 16–h at 4 °C, whereas Pandini et al. [25] used receptors from clones of transfected murine R− fibroblasts and measured binding after 2–h at room temperature, but it is difficult to see why such technical differences would differentially influence the properties of IR-A and IR-B hybrids. The very recent study of Slaaby and colleagues [43] also failed to find any dependence of ligand affinities on IR isoform and also concluded that signalling of insulin through hybrid receptors is unlikely to be physiologically relevant. Indeed, given that IR-A and IR-B homodimers bind insulin with similar affinity, there is no a priori reason to expect that the affinity of hybrid receptors for insulin would depend on IR isoform. Moreover, we found that IR-A/IR-B heterodimers had high affinity for insulin similar to homodimeric IR (Figure 4), indicating that asymmetry of hybrids with respect to exon 11 is not intrinsically inhibitory towards ligand binding.
Heterodimerization between IR-A and IR-B has been investigated in a pancreatic β cell line [41]. It was reported that the isoforms localized to different regions of the plasma membrane and did not form heterodimers. It was thus concluded that the peptide sequence encoded by exon 11 acted as a targeting signal directing IR-A and IR-B to distinct lipid raft microdomains, and that this resulted in differential signalling by the two isoforms. However, several studies have shown that in fibroblasts both IR isoforms heterodimerize efficiently with IGFR [18,25], and the present study confirms that in CHO cells they also readily form hybrids with each other. Moreover, dimerization of pro-receptors and disulfide bond formation between α-subunits takes place as an immediate post-translational event in the endoplasmic reticulum [3]. Segregation of isoforms to prevent heterodimerization would have to occur in this compartment, there being no evidence for disulfide exchange at subsequent stages of receptor maturation and trafficking. We conclude that, if mechanisms do exist to prevent heterodimerization of IR isoforms and to target them to different regions of the plasma membrane, these must be highly specific for certain cell types and are unlikely to be of general importance.
Determination of affinities for ligand–receptor interactions strictly requires analysis of binding data by Scatchard, or similar, plots. In the case of IR and IGFR, such plots are curvilinear, indicating negative co-operativity and/or heterogeneity of binding sites [12], so that no single value can be assigned for ligand affinity. We took IC50 (RLIC assays) or EC50 (BRET assays) as measures of relative affinity, which should approximate to mean dissociation constants under the conditions used. The rank order of IC50 values for insulin binding in RLIC assays performed over 16–h at 4 °C (Table 1) was the same as for the corresponding EC50 values for BRET assays involving incubations for 45–min at 20 °C (Table 2), namely homodimeric IR<IR–IGFR hybrids<homodimeric IGFR. There were, however, some significant differences in absolute values. For instance, the EC50 value for the activation of IGFR by insulin in BRET assays was 70–nM, whereas the IC50 value for insulin binding in RLIC assays was >1000–nM. This may reflect, in part, the high affinity of IGF-I for its own receptor and the need for very high concentrations of insulin to displace IGF-I bound to the receptor.
The IC50 for insulin competition with 125I-IGF-I binding to IR-A–IGFR and IR-B–IGFR hybrids in RLIC assays in the present study (approx 70–nM) was very similar to that reported previously for immunoaffinity purified receptors from human placenta, which should contain both forms of hybrid receptor [24]. In the present study, the quantities of hybrid receptors immunocaptured on microtitre plates were not sufficient to bind a significant fraction of 125I-insulin at tracer concentrations, but in the previous study the different assay format and greater quantities of material available enabled competition studies to be carried out also with 125I-insulin as the tracer. These homogeneous binding assays produced a significantly lower IC50 value for insulin binding to hybrids (3–5–nM), similar to the values that have been reported for isolated half-receptors [44,45], although this was still some 10-fold higher than the value for homodimeric IR.
The dependence of IC50 on the radioligand may, to some extent, be a function of the effectiveness of homologous versus heterologous competition between unlabelled ligand and radiolabelled tracer. However, it is probable that the different tracers bind to different sites on hybrid receptors, 125I-insulin predominantly binds to the IR half and 125I-IGF-I to the IGFR half, so that unlabelled insulin competes directly with 125I-insulin but indirectly with 125I-IGF-I. The question then arises as to which IC50 value is more relevant to activation of the hybrid receptors by insulin. In the present study, the EC50 value for the activation of the hybrids by insulin in BRET assays was similar to the IC50 value for the heterologous competition in RLIC assays, suggesting that the latter is a good measure of productive ligand binding, leading to receptor activation. Although there may be a site on the hybrid receptors with higher affinity for insulin, as revealed in homogeneous radioligand competition assays [24], binding at this site appears not to be productive of receptor activation as assessed by BRET.
A framework for interpreting these anomalies is provided by the binding model of DeMeyts and Whittaker [11], which proposes that high affinity insulin-binding to IR results from a single insulin molecule cross-linking two distinct binding sites, one on each α-subunit. Site 1 on one α-subunit contributes the greater part of the binding energy by interacting with the classical receptor-binding surface of insulin, while site 2 on the other α-subunit has lower intrinsic affinity and interacts with a distinct and more recently defined binding surface of insulin. The model predicts two possible modes of insulin binding to a hybrid receptor, in which the classical binding surface of insulin interacts with either the IR half or the IGFR half (Scheme 1). The former mode would be expected to have the higher intrinsic affinity, but our data suggest that the latter may lead more readily to receptor activation. However, the fact that homodimeric IGFR can be activated by insulin, albeit at high concentrations, indicates that insulin can interact to some degree with both sites 1 and 2 of IGFR. In contrast with insulin, IGFs bind to hybrid and homodimeric IGFRs with similar affinity as measured by IC50 in RLIC assays, although there was a small difference in the respective EC50 values as determined in BRET assays. Thus it would appear either that site 2 interactions for IGF-I can be provided as effectively by IR sequences as by those in IGFR, as also suggested by studies of chimaeric receptors [46], or that such interactions contribute relatively little to IGF binding. In fact it remains uncertain whether the binding mechanisms of insulin and IGF are completely analogous. For instance IR and IGFR exhibit subtle differences in the effect of disulfide reduction on ligand binding [44,45,47,48] and in the concentration dependence of ligand dissociation kinetics [46].
Scheme 1. Model of ligand binding to hybrid receptors.
The model is based on that of DeMeyts and Whittaker [11] and assumes that only a single molecule of ligand binds with high affinity to homodimeric IR or IGFR, by contacting sites on both α-subunits, of which site 1 contributes the greater fraction of binding energy. (A) and (B) represent binding of insulin and IGF to homodimeric IR and IGFR respectively. (C) and (D) represent two potential modes of binding to hybrid receptors, in which site 1 is contributed by the IR or IGFR half respectively. (E) and (F) represent the presumed binding modes of labelled insulin and IGF-I respectively, illustrating that IC50 values for competition by unlabelled insulin will be different for the two labelled ligands. The EC50 for receptor activation by insulin correlates with the IC50 for labelled IGF, suggesting that of the two possible insulin binding modes (D) may be more effective than (C) in leading to receptor activation.
The crystal structures of monomeric N-terminal fragments of IR and IGFR revealed a putative ligand-binding cavity flanked by the L1, cysteine-rich and L2 domains [49,50]. The L1 domain of IR has been identified as a key region conferring affinity and specificity for binding of insulin compared with IGFs [11], whereas the cysteine-rich domain of IGFR makes contributions to binding of IGF-I but not IGF-II [36,51]. However, IR or IGFR constructs consisting only of these three domains do not bind ligand, and a sequence of 16 amino acids from the α-subunit C-terminus (CT domain) is essential for ligand binding [14,52]. The recently reported crystal structure of the disulfide-linked IR ectodomain dimer reveals a folded over conformation that places the ligand binding regions of both α-subunits in juxtaposition [10], and allows descriptions of two ligand contact sites to be made that are fully consistent with the binding model of De Meyts and Whittaker [11]. Site 1, corresponding to the low affinity binding site of half-receptors, involves the L1 domain (and probably the CT domain, although this is not revealed in the crystal structure) and is believed to contact the classical binding surface of insulin. Site 2 involves one or more of the AB, CC' and EF loops between the β-strands of the first FnIII domain (for historical reasons this is variously referred to by different authors as FnIII-0 or FnIII-1) and is believed to contact the hexamerization surface of insulin. In fact the FnIII-1 domain together with the adjacent L2 domain has recently been characterized experimentally as a second insulin-binding site [53]. The structure of the dimeric IR ectodomain thus supports a model of ligand binding involving interaction of insulin with site 1 of one α-subunit and site 2′ of the other, providing an explanation for high affinity, negative co-operativity and conformational changes leading to receptor activation. While the two L1 domains are too far apart to allow insulin to contact both simultaneously, the two L2 domains are much closer together [10]. In the absence of detailed structural information for receptor–ligand complexes, it remains unclear whether the L2 domain contributes to site 1 or site 2, i.e. in cis or in trans relative to the L1 domain.
We addressed this question by studying the properties of hybrid receptors comprised of wt IR together with chimaeric IGFR in which either the L1 or L2/Fn domain had been replaced with the equivalent portion of IR. We found that, within the context of an IR-A–IGFR hybrid receptor, replacing the L2/Fn domain of the IGFR with the equivalent domain from the IR [residues 325–524, including almost the whole L2 domain and β-strands A, B and C of the FnIII-1 domain] increased the affinity for insulin approx. 20-fold (Table 3)]. However, replacement of the same domain within IR-B–IGFR resulted in a much smaller increase in insulin binding affinity of approx. 2-fold. We conclude that in dimeric receptors the IR L2/Fn1 domains contribute to insulin binding in trans from the major insulin contact site in the L1 domain. There is asymmetry between the IR and IGFR in that high affinity insulin binding specifically requires the IR L2/Fn1 region, whereas both the IGFR and IR L2/Fn1 regions support high affinity binding of IGF-I, in the context of chimaeric receptors [46] as well as hybrids. It remains to be determined whether the critical contribution to insulin binding involves residues within the C-terminal portion of the L2 domain or N-terminal portion of the FnIII-1 domain. Moreover, the extent of this contribution is evidently influenced by the presence of the sequence encoded by exon 11 of the IR gene. Alanine scanning of the insulin-binding site has revealed differences between the IR isoforms in the relative energetic contributions of common receptor side-chains to insulin binding, suggesting that insulin employs different modes of interaction with the two isoforms to achieve similar binding, and that there is significant accommodation for structural changes induced by the presence of the extra 12 amino acids in IR-B [54]. In conclusion, our data support a model in which bound insulin contacts both α-subunits within dimeric receptors and suggest that effective cross-linking of α-subunits is essential for receptor activation. Further studies of hybrid receptors incorporating IR/IGFR domain exchanges should allow the contributions of individual binding epitopes, in cis or in trans, to be defined more precisely.
Acknowledgments
We thank Dr Rosemary O'Connor for the gift of IR–GFP and IGF-1R–GFP constructs, Dr Axel Ullrich for the gift of IR/IGFR chimaeric constructs and Dr Henry Lowman (Genentech, South San Francisco, CA, U. S. A.) for the gift of recombinant human IGF-I. We are also grateful for financial support from Diabetes U.K. (for K. H. S.), the Wellcome Trust (for D. H.) and the Algerian Government and Cambridge Overseas Trust (for S. B.).
References
- 1.Ebina Y., Edery M., Ellis L., Standring D., Beaudoin J., Roth R. A., Rutter W. J. Expression of a functional human insulin receptor from a cloned cDNA in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U.S.A. 1985;82:8014–8018. doi: 10.1073/pnas.82.23.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ullrich A., Gray A., Tam A. W., Yang-Feng T., Tsubokawa M., Collins C., Henzel W., Le Bon T., Kathuria S., Chen E., et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson T. S., Bamberger M. J., Lane M. D. Post-translational changes in tertiary and quaternary structure of the insulin proreceptor: correlation with acquisition of function. J. Biol. Chem. 1988;263:7342–7351. [PubMed] [Google Scholar]
- 4.Moxham C. P., Duronio V., Jacobs S. Insulin-like growth factor I receptor β-subunit heterogeneity: evidence for hybrid tetramers composed of insulin-like growth factor I and insulin receptor heterodimers. J. Biol. Chem. 1989;264:13238–13244. [PubMed] [Google Scholar]
- 5.Soos M. A., Siddle K. Immunological relationships between receptors for insulin and insulin-like growth factor I: evidence for structural heterogeneity of insulin-like growth factor I receptors involving hybrids with insulin receptors. Biochem. J. 1989;263:553–563. doi: 10.1042/bj2630553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treadway J. L., Morrison B. D., Goldfine I. D., Pessin J. E. Assembly of insulin/insulin-like growth factor-1 hybrid receptors in vitro. J. Biol. Chem. 1989;264:21450–21453. [PubMed] [Google Scholar]
- 7.Bailyes E. M., Nave B. T., Soos M. A., Orr S. R., Hayward A. C., Siddle K. Insulin receptor/IGF-I receptor hybrids are widely distributed in mammalian tissues: quantification of individual receptor species by selective immunoprecipitation and immunoblotting. Biochem. J. 1997;327:209–215. doi: 10.1042/bj3270209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federici M., Porzio O., Zucaro L., Fusco A., Borboni P., Lauro D., Sesti G. Distribution of insulin/insulin-like growth factor-I hybrid receptors in human tissues. Mol. Cell. Endocrinol. 1997;129:121–126. doi: 10.1016/s0303-7207(97)04050-1. [DOI] [PubMed] [Google Scholar]
- 9.Marino-Buslje C., Martin-Martinez M., Mizuguchi K., Siddle K., Blundell T. L. The insulin receptor: from protein sequence to structure. Biochem. Soc. Trans. 1999;27:715–726. doi: 10.1042/bst0270715. [DOI] [PubMed] [Google Scholar]
- 10.McKern N. M., Lawrence M. C., Streltsov V. A., Lou M. Z., Adams T. E., Lovrecz G. O., Elleman T. C., Richards K. M., Bentley J. D., Pilling P. A., et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature. 2006;443:218–221. doi: 10.1038/nature05106. [DOI] [PubMed] [Google Scholar]
- 11.De Meyts P., Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 12.De Meyts P. The structural basis of insulin and insulin-like growth factor-I receptor binding and negative co-operativity, and its relevance to mitogenic versus metabolic signalling. Diabetologia. 1994;37(Suppl. 2):S135–S148. doi: 10.1007/BF00400837. [DOI] [PubMed] [Google Scholar]
- 13.Schäffer L. A model for insulin binding to the insulin receptor. Eur. J. Biochem. 1994;221:1127–1132. doi: 10.1111/j.1432-1033.1994.tb18833.x. [DOI] [PubMed] [Google Scholar]
- 14.Kristensen C., Wiberg F. C., Andersen A. S. Specificity of insulin and insulin-like growth factor I receptors investigated using chimeric mini-receptors: role of C-terminal of receptor α subunit. J. Biol. Chem. 1999;274:37351–37356. doi: 10.1074/jbc.274.52.37351. [DOI] [PubMed] [Google Scholar]
- 15.Surinya K. H., Molina L., Soos M. A., Brandt J., Kristensen C., Siddle K. Role of insulin receptor dimerization domains in ligand binding, cooperativity, and modulation by anti-receptor antibodies. J. Biol. Chem. 2002;277:16718–16725. doi: 10.1074/jbc.M112014200. [DOI] [PubMed] [Google Scholar]
- 16.Mosthaf L., Grako K., Dull T. J., Coussens L., Ullrich A., McClain D. A. Functionally distinct insulin receptors generated by tissue-specific alternative splicing. EMBO J. 1990;9:2409–2413. doi: 10.1002/j.1460-2075.1990.tb07416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi Y., Flier J. S., Yokota A., Benecke H., Backer J. M., Moller D. E. Functional properties of two naturally occurring isoforms of the human insulin receptor in Chinese hamster ovary cells. Endocrinology. 1991;129:2058–2066. doi: 10.1210/endo-129-4-2058. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi Y., Flier J. S., Benecke H., Ransil B. J., Moller D. E. Ligand-binding properties of the two isoforms of the human insulin receptor. Endocrinology. 1993;132:1132–1138. doi: 10.1210/endo.132.3.8440175. [DOI] [PubMed] [Google Scholar]
- 19.Frasca F., Pandini G., Scalia P., Sciacca L., Mineo R., Costantino A., Goldfine I. D., Belfiore A., Vigneri R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol. Cell. Biol. 1999;19:3278–3288. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louvi A., Accili D., Efstratiadis A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev. Biol. 1997;189:33–48. doi: 10.1006/dbio.1997.8666. [DOI] [PubMed] [Google Scholar]
- 21.Pandini G., Medico E., Conte E., Sciacca L., Vigneri R., Belfiore A. Differential gene expression induced by insulin and insulin-like growth factor-II through the insulin receptor isoform A. J. Biol. Chem. 2003;278:42178–42189. doi: 10.1074/jbc.M304980200. [DOI] [PubMed] [Google Scholar]
- 22.Denley A., Wallace J. C., Cosgrove L. J., Forbes B. E. The insulin receptor isoform exon 11- (IR-A) in cancer and other diseases: a review. Horm. Metab. Res. 2003;35:778–785. doi: 10.1055/s-2004-814157. [DOI] [PubMed] [Google Scholar]
- 23.Kasuya J., Paz I. B., Maddux B. A., Goldfine I. D., Hefta S. A., Fujita-Yamaguchi Y. Characterization of human placental insulin-like growth factor-I/insulin hybrid receptors by protein microsequencing and purification. Biochemistry. 1993;32:13531–13536. doi: 10.1021/bi00212a019. [DOI] [PubMed] [Google Scholar]
- 24.Soos M. A., Field C. E., Siddle K. Purified hybrid insulin/insulin-like growth factor-I receptors bind insulin-like growth factor-I, but not insulin, with high affinity. Biochem. J. 1993;290:419–426. doi: 10.1042/bj2900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandini G., Frasca F., Mineo R., Sciacca L., Vigneri R., Belfiore A. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J. Biol. Chem. 2002;277:39684–39695. doi: 10.1074/jbc.M202766200. [DOI] [PubMed] [Google Scholar]
- 26.Soos M. A., Field C. E., Lammers R., Ullrich A., Zhang B., Roth R. A., Andersen A. S., Kjeldsen T., Siddle K. A panel of monoclonal antibodies for the type I insulin-like growth factor receptor: epitope mapping, effects on ligand binding, and biological activity. J. Biol. Chem. 1992;267:12955–12963. [PubMed] [Google Scholar]
- 27.Soos M. A., Siddle K., Baron M. D., Heward J. M., Luzio J. P., Bellatin J., Lennox E. S. Monoclonal antibodies reacting with multiple epitopes on the human insulin receptor. Biochem. J. 1986;235:199–208. doi: 10.1042/bj2350199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buckley D. A., Loughran G., Murphy G., Fennelly C., O'Connor R. Identification of an IGF-1R kinase regulatory phosphatase using the fission yeast Schizosaccharomyces pombe and a GFP tagged IGF-1R in mammalian cells. Mol. Pathol. 2002;55:46–54. doi: 10.1136/mp.55.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellis L., Clauser E., Morgan D. O., Edery M., Roth R. A., Rutter W. J. Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell. 1986;45:721–732. doi: 10.1016/0092-8674(86)90786-5. [DOI] [PubMed] [Google Scholar]
- 30.Lammers R., Gray A., Schlessinger J., Ullrich A. Differential signalling potential of insulin- and IGF-1-receptor cytoplasmic domains. EMBO J. 1989;8:1369–1375. doi: 10.1002/j.1460-2075.1989.tb03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanquart C., Boute N., Lacasa D., Issad T. Monitoring the activation state of the insulin-like growth factor-1 receptor and its interaction with protein tyrosine phosphatase 1B using bioluminescence resonance energy transfer. Mol. Pharmacol. 2005;68:885–894. doi: 10.1124/mol.105.013151. [DOI] [PubMed] [Google Scholar]
- 32.Boute N., Pernet K., Issad T. Monitoring the activation state of the insulin receptor using bioluminescence resonance energy transfer. Mol. Pharmacol. 2001;60:640–645. [PubMed] [Google Scholar]
- 33.Denley A., Bonython E. R., Booker G. W., Cosgrove L. J., Forbes B. E., Ward C. W., Wallace J. C. Structural determinants for high-affinity binding of insulin-like growth factor II to insulin receptor (IR)-A, the exon 11 minus isoform of the IR. Mol. Endocrinol. 2004;18:2502–2512. doi: 10.1210/me.2004-0183. [DOI] [PubMed] [Google Scholar]
- 34.Frattali A. L., Treadway J. L., Pessin J. E. Transmembrane signaling by the human insulin receptor kinase: relationship between intramolecular β subunit trans- and cis-autophosphorylation and substrate kinase activation. J. Biol. Chem. 1992;267:19521–19528. [PubMed] [Google Scholar]
- 35.Schumacher R., Soos M. A., Schlessinger J., Brandenburg D., Siddle K., Ullrich A. Signaling-competent receptor chimeras allow mapping of major insulin receptor binding domain determinants. J. Biol. Chem. 1993;268:1087–1094. [PubMed] [Google Scholar]
- 36.Whittaker J., Groth A. V., Mynarcik D. C., Pluzek L., Gadsboll V. L., Whittaker L. J. Alanine scanning mutagenesis of a type 1 insulin-like growth factor receptor ligand binding site. J. Biol. Chem. 2001;276:43980–43986. doi: 10.1074/jbc.M102863200. [DOI] [PubMed] [Google Scholar]
- 37.Blanquart C., Gonzalez-Yanes C., Issad T. Monitoring the activation state of insulin/insulin-like growth factor-1 hybrid receptors using bioluminescence resonance energy transfer. Mol. Pharmacol. 2006;70:1802–1811. doi: 10.1124/mol.106.026989. [DOI] [PubMed] [Google Scholar]
- 38.Johansson G. S., Arnqvist H. J. Insulin and IGF-I action on insulin receptors, IGF-I receptors and hybrid insulin/IGF-I receptors in vascular smooth muscle cells. Am. J. Physiol. Endocrinol. Metab. 2006;291:E1124–E1130. doi: 10.1152/ajpendo.00565.2005. [DOI] [PubMed] [Google Scholar]
- 39.Li G., Barrett E. J., Wang H., Chai W., Liu Z. Insulin at physiological concentrations selectively activates insulin but not insulin-like growth factor I (IGF-I) or insulin/IGF-I hybrid receptors in endothelial cells. Endocrinology. 2005;146:4690–4696. doi: 10.1210/en.2005-0505. [DOI] [PubMed] [Google Scholar]
- 40.Sciacca L., Prisco M., Wu A., Belfiore A., Vigneri R., Baserga R. Signaling differences from the A and B isoforms of the insulin receptor (IR) in 32D cells in the presence or absence of IR substrate-1. Endocrinology. 2003;144:2650–2658. doi: 10.1210/en.2002-0136. [DOI] [PubMed] [Google Scholar]
- 41.Uhles S., Moede T., Leibiger B., Berggren P. O., Leibiger I. B. Isoformspecific insulin receptor signaling involves different plasma membrane domains. J. Cell Biol. 2003;163:1327–1337. doi: 10.1083/jcb.200306093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mynarcik D. C., Williams P. F., Schäffer L., Yu G. Q., Whittaker J. Identification of common ligand binding determinants of the insulin and insulin-like growth factor 1 receptors: insights into mechanisms of ligand binding. J. Biol. Chem. 1997;272:18650–18655. doi: 10.1074/jbc.272.30.18650. [DOI] [PubMed] [Google Scholar]
- 43.Slaaby R., Schäffer L., Lautrup-Larsen I., Andersen A. S., Shaw A. C., Mathiasen I. S., Brandt J. Hybrid receptors formed by insulin receptor (IR) and insulin-like growth factor I receptor (IGF-IR) have low insulin and high IGF-1 affinity irrespective of the IR splice variant. J. Biol. Chem. 2006;281:25869–25874. doi: 10.1074/jbc.M605189200. [DOI] [PubMed] [Google Scholar]
- 44.Böni-Schnetzler M., Scott W., Waugh S. M., DiBella E., Pilch P. F. The insulin receptor: structural basis for high affinity ligand binding. J. Biol. Chem. 1987;262:8395–8401. [PubMed] [Google Scholar]
- 45.Sweet L. J., Morrison B. D., Pessin J. E. Isolation of functional αβ heterodimers from the purified human placental α2β2 heterotetrameric insulin receptor complex: a structural basis for insulin binding heterogeneity. J. Biol. Chem. 1987;262:6939–6942. [PubMed] [Google Scholar]
- 46.Christoffersen C. T., Bornfeldt K. E., Rotella C. M., Gonzales N., Vissing H., Shymko R. M., ten Hoeve J., Groffen J., Heisterkamp N., De Meyts P. Negative cooperativity in the insulin-like growth factor-I receptor and a chimeric IGF-I/insulin receptor. Endocrinology. 1994;135:472–475. doi: 10.1210/endo.135.1.8013387. [DOI] [PubMed] [Google Scholar]
- 47.Feltz S. M., Swanson M. L., Wemmie J. A., Pessin J. E. Functional properties of an isolated αβ heterodimeric human placenta insulin-like growth factor 1 receptor complex. Biochemistry. 1988;27:3234–3242. doi: 10.1021/bi00409a017. [DOI] [PubMed] [Google Scholar]
- 48.Tollefsen S. E., Thompson K. The structural basis for insulin-like growth factor I receptor high affinity binding. J. Biol. Chem. 1988;263:16267–16273. [PubMed] [Google Scholar]
- 49.Garrett T. P., McKern N. M., Lou M., Frenkel M. J., Bentley J. D., Lovrecz G. O., Elleman T. C., Cosgrove L. J., Ward C. W. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature. 1998;394:395–399. doi: 10.1038/28668. [DOI] [PubMed] [Google Scholar]
- 50.Lou M., Garrett T. P., McKern N. M., Hoyne P. A., Epa V. C., Bentley J. D., Lovrecz G. O., Cosgrove L. J., Frenkel M. J., Ward C. W. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12429–12434. doi: 10.1073/pnas.0605395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sorensen H., Whittaker L., Hinrichsen J., Groth A., Whittaker J. Mapping of the insulin-like growth factor II binding site of the type I insulin-like growth factor receptor by alanine scanning mutagenesis. FEBS Lett. 2004;565:19–22. doi: 10.1016/j.febslet.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 52.Kristensen C., Andersen A. S., Ostergaard S., Hansen P. H., Brandt J. Functional reconstitution of insulin receptor binding site from non-binding receptor fragments. J. Biol. Chem. 2002;277:18340–18345. doi: 10.1074/jbc.M112249200. [DOI] [PubMed] [Google Scholar]
- 53.Hao C., Whittaker L., Whittaker J. Characterization of a second ligand binding site of the insulin receptor. Biochem. Biophys. Res. Commun. 2006;347:334–339. doi: 10.1016/j.bbrc.2006.06.089. [DOI] [PubMed] [Google Scholar]
- 54.Whittaker J., Sorensen H., Gadsboll V. L., Hinrichsen J. Comparison of the functional insulin binding epitopes of the A and B isoforms of the insulin receptor. J. Biol. Chem. 2002;277:47380–47384. doi: 10.1074/jbc.M208371200. [DOI] [PubMed] [Google Scholar]