Abstract
We previously reported that CS (chondroitin sulfate) GAG (glycosaminoglycan), expressed on MCSP (melanoma-specific CS proteoglycan), is important for regulating MT3-MMP [membrane-type 3 MMP (matrix metalloproteinase)]-mediated human melanoma invasion and gelatinolytic activity in vitro. In the present study, we sought to determine if CS can directly enhance MT3-MMP-mediated activation of pro-MMP-2. Co-immunoprecipitation studies suggest that MCSP forms a complex with MT3-MMP and MMP-2 on melanoma cell surface. When melanoma cells were treated with βDX (p-nitro-β-D-xylopyranoside) to inhibit coupling of CS on the core protein, both active form and proform of MMP-2 were no longer co-immunoprecipitated with either MCSP or MT3-MMP, suggesting a model in which CS directly binds to MMP-2 and presents the gelatinase to MT3-MMP to be activated. By using recombinant proteins, we determined that MT3-MMP directly activates pro-MMP-2 and that this activation requires the interaction of the C-terminal domain of pro-MMP-2 with MT3-MMP. Activation of pro-MMP-2 by suboptimal concentrations of MT3-MMP is also significantly enhanced in the presence of excess C4S (chondroitin 4-sulfate), whereas C6S (chondroitin 6-sulfate) or low-molecular-mass hyaluronan was ineffective. Affinity chromatography studies using CS isolated from aggrecan indicate that the catalytic domain of MT3-MMP and the C-terminal domain of MMP-2 directly bind to the GAG. Thus the direct binding of pro-MMP-2 with CS through the C-domain would present the catalytic domain of pro-MMP-2 to MT3-MMP, which facilitates the generation of the active form of MMP-2. These results suggest that C4S, which is expressed on tumour cell surface, can function to bind to pro-MMP-2 and facilitate its activation by MT3-MMP-expressing tumour cells to enhance invasion and metastasis.
Keywords: chondroitin sulfate, matrix metalloproteinase (MMP), membrane-type 3 MMP (MT3-MMP), proteoglycan, sulfation, tissue inhibitor of metalloproteinases-2 (TIMP-2)
Abbreviations: APMA, 4-aminophenylmercuric acetate; BAP, Escherichia coli bacterial alkaline phosphatase; βDX, p-nitro-β-D-xylopyranoside; ConA, concanavalin A; CS, chondroitin sulfate; CDMMP-2, C-terminal domain of MMP-2; C4S, chondroitin 4-sulfate; ECL, enhanced chemiluminescence; ECM, extracellular matrix; GAG, glycosaminoglycan; HA, hyaluronan; HRP, horseradish peroxidase; IDV, individual values; IPTG, isopropyl β-D-thiogalactoside; MCSP, melanoma-specific CS proteoglycan; MMP, matrix metalloproteinase; MT-MMP, membrane-type MMP; TIMP-2, tissue inhibitor of metalloproteinases-2
INTRODUCTION
MMPs (matrix metalloproteinases) are zinc-dependent endopeptidases, which are capable of degrading a wide variety of substrates, including ECM (extracellular matrix) proteins, latent growth factors and other bioactive molecules [1]. The increased expression and activation of MMPs are associated with the progression of many tumour types [2–5]. Among soluble MMPs, MMP-2 (gelatinase A) has been extensively studied for the activation mechanisms [6–10], since immunohistochemical studies indicated that the localized activation of pro-MMP-2 on tumour cell surfaces facilitates focal proteolysis at the invasive edge of tumours [11]. MMP-2 is produced as a catalytically inactive zymogen form (pro-MMP-2), which needs to be activated by removing the prodomain of the zymogen. The active form of MMP-2 can facilitate invasion through basement membranes and connective tissue by degrading type IV collagen, partially denatured type I collagen and other ECM proteins [6–10,12,13]. Thus the characterization of the factors that facilitate pro-MMP-2 activation is an important consideration to understand further the mechanisms of tumour invasion and metastasis.
MT-MMPs (membrane-type MMPs) are thought to constitute major contributors to the mechanism of pro-MMP-2 activation. MT1-MMP (membrane-type 1 MMP) was first described as an activator of pro-MMP-2 expressed on the cell surface [6]. Since that original observation, several other MT-MMPs (e.g. MT2-, MT3- and MT5-MMP) have been shown to activate pro-MMP-2 by similar, but distinct, mechanisms [14–17]. MT3-MMP (MMP-16) shows relatively restricted expression patterns; normal tissue distributions are mainly limited to brain, placenta and lungs [17,18]. This protease is expressed at high levels in oral melanomas, renal cell carcinomas and astrocytomas [19,20], indicating that it may be important in malignant progression of certain neoplasms. MT3-MMP also facilitates tube formation in vitro by specific populations of human microvessel cells [21], suggesting it may also be important in regulating angiogenesis. We previously showed [22] that MT3-MMP expressed on VGP (vertical growth phase) melanoma cells is important for cellular invasion into type I collagen gel in vitro. Furthermore, removal of CS (chondroitin sulfate) GAGs (glycosaminoglycans) from the cell surface significantly inhibited invasion into type I collagen gels and resulted in decreased gelatinolytic activity by these cells [22]. These results suggest a model in which CS proteoglycans associated with melanoma cells may regulate the catalytic activity and/or cell surface localization of MT3-MMP and MMP-2.
CS is a polyanionic ubiquitous component of most ECMs and it interacts with a number of ECM proteins and certain growth factors [23,24]. CS synthesis occurs in the Golgi, where polymers are synthesized on core proteins and sulfated at specific sites to generate specific patterns. There is increasing evidence that suggests that the sulfation pattern of CS is important for dictating specificity for the binding to proteins. For example, C4S (chondroitin 4-sulfate) directly interacts with the cysteine protease, cathepsin K, which leads to changes in the substrate specificity of the protease [25,26]. CSs with distinct sulfation patterns have also been shown to bind chemokines, thereby stabilizing tissue gradients of these factors and regulating their biological functions [27].
In the present study, we demonstrate that cell surface CS, attached to the MCSP core protein, plays a key role in the activation of pro-MMP-2 by MT3-MMP. Further studies using recombinant proteases and purified CS demonstrated that C4S but not C6S (chondroitin 6-sulfate) or HA (hyaluronan) significantly enhances the MT3-MMP-mediated activation of pro-MMP-2. These results suggest that cell surface CS concentrates the protease activation complexes on cell surface to facilitate tumour invasion and pericellular matrix degradation. Thus disruption of this complex may be part of a therapeutic strategy to inhibit the invasion and metastasis of malignant melanomas.
MATERIALS AND METHODS
Reagents
Escherichia coli BL21(DE3)plysS Singles™ were purchased from Novagen (Madison, WI, U.S.A.). IPTG (isopropyl β-D-thiogalactoside) was purchased from Invitrogen (Carlsbad, CA, U.S.A.). The bacterial expression vector pFLAG-CTC, HRP (horseradish peroxidase)-conjugated anti-FLAG M2 antibody, anti-FLAG (M2)–agarose, anti-FLAG (M2) antibody, βDX (p-nitro-β-D-xylopyranoside), C-terminal FLAG–BAP (E. coli bacterial alkaline phosphatase) fusion protein and heparan sulfate (bovine kidney) were purchased from Sigma (St. Louis, MO, U.S.A.). C4S (sturgeon notochord: C4S is over 90%), C6S (shark cartilage: molar ratio of C6S and C4S was 9:1) and anti-C4S antibody (2H6) were purchased from Seikagaku America (Cape Cod, MA, U.S.A.). HA (average molecular mass: 10000 Da) was purchased from LifeCore Biomedical (Chaska, MN, U.S.A.). GelcodeR was purchased from Pierce (Rockford, IL, U.S.A.). Mouse-anti-human MT3-MMP (IM50L) and pro-MMP-2 were purchased from Calbiochem (Palo Alto, CA, U.S.A.). Recombinant TIMP-2 (tissue inhibitor of metalloproteinases-2), rabbit anti-human MT3-MMP (AB19088) and anti-human MMP-2 antibody (AB807), and anti-αv integrin antibodies (MAB1980 and AB1930) were purchased from Chemicon (Temecula, CA, U.S.A.). Pefabloc® SC (a potent inhibitor for serine protease) [4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride] was purchased from Roche (Mannheim, Germany). Other chemicals were obtained from Sigma. An ECL (enhanced chemiluminescence) kit was purchased from Amersham Biosciences (Little Chalfont, Bucks., U.K.).
Co-immunoprecipitations
WM1341D melanoma cells (generously provided by Dr Meenhard Herlyn, The Wistar Philadelphia, PA, U.S.A.) were cultured in RPMI 1640 containing 10% FBS (foetal bovine serum). Cells were washed with PBS five or six times and lysed in 100 mM Tris/HCl (pH 7.5) containing 1% Brij 35, 140 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM MnCl2, 1 mM N-ethylmaleimide and 1 mM PMSF. The cell lysates were cleared by centrifugation at 16000 g for 20 min. The supernatants were collected, precleared and immunoprecipitated with anti-MCSP antibody (9.2.27), anti-MT3-MMP (IM50L), or anti-αV integrin (MAB1980)-conjugated agarose beads. The immunoprecipitated proteins were released from the beads in sample buffer (80 mM Tris/HCl, 3% SDS, 15% glycerol and 0.01% Bromophenol Blue) and then heated at 90 °C for 5 min under reducing conditions for detecting MCSP and MT3-MMP by Western blots. Alternatively, proteins were released using non-reducing conditions for detecting MMP-2 in zymography. The proteins were separated on SDS/PAGE and transferred on to Immobilon-P membranes (Millipore, Bedford, MA, U.S.A.). αV Integrin was detected with polyclonal antibody (AB1930) generated against its cytoplasmic domain. MCSP and MT3-MMP were detected with 9.2.27 and AB19088 respectively with appropriate HRP-conjugated secondary antibodies followed by ECL systems. MMP-2 was detected by gelatin zymography as described below.
For detecting C4S on MCSP core protein, components within total cell lysates were separated by SDS/PAGE under reducing conditions and then transferred on to Immobilon-P membranes. The membranes were blocked and incubated at 4 °C overnight with either anti-MCSP antibody or anti-C4S antibody. In some experiments, C4S chain was digested on the membrane by incubating with chondroitinase ABC (1 unit/ml) in PBS+3% (w/v) BSA for 2 h at 37 °C. After extensive washing of membranes with PBS to remove digested C4S, membranes were incubated with anti-C4S antibody as described above. The immunoreactive proteins were visualized with appropriate HRP-conjugated secondary antibodies followed by ECL.
Zymography
Samples were mixed with SDS sample buffer [6% (w/v) SDS, 0.16 M Tris/HCl, 30% (v/v) glycerol and 0.2% Bromophenol Blue] and then separated on SDS/PAGE containing 1 mg/ml gelatin. The gels were washed twice with 2.5% (v/v) Triton X-100 and then incubated in a buffer containing 20 mM Tris/HCl (pH 7.5), 1.25% (v/v) Triton X-100, 5 mM CaCl2 and 1 μM ZnCl2 overnight at 37 °C. The gels were stained with Coomassie Blue and destained. As a standard, serum-free conditioned media prepared from ConA (concanavalin A)-stimulated HT1080 cells were used to localize proform (P), intermediate form (I) and active form (A) of MMP-2.
Construction of plasmids and expression of the proteins
Full-length MT3-MMP (GenBank® accession number D85511) and MT1-MMP (GenBank® accession number P50281) were obtained from mRNA of human melanoma cells (WM1341D) by RT (reverse transcriptase)–PCR and constructed in pIRES2-EGFP. The soluble form of MT3-MMP (Tyr120–Val574) consists of the catalytic, hinge and haemopexin domains, and its fragment harbouring the catalytic domain and the hinge domain (Tyr120–Ile353) and that of the MT3-MMP haemopexin domain (Cys354–Val574) were ligated into HindIII/SalI sites of pFLAG-CTC vector. The catalytically inactive mutant, in which Glu247 was converted into alanine (E/AMT3-MMP), was generated using a site-directed mutagenesis kit (Stratagene, Cedar Creek, TX, U.S.A.) [28]. The soluble form of MT1-MMP (Tyr112–Val537) was ligated into the same vector using HindIII/KpnI site. Sequences were verified by the MicroChemical Facility of the University of Minnesota (Minneapolis, MN, U.S.A.). The constructs prepared as described above were expressed in E. coli BL21(DE3)plysS Single™ and IPTG was used to induce protein expression. Soluble forms of MT1-MMP and MT3-MMP were isolated and purified by the methods reported previously with brief modifications [29,30]. Briefly, transformed bacteria were grown until the D600 (attenuance) reached 0.4–0.5. The protein expression was induced for 60 min in the presence of IPTG (1 mM). Cells were collected by centrifugation at 14000 g for 10 min at 4 °C and lysed by sonication (10 s, four times) in 10 ml of PBS containing 1% (v/v) Triton X-100, 10 mM EDTA and one tablet of Pefabloc® SC. Cell lysates were centrifuged at 9000 g for 15 min at 4 °C. Aliquots of the supernatant were snap-frozen in liquid nitrogen and then stored at −80 °C until used.
Purification of recombinant proteins
Anti-FLAG (M2) antibody-conjugated agarose beads were washed as recommended by the manufacturer's protocol. The washed beads were mixed with cell lysates prepared as described above and then incubated for 4 h at 4 °C in the presence of additional 1 mM Pefabloc® SC. The beads were washed three times with PBS containing 0.4 M NaCl and 10 mM EDTA and then washed three times with PBS containing 10 mM EDTA. In order to determine the amounts of the recombinant proteins immobilized on the beads, purified recombinant proteins eluted by SDS-sample buffer under reducing conditions and serial amounts of BAP–FLAG fusion protein were separated on the same SDS/PAGE and transferred on to a membrane. The purified proteins were also separated in parallel on SDS/PAGE and were stained with GelcodeR according to the manufacturer's protocol. The membrane was blotted with HRP-conjugated anti-FLAG (M2) monoclonal antibody and developed using ECL systems. Densitometry analyses were performed on the X-ray film of the blot using the program Molecular Analyst (BioRad). The area (D/mm) of each spot on the blot was measured. A standard curve of area versus amounts (ng) of BAP–FLAG fusion protein was created, and we confirmed that the curve was linear in this range without saturation. This plot was used to determine the amounts of each recombinant protein eluted from beads. Analysis was performed three times from one preparation of the bacterial cell lysates with less than 10% difference between analyses for the amounts of purified proteins. Moreover, since the amounts of recovered recombinant proteins varied from experiment to experiment in each batch of lysate, this procedure was repeated every time a new lysate preparation was made to ensure the correct molar ratios for each experiment. Although the amount of the purified recombinant proteins differed somewhat from experiment to experiment, the molar ratios between pro-MMP-2, MT-MMPs and TIMP-2 were kept constant. C-terminal domain of MMP-2 (CDMMP-2) was purified as described previously [31].
Activation of pro-MMP-2 by MT-MMPs
Pro-MMP-2 was incubated with varying molar ratios of MT-MMPs immobilized on beads in activation buffer (20 mM Tris/HCl, pH 7.3, 0.14 M NaCl, 5 mM CaCl2, 0.1 mM ZnCl2, 0.02% Brij 35 and 0.02% NaN3) for 1 h by rotating at 37 °C. When TIMP-2 was used in the present study, the concentration of TIMP-2 was determined using a molar absorption coefficient at 280 nm of 39600 M−1·cm−1 as described previously [32]. CDMMP-2 was first incubated with MT3-MMP-immobilized beads and washed to remove excess amounts of the protein. GAGs were incubated with the MT3-MMP/pro-MMP-2 reaction mixture. As a standard, APMA (4-aminophenylmercuric acetate)-activated pro-MMP-2 was used to localize the pro- and active forms of this enzyme. The reaction was stopped by the addition of SDS-sample buffer under reducing conditions. The reaction mixture was heated at 90 °C and then separated by SDS/PAGE. MMP-2 was detected by Western blotting using polyclonal anti-MMP-2 antibody as described in standard protocols.
Densitometry analysis
In order to evaluate the activation of MMP-2, we calculated the ratio between density of the active form (A) and the proform (P) of MMP-2 using the AlphaEaseR FC Imaging System (AlphaImager™ IS-3400; Alpha Innotech, San Leandro, CA, U.S.A.). Briefly, the areas of interest were selected including the background, and the average of the lowest ten pixel values in each individual object was assigned as BACK. Then, IDV (individual values) of each area was calculated as a sum of all pixel values after subtraction of the BACK value. Each experiment was repeated three times. The A/P ratio was calculated by the following formula:
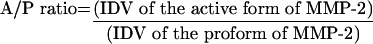 |
where IDV=Σ(each pixel value–BACK).
CS-affinity chromatography
CS was immobilized on agarose beads according to the methods described previously [22,33]. Briefly, freeze-dried rat aggrecan (100 mg dry weight) was digested with trypsin and proteinase K in 50 mM Tris/HCl (pH 7.5) containing 100 mM sodium acetate and mixed with equal volume of 50 mM Tris/HCl (pH 7.5) with 8 M guanidine. CsCl was added to the mixture to give a final density of 1.56 and centrifuged at 40000 rev./min for 48 h. The CS-modified core protein fragments were recovered from the bottom fractions and extensively dialysed against 0.25 M sodium acetate followed by PBS. CNBr-activated agarose beads (1 ml gel volume) were incubated with the purified CS according to the manufacturer's protocol. Under these conditions, 1 mg of uronic acid was immobilized on 1 ml of agarose beads. The same amount of ectodomain, the catalytic/hinge domain and the haemopexin domain of MT3-MMP was incubated with CS-beads or mock-beads in 50 mM Tris/HCl (pH 7.5) containing 0.14 M NaCl and 10 mM CaCl2 overnight at 4 °C. Purified MMP-2 (500 ng) or CDMMP-2 (500 ng) was incubated with CS-beads or mock-beads in the same manner. The beads were washed three times with the same buffer and the bound proteins were extracted by incubation with SDS-sample buffer under non-reducing conditions (pro-MMP-2) or reducing conditions (CDMMP-2) and detected by gelatin zymography (pro-MMP-2) or Western blotting with anti-MMP-2 antibody (CDMMP-2) as described above. The fragments of MT3-MMP were eluted from the beads and then detected by Western blotting using HRP-conjugated anti-FLAG monoclonal antibody.
RESULTS
MCSP (melanoma-specific CS proteoglycan) forms a complex with MT3-MMP and MMP-2 on melanoma cell surface
Previous studies showed that MT3-MMP directly interacts with MCSP, but not αvβ3 integrin, in WM1341D melanoma cells [22]. Furthermore, inhibition of CS addition to the core protein of MCSP inhibited tumour cell invasion and gelatinolytic activity of these melanoma cells in vitro, suggesting that CS can regulate activation of gelatinases such as MMP-2. Experiments were therefore performed to determine if CS on the core protein of MCSP might interact with MT3-MMP and MMP-2 in a ternary complex on melanoma cells.
When cell lysates were immunoprecipitated with anti-MCSP or anti-αv integrin antibody, MT3-MMP was co-precipitated with MCSP, but not with αv integrin (lanes 2 and 4 in Figure 1A). The upper band of the MCSP represents the proteoglycan form of the core protein, whereas the lower band represents 250 kDa core protein that has not been modified with CS (lanes 2 and 4 in Figure 1A). Furthermore, anti-MCSP antibody immunoprecipitated both pro- and active forms of MMP-2 (lane 2 in Figure 1A), suggesting that pro-MMP-2 is activated in a ternary complex containing MCSP and MT3-MMP. By contrast, no MMP-2 was detected in the anti-αv immunoprecipitates (lane 4 in Figure 1A). Several antibodies that recognize αv integrin also failed to co-immunoprecipitate MMP-2 and MT3-MMP (results not shown), indicating that the lack of association between the enzymes and this integrin was not due to the antibody-induced dissociation of the complex during immunoprecipitation.
Figure 1. MT3-MMP forms a molecular complex with MCSP and MMP-2.
Melanoma cells (WM1341D) were cultured overnight at 37 °C. In order to inhibit CS modification of the core protein of MCSP, cells were incubated overnight in the presence of 1 mM βDX to block the coupling of CS with the core protein of MCSP. Cell lysates were prepared and cleared by centrifugation at 16000 g for 20 min. The supernatants were collected, precleared and immunoprecipitated (IP) with anti-MCSP or anti-αv integrin antibody (A) or anti-MT3-MMP antibody (B). The immunoprecipitated proteins were released from the beads heated at 90 °C for 5 min under reducing conditions for detecting MCSP and MT3-MMP. αv Integrin and MMP-2 were liberated from beads by incubating at room temperature (25 °C) for 15 min under non-reducing conditions. Proteins were separated on SDS/PAGE. MSCP, αv integrin and MT3-MMP were detected with 9.2.27, AB1930 and AB19088 respectively. MMP-2 was detected by gelatin zymography. Molecular mass markers are shown (kDa). Pro- (P) and active (A) forms of MMP-2 were localized by using conditioned media prepared from ConA-stimulated HT1080 cells. Note the presence of non-specific binding of pro-MMP-2 in the agarose beads used for the immunoprecipitation studies (A). cIgG, control IgG.
When melanoma cells were pretreated with βDX to prevent coupling of CS with the core protein of proteoglycans, the amount of CS-modified MCSP was significantly reduced, and the major species identified by the anti-MCSP antibody was the unmodified core protein (lane 3 in Figure 1A). Importantly, anti-MCSP antibody immunoprecipitates from βDX-treated cells contained reduced amounts of MMP-2 and MT3-MMP, despite the fact that the amount of core protein is the same in cells whether or not they have been treated with βDX (lane 3 in Figure 1). An anti-MT3-MMP antibody also precipitated both CS-modified and unmodified MCSP from control melanoma cells; however, only the core protein of MCSP was detected in βDX-treated cells with the antibody (lanes 2 and 3 in Figure 1B). The presence of MCSP core protein in anti-MT3-MMP antibody immunoprecipitates from βDX-treated melanoma cells implies that the MSCP core protein can directly or indirectly interact with MT3-MMP in a CS-independent manner. MMP-2 was not detected in immunoprecipitates obtained using anti-MCSP (lane 3 in Figure 1A) or anti-MT3-MMP (lane 3 in Figure 1B) antibodies from βDX-treated melanoma cells. Collectively, these results suggest a model in which MCSP/pro-MMP-2/MT3-MMP form a ternary complex on the surface of melanoma cells, which facilitates activation of pro-MMP-2 by a mechanism involving both CS and MT3-MMP.
MT3-MMP directly interacts with and activates pro-MMP-2 in the absence of TIMP-2
To characterize the nature of the interactions that form this ternary complex, we used recombinant MT3-MMP and pro-MMP-2 to determine if there is a direct interaction between these two MMPs. For these studies, we utilized a model system in which MT3-MMP was affinity immobilized on agarose beads through its C-terminal domain so as to mimic the way by which MT3-MMP might be presented on cell surfaces as described previously [29]. To check the initial purity of the MT3-MMP, the beads were treated with sample buffer and the eluates were subjected to SDS/PAGE. GelcodeR staining of the gel revealed that the purified MT3-MMP preparation contains a major component with a relative mobility of approx. 63 kDa and a minor component with a relative mobility of 60 kDa (lane 1 in Figure 2A). Using an HRP-conjugated anti-FLAG (M2) monoclonal antibody as a probe, the FLAG-tagged MT3-MMP was detected as a single band at 63 kDa (lane 2 in Figure 2A). These results suggest that the degradation of MT3-MMPP was minimal during the purification of the antibody-conjugated beads.
Figure 2. MT3-MMP directly activates pro-MMP-2.
(A) The construct harbouring the soluble form of FLAG-tagged MT3-MMP consisting of the catalytic, hinge and haemopexin domains was expressed in E. coli strain BL21(DE3)plysS Singles™ and IPTG was used to induce protein expression. The protein was isolated by immunoprecipitating with anti-FLAG (M2)-conjugated agarose beads and washed as described in the Materials and methods section. The immunoprecipitated MT3-MMP protein was separated by SDS/PAGE and stained with GelCodeR Blue Stain (lane 1). The same sample was run on SDS/PAGE and transferred on to Immobilon-P membrane and detected by immunoblotting with HRP-conjugated anti-FLAG (M2) antibody (lane 2). Apparent molecular masses (kDa) are shown. The difference in the mobility of molecular mass markers between lane 1 and lane 2 was the result of the shrinkage of SDS/polyacrylamide gel during transferring of proteins to Immobilon-P membrane. (B) MT3-MMP-immmobilized agarose beads were incubated with pro-MMP-2 at a molar ratio of 1:1 for 2 h at 37 °C. The proteins were separated on SDS/PAGE and transferred on to Immobilon-P membrane. As a control, APMA-activated MMP-2 was separated on the same gel. The membranes were immunoblotted with anti-MMP-2 antibody followed by HRP-conjugated anti-rabbit IgG. Apparent molecular masses (kDa) are shown. Proform of MMP-2 (P) and active form of MMP-2 (A) are shown. (C) A constant amount of pro-MMP-2 was incubated with various molar ratios of MT3-MMP-immoblized agarose beads for 2 h at 37 °C. Proteins were detected by immunoblotting with anti-MMP-2 antibody. Apparent molecular masses (kDa) are shown. Proform (P), intermediate form (I) and active form of MMP-2 (A) are shown. (D) MT3-MMP or its catalytically inactive mutant E/AMT3-MMP was incubated with pro-MMP-2 at a molar ratio of 1:1 for 2 h at 37 °C. Proteins were separated and detected as described above. Apparent molecular masses (kDa) are shown. Proform (P) and active forms (A) of MMP-2 are shown.
Beads containing bound MT3-MMP were then mixed with recombinant pro-MMP-2 at a molar ratio of 1:1 and incubated for 2 h at 37 °C. The reaction mixture was separated and transferred on to Immobilon-P membranes. As a positive control, APMA-activated MMP-2 was also included in this gel in order to identify the active form of this protease. MT3-MMP-coated beads cleaved pro-MMP-2 to generate a smaller species which migrated at 64 kDa, identical with the apparent molecular mass of active MMP-2 generated by APMA treatment (Figure 2B). In addition, MT3-MMP activated pro-MMP-2 in a dose-dependent manner with a corresponding decrease in the amounts of zymogen form of MMP-2 at lower molecular ratios (Figure 2C). At a molar ratio of 1:2 (pro-MMP-2/MT3-MMP), almost all pro-MMP-2 was converted into active MMP-2 (Figure 2C). In contrast, a catalytically inactive form of MT3-MMP, in which Glu247 was converted into alanine (E/AMT3-MMP), was not able to convert pro-MMP-2 into an active form (Figure 2D). These results indicate that the catalytic activity of MT3-MMP is required to cleave and activate pro-MMP-2.
We next generated and purified a soluble form of MT1-MMP using the same protocol as for MT3-MMP. The purified FLAG-tagged MT1-MMP recombinant protein migrated as a single polypeptide of 60 kDa (results not shown). As reported previously [29,34], the activation of pro-MMP-2 by MT1-MMP required TIMP-2, but higher levels (5 molar excess of TIMP-2 over MT1-MMP) blocked this activation (Figure 3A). By contrast, exogenous TIMP-2 only marginally enhanced MT3-MMP-mediated activation of proMMP-2, due to the fact that MT3-MMP was already effective at activating pro-MMP-2 in the absence of TIMP-2 (Figure 3B). Higher relative molar ratios of TIMP-2 (ranging from 2 to 5 molar excess of TIMP-2 added to a 1:1 molar complex of MT3-MMP–pro-MMP-2) caused inhibition of pro-MMP-2 activation (Figure 3B), which is consistent with previous studies [28]. These results indicate that MT3-MMP can directly activate pro-MMP-2 in the absence of TIMP-2.
Figure 3. Differences in the requirement for TIMP-2 in MT1-MMP- versus MT3-MMP-mediated pro-MMP-2 activation.
Immobilized MT1-MMP (A) or MT3-MMP (B) was incubated with pro-MMP-2 at a molar ratio of 1:1 for 1 h at 37 °C in the presence or absence of various amounts of TIMP-2. As a negative control (C), pro-MMP-2 was incubated with anti-FLAG (M2) monoclonal antibody-conjugated agarose beads for the same period (lane 1). Pro-MMP-2 was incubated with MT1-MMP alone (lane 2) or MT3-MMP alone (lane 8) for 1 h at 37 °C. The molar ratios of TIMP-2 in the remaining samples were 0.25 (lanes 3 and 9), 0.5 (lanes 4 and 10), 1 (lanes 5 and 11), 2 (lanes 6 and 12) and 5 (lanes 7 and 13) times greater than the amount of MT-MMPs and pro-MMP-2 (1:1). The reactions were stopped by adding SDS-sample buffer under reducing conditions. The proteins were separated on SDS/PAGE and transferred on to Immobilon-P membranes. The membranes were immunoblotted with anti-MMP-2 antibody followed by HRP-conjugated sheep anti-rabbit IgG. Apparent molecular masses (kDa) are shown in the left column. The graph was generated by measuring the D values of the active form (A) and proform (P) of MMP-2. The plot was generated by calculating the ratio of the active form to the proform (A/P ratio) as described in the Materials and methods section. Each bar represents the standard deviation of the mean.
Recombinant C-domain of MMP-2 inhibits MT3-MMP-mediated activation of pro-MMP-2
In order to define further the structural basis for MT3-MMP–pro-MMP-2 interactions, we next tested the ability of the C-terminal domain of pro-MMP-2 to inhibit activation of the zymogen. Previous studies have shown that the C-domain of pro-MMP-2 is important in MT1- or MT2-MMP-mediated activation of pro-MMP-2 [29,31,34]. Therefore we used a recombinant C-domain of MMP-2 (CDMMP-2), which contains the hinge and haemopexin domains of the protease, in an attempt to inhibit MT3-MMP-mediated activation of pro-MMP-2 [31]. MT3-MMP-immobilized agarose beads were pre-incubated with various concentrations of CDMMP-2 for 1 h and washed extensively to remove excess CDMMP-2. The beads were then incubated with pro-MMP-2 for an additional 1 h. Although lower molar ratios of CDMMP-2 (1- and 5-fold over 1:1 molar complexes of MT3-MMP–pro-MMP-2) had no effect on activation of pro-MMP-2, higher molar ratios (10-fold excess of CDMMP-2) partially inhibited activation, and inhibition was complete at the highest concentration tested (20-fold) (Figure 4A). High concentrations of CDMMP-2 are also required to inhibit MT2-MMP-mediated activation of pro-MMP-2, which is also TIMP-2-independent [31].
Figure 4. Excess recombinant C-terminal domain of MMP-2 (CDMMP-2) inhibits MT3-MMP-mediated activation of pro-MMP-2.
(A) Immobilized MT3-MMP on the agarose beads was incubated with various amounts of CDMMP-2 for 1 h at 37 °C, washed and then incubated with pro-MMP-2 for an additional 1 h. The molar ratio between MT3-MMP and pro-MMP-2 was 1:1. As a negative control, pro-MMP-2 was incubated with anti-FLAG (M2) monoclonal antibody-conjugated agarose beads (lane 1). As a positive control, MT3-MMP-immobilized beads were incubated with pro-MMP-2 (lane 2). CDMMP-2 was used at 1 (lane 3), 5 (lane 4), 10 (lane 5) and 20 (lane 6) times molar excess over MT3-MMP and pro-MMP-2 (1:1). The reactions were stopped by adding SDS-sample buffer under reducing conditions. Immobilon-P membranes were immunoblotted with anti-MMP-2 antibody followed by HRP-conjugated sheep anti-rabbit IgG. Apparent molecular masses (kDa) are shown in the left column. The D values of the proform (P) and active form (A) of MMP-2 were used to calculate the A/P ratio as described in the Materials and methods section. Each bar represents the standard deviation of the mean. (B) Immobilized MT1-MMP, MT3-MMP or anti-FLAG (M2)-antibody-conjugated agarose beads were incubated with 100 ng of CDMMP-2 for 1 h at 37 °C. The beads were washed with the reaction buffer (20 mM Tris/HCl, pH 7.3, 0.14 M NaCl, 5 mM CaCl2, 0.1 mM ZnCl2, 0.02% Brij35 and 0.02% NaN3) three times and then eluted with SDS sample buffer. The proteins were separated on SDS/PAGE and transferred on to Immobilon-P membrane, which were then immunoblotted with anti-MMP-2 antibody followed by HRP-conjugated sheep anti-rabbit IgG. Lane 1: anti-FLAG (M2) antibody-conjugated agarose beads; lane 2: MT1-MMP immobilized beads; lane 3: MT3-MMP immobilized beads. Note that the CDMMP-2 migrates at 27 kDa. Apparent molecular masses (kDa) are shown in the left column.
We next incubated agarose beads-immobilized MT3-MMP or MT1-MMP with soluble CDMMP-2 to determine if these membrane-associated MMPs can directly bind this fragment (Figure 4B). CDMMP-2 was incubated with mock, MT1-MMP- or MT3-MMP-immobilized beads for 1 h. After extensive washing to remove excess CDMMP-2, bound CDMMP-2 was eluted from beads using sample buffer and detected by Western blot (Figure 4B). Although CDMMP-2 was eluted from MT3-MMP-immobilized beads (lane 3 in Figure 4B), it was not eluted from MT1-MMP beads (lane 2 in Figure 4B). Mock-coupled agarose beads were included in this assay as a negative control (lane 1 in Figure 4). These results indicate that MT3-MMP specifically binds to the C-terminal region of pro-MMP-2 and directly activates it, which is distinct from the mechanism by which MT1-MMP interacts with pro-MMP-2 that requires TIMP-2 [34].
MT3-MMP and pro-MMP-2 directly bind to CS, leading to enhanced activation of pro-MMP-2
Experiments were performed using CS-conjugated agarose beads to determine if CS could interact with either MT3-MMP or pro-MMP-2. The recombinant fragments of ectodomain of MT3-MMP (MT3-MMP), the catalytic and hinge domains [MT3 (Cat/Hinge)] and the haemopexin domain [MT3 (Hex)] were expressed as single polypeptides (Figure 5A). The same amounts of these recombinant proteins were applied on CS-coupled or mock-beads. As we have previously described [22], CS-conjugated beads (but not mock-coupled beads) could directly bind soluble intact MT3-MMP (lanes 1 and 2 in Figure 5B). Our results indicate that MT3 (Cat+Hinge) directly bound to CS-conjugated beads, while MT3 (Hex) failed to bind these beads (Figure 5B). Pro-MMP-2 and the CDMMP-2 fragment directly bound to CS-conjugated beads, but not to mock-beads (Figure 5C). These results indicate that that CS can interact with CD domain of pro-MMP-2, consistent with a model in which the pro- and catalytic domains of pro-MMP-2 would be accessible to be cleaved by MT3-MMP.
Figure 5. Pro-MMP-2 and MT3-MMP directly bind to CS-coupled beads.
(A) The constructs harbouring the soluble form of FLAG-tagged MT3-MMP, the catalytic and hinge domains of MT3-MMP [MT3 (Cat+Hinge)] and the haemopexin domain [MT3 (Hex)] were expressed in E. coli as described in the Materials and methods section. The proteins were isolated by immunoprecipitating with anti-FLAG (M2)-conjugated agarose beads and washed as described in the Materials and methods section. The purity and integrity of the recombinant proteins were assessed by immunoblotting with HRP-conjugated anti-FLAG (M2) antibody. Apparent molecular masses (kDa) are shown. (B) The same amounts of the bacterial cell lysates of MT3-MMP (lanes 1 and 2), MT3 (Cat+Hinge) (lanes 3 and 4) or MT3 (Hex) (lanes 5 and 6) were incubated with CS-coupled beads (100 μg of uronic acid) (lanes 1, 3 and 5) or mock (M) (lanes 2, 4 and 6) beads, which were not coupled with CS, overnight at 4 °C. The beads were extensively washed and the bound protein detected by immunoblotting with HRP-conjugated anti-FLAG (M2) antibody. Apparent molecular masses (kDa) are shown in the right column. (C) Pro-MMP-2 (500 ng) was incubated with CS-coupled beads (100 μg of uronic acid) or mock-coupled beads overnight at 4 °C. The bound pro-MMP-2 was detected by gelatin zymography (left panel). Note that trace amounts of intermediate form of MMP-2, which is a contaminant of the original pro-MMP-2 preparation, were detected under these experimental conditions. CDMMP-2 (500 ng) was also incubated with CS-coupled beads (100 μg of uronic acid) or mock-beads overnight at 4 °C. The beads were extensively washed and the bound CDMMP-2 was detected by immunoblotting using anti-MMP-2 antibody (right panel).
In order to evaluate the potential of CS to regulate the activation of pro-MMP-2 by MT3-MMP, we first focused on C4S, since previous studies have shown that melanoma cells express C4S on the core protein of MCSP [35]. Western blotting analysis demonstrated that anti-MCSP antibody detected two molecular species of the proteoglycan (lane 1 in Figure 6) in WM1341D melanoma cells, consistent with the results of Figure 1. On the other hand, only the higher molecular mass band of MCSP, which was at the top of the gel, was detected by anti-C4S antibody (2H6) (lane 2 in Figure 6). Furthermore, chondroitinase ABC treatment of the membrane significantly diminished the reactivity of anti-C4S with the higher molecular mass molecule of MCSP (lane 3 in Figure 6), indicating that this is identical with MCSP proteoglycan modified by C4S. The lower molecular mass of MCSP was detected by anti-MCSP (lane 1 in Figure 6) but not by anti-C4S antibody (lane 2 in Figure 6), suggesting that this MCSP molecule is expressed as an unmodified form of the core protein. These results suggest that MCSP is expressed as a ‘part-time proteoglycan’ that is modified by C4S in this melanoma cell line.
Figure 6. MCSP is expressed as a part-time proteoglycan modified by C4S.
Total cell lysates of WM1341 cells (5×106 cells) were separated on SDS/PAGE and transferred on to Immobilon-P membrane. Membranes were blocked and then blotted with anti-MCSP (lane 1) or anti-C4S (lanes 2 and 3) antibody. The membrane of anti-C4S antibody blotting (lane 2) was stripped, blocked and then incubated with chondroitinase ABC (1 unit/ml). Membrane was probed with anti-C4S antibody (lane 3) as described in the Materials and methods section. Apparent molecular mass (250 kDa) is shown.
MT3-MMP-immobilized beads were incubated with pro-MMP-2 at a suboptimal molar ratio of 0.5:1 (MT3-MMP versus pro-MMP-2) in the presence of various amounts of soluble C4S. Under these conditions, MT3-MMP only minimally activated pro-MMP-2 (lane 2 in Figure 7A and Figure 2B) in the absence of C4S. However, increasing concentrations of C4S in the reaction mixture resulted in increased pro-MMP-2 activation (Figure 7A). This activation was observed even at low concentrations of C4S (1 ng/ml) and was maximal in the range of 1–10 μg/ml. Higher concentrations of C4S (100 μg/ml) resulted in inhibition of activation of pro-MMP-2 from these maximal levels (Figure 7A), indicating that the stoichiometry of the three components is critical for determining the outcome of these activation assays.
Figure 7. C4S specifically enhances MT3-MMP-mediated activation of pro-MMP-2.
Pro-MMP-2 was incubated with immobilized MT3-MMP at a suboptimal molar ratio of 1:0.5 in the presence of various amounts of C4S (A) or C6S (B) for 1 h at 37 °C. The reactions were stopped by adding SDS-sample buffer under reducing conditions. The proteins were separated on SDS/PAGE, transferred on to Immobilon-P membranes and immunoblotted with anti-MMP-2 antibody followed by HRP-conjugated sheep anti-rabbit IgG. (A, B) Pro-MMP-2 alone (lane 1), pro-MMP-2 and MT3-MMP in the absence (lane 2) or presence of CS at 100 μg/ml (lane 3), 10 μg/ml (lane 4), 1 μg/ml (lane 5), 100 ng/ml (lane 6), 10 ng/ml (lane 7), and 1 ng/ml (lane 8). Apparent molecular masses (kDa) are shown in the left column. Proform (P) and active forms (A) of MMP-2 are shown. (C) Pro-MMP-2 was incubated with immobilized MT3-MMP at a suboptimal molar ratio of 1:0.5 in the presence of various amounts of HA for 1 h at 37 °C. Pro-MMP-2 alone (lane 1); pro-MMP-2 and MT3-MMP in the absence (lane 2) or presence (lanes 3–8) of HA: 100 μg/ml (lane 3), 10 μg/ml (lane 4), 1 μg/ml (lane 5), 100 ng/ml (lane 6), 10 ng/ml (lane 7). Apparent molecular masses (kDa) are shown in the left column. Proform (P) and active form (A) of MMP-2 are shown. (D) The A/P ratio was calculated from the D values of pro-MMP-2 and active MMP-2 as described in the Materials and methods section. Each bar represents the standard deviation.
Additional studies were performed to determine the structural specificity of CS for the activation of pro-MMP-2 by MT3-MMP. There was minimal enhancement of MT3-MMP-mediated pro-MMP-2 activation at all concentrations of C6S (Figure 7B). Similar results were observed with low-molecular-mass HA (10000 Da; Figure 7C). These results indicate that the ability of C4S to enhance MT3-MMP-mediated activation of pro-MMP-2 was not due to a non-specific anionic effect of the carbohydrate, and that the position of the sulfate on the N-acetylgalactosamine residues is important for this activity. We also incubated pro-MMP-2 with the same concentrations of C4S to determine if this carbohydrate could directly cause auto-activation of this gelatinase. Pro-MMP-2 could not be activated in the presence of C4S alone (results not shown), indicating that C4S-enhanced activation of pro-MMP-2 is MT3-MMP-dependent. Collectively, the results support a model in which the formation of ternary complexes of MCSP–MT3-MMP–pro-MMP2 results in the enhanced invasion and matrix degradation of melanoma cells by increasing pro-MMP-2 activation on the melanoma cell surface.
DISCUSSION
We previously reported that cell surface CS GAG plays a key role in MT3-MMP-mediated human melanoma invasion and gelatinolysis [22]. In the present study, we can propose a mechanism by which CS facilitates MT3-MMP-mediated pro-MMP-2 activation by melanoma cells. We demonstrated the presence of molecular complexes consisting of MCSP, MT3-MMP and MMP-2 on the melanoma cell surface. In further studies utilizing in vitro protease activation assays, we showed that C4S (but not C6S or HA) enhances MT3-MMP-mediated activation of pro-MMP-2. These results suggest an alternative mechanism for activating pro-MMP-2 in tumour microenvironments with elevated levels of specific GAGs such as C4S. Expression of CS is enhanced in certain chronically inflamed tissues (e.g. arthritic joints and atherosclerotic plaques) as well as many malignant tumours [23,36,37]. Thus our results suggest that C4S may serve to limit and focus proteolytic activities on cell surfaces to regulate pericellular matrix degradation and turnover in tumour invasion.
We previously showed that MT3-MMP co-precipitates with MCSP [22]. The present studies further support the conclusion by demonstrating the presence of ternary molecular complexes of MCSP–MT3-MMP–MMP-2. While it was somewhat unexpected that pro-MMP-2 did not associate with αvβ3 integrin in our experimental system, it may suggest yet another mechanism for the association of these molecules on cell surface. Indeed, previous studies demonstrated that αvβ3 integrin binds to MMP-2 and co-localizes with degraded type I collagen [38–40]. Previous studies showed that αvβ3 integrin physically associates with MT1-MMP [41]. Thus αvβ3 integrin may function to localize these MMPs in close proximity in order to enhance pericellular degradation of ECM proteins. In contrast, melanoma cells that we used in the previous and present studies preferentially express MT3-MMP with undetectable level of MT1-MMP protein [22]. Thus the mechanisms by which pro-MMP-2 associates on cell surface may be cell-type-specific and dependent on the expression of specific binding partners. Our results suggest that CS expressed on the core protein of MCSP binds to pro-MMP-2, while the core protein could associate with MT3-MMP. Although MCSP core protein was co-immunoprecipitated with MT3-MMP, anti-MCSP antibody failed to co-precipitate MT3-MMP in βDX-treated melanoma cells. This may reflect the fact that the expression level of MCSP is much higher than that of MT3-MMP, making the detection of MT3-MMP difficult when MCSP antibodies are used to immunoprecipitate the complex from βDX-treated melanoma cells. Collectively, our results suggest that the MCSP expressed as a CS-modified form functions in a complex including MCSP–MT3-MMP–MMP-2 to promote tumour invasion and growth in vivo, as reported previously [22,42–44].
Although the mechanisms by which MT3-MMP and MCSP interact are not clear from these studies, it is possible that the ectodomain of MCSP could interact directly (or indirectly) with MT3-MMP. Previous studies showed that MT1-MMP directly interacts with CD44 through its haemopexin domain, serving to promote tumour cell migration [45,46]. In those studies, it was shown that the GAG of CD44 is not important for MT1-MMP–CD44 complex formation [45,46]. Given the fact that there is significant homology between the haemopexin domains of MT1-MMP and MT31-MMP [47], it is possible that the haemopexin domain of MT3-MMP is important for promoting interaction with the MCSP core protein, although this is yet to be proven. The studies collectively demonstrate that both the core protein and CS moiety of MCSP are important for contributing to the formation of a protease activation complex important for invasion.
Our results demonstrate that MT3-MMP immobilized on agarose beads activates pro-MMP-2 in a TIMP-2-independent manner, consistent with the previous studies demonstrating that soluble ectodomain of MT3-MMP directly activates pro-MMP-2 [28,30]. Further studies demonstrate that the haemopexin domain of pro-MMP-2 competitively inhibits MT3-MMP-mediated activation of pro-MMP-2. Previous studies showed the haemopexin domain-dependent activation mechanisms in the activation of pro-MMP-2 by MT2-MMP and the activation of pro-MMP-13 by MT1-MMP [31,48]. The haemopexin domains of these soluble MMPs appear to be important in their activation, because they allow the MMP molecule to associate with cell surface where they can be activated [31,48]. Our results suggest that structural features found in MT3-MMP, but not in MT1-MMP, enable it to bind directly to the C-terminal domain of MMP-2. Thus identifying the structural features of MT3- and MT2-MMPs that directly interact with pro-MMP-2 may lead to the development of novel compounds that can inhibit TIMP-independent activation of pro-MMP-2.
Pull downs using immobilized CS demonstrate that both MT3-MMP and pro-MMP-2 bind to CS, suggesting that CS may work by acting to bridge the two MMPs. Consistent with this model, inhibiting CS synthesis prevented the formation of this complex on melanoma cell surfaces (Figure 1). Despite the fact that the similarity in the domain structures between pro-MMP-2 and MT3-MMP, CS binds to the C-terminal domain of pro-MMP-2 and to MT3-MMP via domains containing the catalytic and hinge regions. These results suggest that specific primary and/or secondary structures within these domains of each MMP specifically bind CS. This may be particularly important on melanoma cell surfaces where the surface density of MT3-MMP is relatively low, thus CS could focally regulate the localization and activation of pro-MMP-2.
Finally, we have demonstrated that the sulfation pattern on the CS polymer is important to facilitate activation of pro-MMP2 by MT3-MMP. The enhancement mediated by C4S was optimal in the presence of 1–10 μg/ml of this GAG in the reaction mixture. None among exogenous HA, C6S and heparan sulfate was effective at promoting MT3-MMP-mediated activation of pro-MMP-2 at all concentrations tested, suggesting that the effects of C4S are not due to polyanionic properties of this GAG. Although we determined that pro-MMP-2 directly binds to a heparin-Sepharose column (as has been previously shown [49]), heparin had no effect on the activation of pro-MMP-2 by MT3-MMP (results not shown). These results suggest that pro-MMP-2 can bind multiple anionic ligands; however, the specific combination of disaccharide units and sulfation pattern found in C4S is required both to bind and to activate pro-MMP-2 in the presence of MT3-MMP.
C4S has previously been shown to enhance the collagenolytic activity of other non-MMP proteases such as cathepsin K [25,26]. In addition to emphasizing the importance of CS sulfation patterns in the microenvironment for enhancing protease activation, the results suggest that specific binding structures exist within the CS-binding domain(s) of these proteases. Understanding this specificity could lead to the development of specific small molecule inhibitors to inhibit proteolysis and tumour invasion. Alternatively, identifying the biosynthetic pathways that generate specific sulfation patterns in CS may lead to new strategies to interfere with the interaction with MMPs and growth factors to inhibit tumour invasion, growth and metastasis.
Acknowledgments
This study is supported by NIH (National Institutes of Health) grants RO1 CA92222 and RO1CA82295.
References
- 1.McCawley L. J., Matrisian L. M. Matrix metalloproteinases: they're not just for matrix anymore! Curr. Opin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 2.Lynch C. C., Matrisian L. M. Matrix metalloproteinases in tumor–host cell communication. Differentiation. 2002;70:561–573. doi: 10.1046/j.1432-0436.2002.700909.x. [DOI] [PubMed] [Google Scholar]
- 3.MacDougall J. R., Matrisian L. M. Contributions of tumor and stromal matrix metalloproteinases to tumor progression, invasion and metastasis. Cancer Metastasis Rev. 1995;14:351–362. doi: 10.1007/BF00690603. [DOI] [PubMed] [Google Scholar]
- 4.Hornebeck W., Emonard H., Monboisse J. C., Bellon G. Matrix-directed regulation of pericellular proteolysis and tumor progression. Semin. Cancer Biol. 2002;12:231–241. doi: 10.1016/s1044-579x(02)00026-3. [DOI] [PubMed] [Google Scholar]
- 5.Rudek M. A., Venitz J., Figg W. D. Matrix metalloproteinase inhibitors: do they have a place in anticancer therapy? Pharmacotherapy. 2002;22:705–720. doi: 10.1592/phco.22.9.705.34062. [DOI] [PubMed] [Google Scholar]
- 6.Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature. 1994;370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 7.Sounni N. E., Baramova E. N., Munaut C., Maquoi E., Frankenne F., Foidart J. M., Noel A. Expression of membrane type 1 matrix metalloproteinase (MT1-MMP) in A2058 melanoma cells is associated with MMP-2 activation and increased tumor growth and vascularization. Int. J. Cancer. 2002;98:23–28. doi: 10.1002/ijc.10134. [DOI] [PubMed] [Google Scholar]
- 8.Will H., Atkinson S. J., Butler G. S., Smith B., Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP-2 and TIMP-3. J. Biol. Chem. 1996;271:17119–17123. doi: 10.1074/jbc.271.29.17119. [DOI] [PubMed] [Google Scholar]
- 9.Sato T., Kondo T., Seiki M., Ito A. Cell type-specific involvement of furin in membrane type 1 matrix metalloproteinase-mediated progelatinase A activation. Ann. N. Y. Acad. Sci. 1999;878:713–715. doi: 10.1111/j.1749-6632.1999.tb07770.x. [DOI] [PubMed] [Google Scholar]
- 10.Monea S., Lehti K., Keski-Oja J., Mignatti P. Plasmin activates pro-matrix metalloproteinase-2 with a membrane-type 1 matrix metalloproteinase-dependent mechanism. J. Cell. Physiol. 2002;192:160–170. doi: 10.1002/jcp.10126. [DOI] [PubMed] [Google Scholar]
- 11.Seiki M., Koshikawa N., Yana I. Role of pericellular proteolysis by membrane-type 1 matrix metalloproteinase in cancer invasion and angiogenesis. Cancer Metastasis Rev. 2003;22:129–143. doi: 10.1023/a:1023087113214. [DOI] [PubMed] [Google Scholar]
- 12.Birkedal-Hansen H. Proteolytic remodeling of extracellular matrix. Curr. Opin. Cell Biol. 1995;7:728–735. doi: 10.1016/0955-0674(95)80116-2. [DOI] [PubMed] [Google Scholar]
- 13.Basbaum C. B., Werb Z. Focalized proteolysis: spatial and temporal regulation of extracellular matrix degradation at the cell surface. Curr. Opin. Cell Biol. 1996;8:731–738. doi: 10.1016/s0955-0674(96)80116-5. [DOI] [PubMed] [Google Scholar]
- 14.Will H., Hinzmann B. cDNA sequence and mRNA tissue distribution of a novel human matrix metalloproteinase with a potential transmembrane segment. Eur. J. Biochem. 1995;231:602–608. doi: 10.1111/j.1432-1033.1995.tb20738.x. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka M., Sato H., Takino T., Iwata K., Inoue M., Seiki M. Isolation of a mouse MT2-MMP gene from a lung cDNA library and identification of its product. FEBS Lett. 1997;402:219–222. doi: 10.1016/s0014-5793(96)01537-2. [DOI] [PubMed] [Google Scholar]
- 16.Pei D. Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 1999;274:8925–8932. doi: 10.1074/jbc.274.13.8925. [DOI] [PubMed] [Google Scholar]
- 17.Shofuda K., Yasumitsu H., Nishihashi A., Miki K., Miyazaki K. Expression of three membrane-type matrix metalloproteinases (MT-MMPs) in rat vascular smooth muscle cells and characterization of MT3-MMPs with and without transmembrane domain. J. Biol. Chem. 1997;272:9749–9754. doi: 10.1074/jbc.272.15.9749. [DOI] [PubMed] [Google Scholar]
- 18.Takino T., Sato H., Shinagawa A., Seiki M. Identification of the second membrane-type matrix metalloproteinase (MT-MMP-2) gene from a human placenta cDNA library. MT-MMPs form a unique membrane-type subclass in the MMP family. J. Biol. Chem. 1995;270:23013–23020. doi: 10.1074/jbc.270.39.23013. [DOI] [PubMed] [Google Scholar]
- 19.Ohnishi Y., Tajima S., Ishibashi A. Coordinate expression of membrane type-matrix metalloproteinases-2 and 3 (MT2-MMP and MT3-MMP) and matrix metalloproteinase-2 (MMP-2) in primary and metastatic melanoma cells. Eur. J. Dermatol. 2001;11:420–423. [PubMed] [Google Scholar]
- 20.Kitagawa Y., Kunimi K., Uchibayashi T., Sato H., Namiki M. Expression of messenger RNAs for membrane-type 1, 2, and 3 matrix metalloproteinases in human renal cell carcinomas. J. Urol. 1999;162:905–909. doi: 10.1097/00005392-199909010-00088. [DOI] [PubMed] [Google Scholar]
- 21.Plaisier M., Kapiteijn K., Koolwijk P., Fijten C., Hanemaaijer R., Grimbergen J. M., Mulder-Stapel A., Quax P. H., Helmerhorst F. M., van Hinsbergh V. W. Involvement of membrane-type matrix metalloproteinases (MT-MMPs) in capillary tube formation by human endometrial microvascular endothelial cells: role of MT3-MMP. J. Clin. Endocrinol. Metab. 2004;89:5828–5836. doi: 10.1210/jc.2004-0860. [DOI] [PubMed] [Google Scholar]
- 22.Iida J., Pei D., Kang T., Simpson M. A., Herlyn M., Furcht L. T., McCarthy J. B. Melanoma chondroitin sulfate proteoglycan regulates matrix metalloproteinase-dependent human melanoma invasion into type I collagen. J. Biol. Chem. 2001;276:18786–18794. doi: 10.1074/jbc.M010053200. [DOI] [PubMed] [Google Scholar]
- 23.Wegrowski Y., Maquart F. X. Involvement of stromal proteoglycans in tumour progression. Crit. Rev. Oncol. Hematol. 2004;49:259–268. doi: 10.1016/j.critrevonc.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Li Z., Hou W. S., Bromme D. Collagenolytic activity of cathepsin K is specifically modulated by cartilage-resident chondroitin sulfates. Biochemistry. 2000;39:529–536. doi: 10.1021/bi992251u. [DOI] [PubMed] [Google Scholar]
- 26.Li Z., Yasuda Y., Li W., Bogyo M., Katz N., Gordon R. E., Fields G. B., Bromme D. Regulation of collagenase activities of human cathepsins by glycosaminoglycans. J. Biol. Chem. 2004;279:5470–5479. doi: 10.1074/jbc.M310349200. [DOI] [PubMed] [Google Scholar]
- 27.Hirose J., Kawashima H., Yoshie O., Tashiro K., Miyasaka M. Versican interacts with chemokines and modulates cellular responses. J. Biol. Chem. 2001;276:5228–5234. doi: 10.1074/jbc.M007542200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao H., Bernardo M. M., Osenkowski P., Sohail A., Pei D., Nagase H., Kashiwagi M., Soloway P. D., DeClerck Y. A., Fridman R. Differential inhibition of MT3-MMP and MT1-MMP by TIMP-2 and TIMP-3 regulates Pro-MMP-2 activation. J. Biol. Chem. 2004;279:8592–8601. doi: 10.1074/jbc.M308708200. [DOI] [PubMed] [Google Scholar]
- 29.Kinoshita T., Sato H., Okada A., Ohuchi E., Imai K., Okada Y., Seiki M. TIMP-2 promotes activation of progelatinase A by membrane-type 1 matrix metalloproteinase immobilized on agarose beads. J. Biol. Chem. 1998;273:16098–16103. doi: 10.1074/jbc.273.26.16098. [DOI] [PubMed] [Google Scholar]
- 30.Shimada T., Nakamura H., Ohuchi E., Fujii Y., Murakami Y., Sato H., Seiki M., Okada Y. Characterization of a truncated recombinant form of human membrane type 3 matrix metalloproteinase. Eur. J. Biochem. 1999;262:907–914. doi: 10.1046/j.1432-1327.1999.00459.x. [DOI] [PubMed] [Google Scholar]
- 31.Morrison C. J., Butler G. S., Bigg H. F., Roberts C. R., Soloway P. D., Overall C. M. Cellular activation of MMP-2 (gelatinase A) by MT2-MMP occurs via a TIMP-2-independent pathway. J. Biol. Chem. 2001;276:47402–47410. doi: 10.1074/jbc.M108643200. [DOI] [PubMed] [Google Scholar]
- 32.DeClerck Y. A., Yean T. D., Lu H. S., Ting J., Langley K. E. Inhibition of autoproteolytic activation of interstitial procollagenase by recombinant metalloproteinase inhibitor MI/TIMP-2. J. Biol. Chem. 1991;266:3893–3899. [PubMed] [Google Scholar]
- 33.Iida J., Meijne A. M., Oegema T. R., Jr, Yednock T. A., Kovach N. L., Furcht L. T., McCarthy J. B. A role of chondroitin sulfate glycosaminoglycan binding site in alpha4beta1 integrin-mediated melanoma cell adhesion. J. Biol. Chem. 1998;273:5955–5962. doi: 10.1074/jbc.273.10.5955. [DOI] [PubMed] [Google Scholar]
- 34.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995;270:5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 35.Harper J. R., Quaranta V., Reisfeld R. A. Ammonium chloride interferes with a distinct step in the biosynthesis and cell surface expression of human melanoma-type chondroitin sulfate proteoglycan. J. Biol. Chem. 1986;261:3600–3606. [PubMed] [Google Scholar]
- 36.Honke K., Taniguchi N. Sulfotransferases and sulfated oligosaccharides. Med. Res. Rev. 2002;22:637–654. doi: 10.1002/med.10020. [DOI] [PubMed] [Google Scholar]
- 37.Shinmei M., Miyauchi S., Machida A., Miyazaki K. Quantitation of chondroitin 4-sulfate and chondroitin 6-sulfate in pathologic joint fluid. Arthritis Rheum. 1992;35:1304–1308. doi: 10.1002/art.1780351110. [DOI] [PubMed] [Google Scholar]
- 38.Brooks P. C., Silletti S., von Schalscha T. L., Friedlander M., Cheresh D. A. Disruption of angiogenesis by PEX, a noncatalytic metalloproteinase fragment with integrin binding activity. Cell. 1998;92:391–400. doi: 10.1016/s0092-8674(00)80931-9. [DOI] [PubMed] [Google Scholar]
- 39.Brooks P. C., Stromblad S., Sanders L. C., von Schalscha T. L., Aimes R. T., Stetler-Stevenson W. G., Quigley J. P., Cheresh D. A. Localization of matrix metalloproteinase MMP-2 to the surface of invasive cells by interaction with integrin alpha v beta 3. Cell. 1996;85:683–693. doi: 10.1016/s0092-8674(00)81235-0. [DOI] [PubMed] [Google Scholar]
- 40.Petitclerc E., Stromblad S., von Schalscha T. L., Mitjans F., Piulats J., Montgomery A. M., Cheresh D. A., Brooks P. C. Integrin alpha(v)beta3 promotes M21 melanoma growth in human skin by regulating tumor cell survival. Cancer Res. 1999;59:2724–2730. [PubMed] [Google Scholar]
- 41.Deryugina E. I., Ratnikov B. I., Postnova T. I., Rozanov D. V., Strongin A. Y. Processing of integrin alpha(v) subunit by membrane type 1 matrix metalloproteinase stimulates migration of breast carcinoma cells on vitronectin and enhances tyrosine phosphorylation of focal adhesion kinase. J. Biol. Chem. 2002;277:9749–9756. doi: 10.1074/jbc.M110269200. [DOI] [PubMed] [Google Scholar]
- 42.Brekke C., Lundervold A., Enger P. O., Brekken C., Stalsett E., Pedersen T. B., Haraldseth O., Kruger P. G., Bjerkvig R., Chekenya M. NG2 expression regulates vascular morphology and function in human brain tumours. NeuroImage. 2005;29:965–976. doi: 10.1016/j.neuroimage.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 43.Ozerdem U., Grako K. A., Dahlin-Huppe K., Monosov E., Stallcup W. B. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev. Dyn. 2001;222:218–227. doi: 10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- 44.Ozerdem U., Monosov E., Stallcup W. B. NG2 proteoglycan expression by pericytes in pathological microvasculature. Microvasc. Res. 2002;63:129–134. doi: 10.1006/mvre.2001.2376. [DOI] [PubMed] [Google Scholar]
- 45.Suenaga N., Mori H., Itoh Y., Seiki M. CD44 binding through the hemopexin-like domain is critical for its shedding by membrane-type 1 matrix metalloproteinase. Oncogene. 2005;24:859–868. doi: 10.1038/sj.onc.1208258. [DOI] [PubMed] [Google Scholar]
- 46.Mori H., Tomari T., Koshikawa N., Kajita M., Itoh Y., Sato H., Tojo H., Yana I., Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagase H., Woessner J. F., Jr Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 48.Knauper V., Will H., Lopez-Otin C., Smith B., Atkinson S. J., Stanton H., Hembry R. M., Murphy G. Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase a (MMP-2) are able to generate active enzyme. J. Biol. Chem. 1996;271:17124–17131. doi: 10.1074/jbc.271.29.17124. [DOI] [PubMed] [Google Scholar]
- 49.Crabbe T., Ioannou C., Docherty A. J. Human progelatinase A can be activated by autolysis at a rate that is concentration-dependent and enhanced by heparin bound to the C-terminal domain. Eur. J. Biochem. 1993;218:431–438. doi: 10.1111/j.1432-1033.1993.tb18393.x. [DOI] [PubMed] [Google Scholar]









