Abstract
Understanding pathways controlling cardiac development may offer insights that are useful for stem cell-based cardiac repair. Developmental studies indicate that the Wnt/β-catenin pathway negatively regulates cardiac differentiation, whereas studies with pluripotent embryonal carcinoma cells suggest that this pathway promotes cardiogenesis. This apparent contradiction led us to hypothesize that Wnt/β-catenin signaling acts biphasically, either promoting or inhibiting cardiogenesis depending on timing. We used inducible promoters to activate or repress Wnt/β-catenin signaling in zebrafish embryos at different times of development. We found that Wnt/β-catenin signaling before gastrulation promotes cardiac differentiation, whereas signaling during gastrulation inhibits heart formation. Early treatment of differentiating mouse embryonic stem (ES) cells with Wnt-3A stimulates mesoderm induction, activates a feedback loop that subsequently represses the Wnt pathway, and increases cardiac differentiation. Conversely, late activation of β-catenin signaling reduces cardiac differentiation in ES cells. Finally, constitutive overexpression of the β-catenin-independent ligand Wnt-11 increases cardiogenesis in differentiating mouse ES cells. Thus, Wnt/β-catenin signaling promotes cardiac differentiation at early developmental stages and inhibits it later. Control of this pathway may promote derivation of cardiomyocytes for basic research and cell therapy applications.
Keywords: heart development, mesoderm, Dickkopf-1, regeneration
Embryonic stem (ES) cells are pluripotent and spontaneously differentiate into cardiomyocytes, among other cell types. Cardiomyocytes derived from mouse ES cells express contractile proteins and show expected electrophysiologic phenotypes (1), providing a model for cardiac development and a promising source for cell-based cardiac therapy (2). At present, however, only a small percentage of ES cells spontaneously differentiate into cardiomyocytes through serum induction of embryoid bodies (EBs). We sought to discover pathways controlling cardiac differentiation and thereby to find the optimal way to guide differentiation for cardiac repair.
Wnt/β-catenin signaling is critical for vertebrate cardiac development. Overactivation of Wnt/β-catenin signaling suppresses heart formation in Xenopus and chick embryos (3–6) and reduces cardiac differentiation in mouse ES cells (7, 8). Conversely, inhibition of Wnt/β-catenin signaling by overexpression of dickkopf-1 (dkk-1), crescent, or glycogen synthase kinase-3β induces ectopic heart formation in Xenopus and chick embryos (3, 4). Furthermore, deletion of β-catenin in mouse embryonic endoderm results in formation of multiple ectopic hearts (9). These data show that Wnt/β-catenin signaling inhibits cardiogenesis. However, Wnt/β-catenin signaling has been shown recently to activate cardiogenesis in pluripotent P19CL6 cells, indicating that this pathway can be pro-cardiac in some contexts (10). Furthermore, the heart forms from lateral mesoderm (11), a tissue that is known to require Wnt/β-catenin signaling for its specification in Xenopus and zebrafish embryos (12), as well as ES cells (13). Finally, multiple reports indicate that β-catenin-independent Wnt family members, such as Wnt-11, can act to enhance cardiac differentiation (14, 15). We thus hypothesized that Wnt/β-catenin signaling plays multiple, temporally distinct roles in cardiac development and that by defining these processes in zebrafish embryos, we would gain insights into enhancing cardiac differentiation in ES cells.
Results
To test whether Wnt/β-catenin signaling has different effects on heart specification at different stages of vertebrate development, we used zebrafish that were transgenic for heat-shock-inducible activators and inhibitors of the pathway. Activation of Wnt/β-catenin signaling was achieved by overexpression of Wnt8 (hsWnt8 transgenic line), which causes the characteristic phenotypes associated with elevated Wnt/β-catenin signaling (16). To interfere with endogenous Wnt/β-catenin signaling, we overexpressed Dickkopf-1 (17) (hsDkk1 transgenic line), which specifically inhibits Wnt/β-catenin signaling (18).
Wnt/β-Catenin Signaling Promotes Cardiogenesis Before Gastrulation and Inhibits Heart Specification Later.
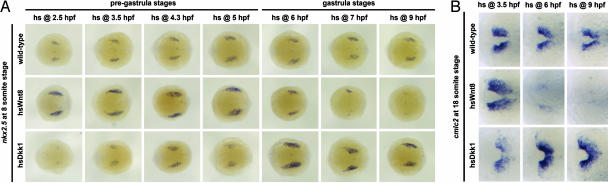
We heat-shocked WT, hsWnt8, and hsDkk1 transgenic embryos for 30 min at different stages of development, fixed them at early somitogenesis stages, and assayed for expression of nkx2.5, a transcription factor that marks early heart progenitors (Fig. 1A). Activation of Wnt/β-catenin signaling at pregastrula stages [up to 5 h postfertilization (5 hpf)] caused an increase in nkx2.5 expression, whereas activation during gastrulation (from 6 to 9 hpf) suppressed nkx2.5 expression (Fig. 1A). Conversely, inhibition of Wnt/β-catenin signaling in hsDkk1 transgenic embryos at pregastrula stages (up to 5 hpf) inhibited nkx2.5 expression, whereas expression of Dkk1 during gastrulation (from 6 to 9 hpf) caused an expansion of nkx2.5 expression (Fig. 1A). Thus, Wnt/β-catenin signaling at pregastrula stages is required for subsequent heart precursor specification, whereas this pathway inhibits heart formation during gastrulation.
Fig. 1.
Wnt/β-catenin signaling promotes zebrafish heart specification at pregastrula stages but inhibits heart formation at gastrula stages. WT, hsWnt8, or hsDkk1 transgenic embryos were heat-shocked for 30 min starting at the indicated times. (A) The embryos were fixed at 13 hpf (eight-somite stage) and processed for nkx2.5 in situ hybridization. The embryos are shown in dorsal view (anterior left); n > 12 for each experimental point. The experiment was repeated twice each for heat shock at 3.5, 6, and 9 hpf. The combined percentages of affected embryos from these three experiments were as follows: Wnt8, 3.5 hpf, up: 86% (n = 43); Wnt8, 6 hpf, up: 36% (n = 59); Wnt8, 9 hpf, down: 100% (n = 28); Dkk1, 3.5 hpf, down: 100% (n = 52); Dkk1, 6 hpf, up: 73% (n = 40); and Dkk1, 9 hpf, up: 18% (n = 61). (B) The embryos were fixed at 18 hpf (18-somite stage) and processed for in situ hybridization with cardiac myosin light chain 2 (cmlc2). Close-up views of cmlc2 expression are shown (anterior left). Wnt8 overexpression at 3.5 hpf results in expansion of cmlc2 expression (24 of 29 embryos) but represses cmlc2 expression when activated at 6 hpf (15 of 19) or 9 hpf (13 of 14). Conversely, Dkk1 overexpression at 3.5 hpf weakly represses cmlc2 (7 of 20) but increases cmlc2 expression when activated at 6 hpf (11 of 12) or 9 hpf (8 of 19).
Remarkably, the response of heart precursors to Wnt/β-catenin signaling switched from positive to negative over a 1-h window at the beginning of gastrulation (compare heat shocks at 5 hpf with those at 6 hpf in Fig. 1A). The same temporally opposing effects of Wnt/β-catenin signaling were observed by monitoring the expression of cardiac myosin light chain 2 (cmlc2), a myofibrillar protein that is expressed later in cardiac differentiation (Fig. 1B). This indicates that Wnt/β-catenin signaling does not just induce a transient effect on heart precursor cells but, rather, induces effects that persist through later stages of development. Overexpression of Wnt5, which activates β-catenin-independent signaling pathways during early zebrafish development, with the use of the same heat-shock induction system had no discernible effect on the size of the heart-forming field (data not shown).
These data point to an early role for Wnt/β-catenin signaling in the positive regulation of heart formation. We therefore asked whether induction of heart precursors was accompanied by an induction of all mesodermal cell types. To test this, we heat-shocked transgenic embryos at 3.5 hpf and assayed them at three stages that covered a time window from late blastula to mid-gastrula stages for expression of the pan-mesodermal marker ntl (ortholog of murine brachyury T; Fig. 2A), and the markers antivin and wnt11 that at these stages are specifically expressed in all mesodermal progenitors. After overexpression of Wnt8, we found no increase in mesodermal marker gene expression at any stage (Fig. 2A and data not shown). In contrast, induction of Dkk1 reduced the expression of all three markers at all times (Fig. 2A and data not shown). Thus, although Wnt/β-catenin signaling appears to be required for full mesoderm formation, the induction of heart precursors by overexpression of Wnt8 at pregastrula stages cannot be explained by an overall increase in mesoderm.
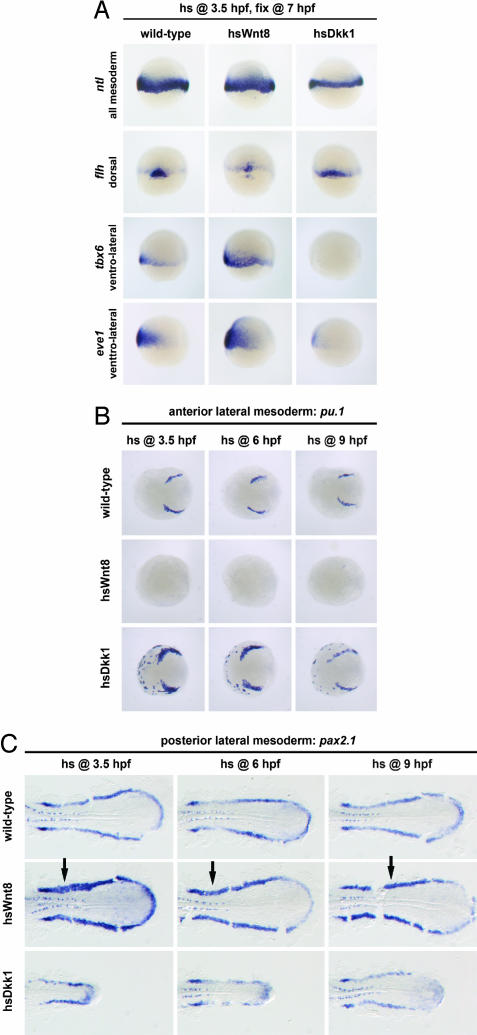
Fig. 2.
Effect of manipulating Wnt/β-catenin signaling on mesoderm induction and mesoderm patterning. WT, hsWnt8, or hsDkk1 transgenic embryos were heat-shocked at 3.5 hpf for 30 min, fixed at 7 hpf, and processed for in situ hybridization with the marker genes indicated in the figure. (A) Wnt8 overexpression is not sufficient to expand expression of the pan-mesodermal marker ntl, but Dkk1 overexpression results in decreased ntl expression (44 of 44 embryos). Wnt8 overexpression reduces expression of the dorsal marker flh (13 of 20) and expands the ventrolateral markers tbx6 (13 of 18) and eve1 (30 of 31). Conversely, Dkk1 overexpression expands flh (32 of 34) and suppresses tbx6 (15 of 15) and eve1 (30 of 30) expression. (B and C) Wnt/β-catenin signaling at pregastrula and gastrula stages suppresses anterior lateral mesoderm and expands posterior lateral mesoderm. WT, hsWnt8, or hsDkk1 transgenic embryos were heat-shocked for 30 min at the times indicated in the figure, fixed at 13 hpf (eight-somite stage), and processed for in situ hybridization. (B) pu.1 expression in anterior lateral mesoderm (myeloid precursors) is repressed by Wnt8 overexpression at 3.5 hpf (13 of 13 embryos), 6 hpf (16 of 16), and 9 hpf (17 of 17), whereas Dkk1 overexpression results in expanded and ectopic pu.1 expression (3.5 hpf, 19 of 20; 6 hpf, 22 of 22; 9 hpf, 10 of 13). Note that the anterior pole of the embryos is shown to demonstrate the ectopic pu.1 expression in hsDkk1 embryos, which, in other views, would not have been visible. (C) pax2.1 expression in pronephric mesoderm (arrows) is expanded by Wnt8 overexpression at 3.5 hpf (11 of 17 embryos), 6 hpf (26 of 42), or 9 hpf (10 of 10), whereas Dkk1 overexpression results in severe shortening of posterior pax2.1-expressing mesodermal regions (3.5 hpf, 29 of 29; 6 hpf, 21 of 21; 9 hpf, 18 of 18). The posterior half of flat-mounted embryos (anterior left) is shown.
Other Lateral Mesodermal Cell Fates Do Not Show a Biphasic Response to Wnt Signaling.
It is well known that Wnt/β-catenin signaling patterns mesoderm along the dorsal–ventral axis during pregastrula stages by promoting lateral mesodermal fates (12, 19, 20). Because heart precursors are found in lateral regions of the pregastrula embryo (11), we asked whether the early cardiac induction by Wnt signaling reflects a general expansion of cells with lateral mesodermal fates. Overexpression of Wnt8 at 3.5 hpf reduced the dorsal marker flh and expanded the ventrolateral markers tbx6 and eve1, whereas overexpression of Dkk1 had the opposite effect: it expanded flh and suppressed tbx6 and eve1 (Fig. 2A). Thus, Wnt/β-catenin signaling at pregastrula stages is required for formation of ventral and lateral mesodermal fates, and overactivation of Wnt signaling is sufficient to expand these cell types.
These results are consistent with a model in which the early heart-inducing role of Wnt signaling is due to a general expansion of lateral mesodermal fates. To test this, we assayed other lateral mesodermal cell types at the same stages analyzed for nkx2.5 in Fig. 1A. Interestingly, in contrast to the biphasic roles of Wnt/β-catenin signaling on heart markers, we found that all other tested lateral mesodermal markers behaved in a consistent manner, regardless of whether Wnt signaling was manipulated at early or late stages. Specifically, overexpression of Wnt8 at 3.5 hpf (pregastrula), 6 hpf (early gastrula), or 9 hpf (late gastrula stage) completely repressed pu.1 in anterior myeloid precursors (Fig. 2B). Conversely, overexpression of Dkk1 at early or late stages caused expansion and ectopic expression of pu.1 (Fig. 2B). Likewise, expression of flk1 in anterior endothelial precursors and of nkx2.7 in anterior lateral plate mesoderm was repressed by Wnt8 and enhanced by Dkk1, no matter when the transgene was induced [see supporting information (SI) Fig. 6 A and B]. In addition, overexpression of Dkk1 resulted in ectopic expression of the anterior lateral mesodermal markers gata4 and nkx2.7 in the ventral (SI Fig. 7 A and C) and posterior (SI Fig. 7 B and D) embryo. We conclude that Wnt signaling is required to restrict expression of these genes to anterior and lateral regions and that anterior lateral mesodermal fates are repressed by Wnt/β-catenin signaling from pregastrula stages until the end of gastrulation.
In contrast, the more posteriorly expressed gene pax2.1, which marks precursors of the pronephric tubules and ducts, is expanded by overexpression of Wnt8 (Fig. 2C), regardless of timing. Conversely, overexpression of Dkk1 reduced pax2.1 expression in pronephric mesoderm (Fig. 2C). Thus, Wnt/β-catenin signaling represses anterior lateral mesodermal fates and promotes posterior lateral mesodermal fates. Consequently, induction of heart fate by Wnt signaling at pregastrula stages cannot be explained by a general expansion of lateral mesodermal fates.
Heart precursors appear to be unique among lateral mesoderm in that Wnt/β-catenin signaling has temporally distinct effects on their specification. Whereas Wnt signaling represses anterior lateral mesodermal cell types and induces posterior cell types, heart precursors are induced early and repressed late. This prompted us to map the position of heart precursors along the anteroposterior axis. In WT embryos, pu.1 is expressed more anteriorly than nkx2.5, whereas the mesodermal expression of pax2.1 is more posterior than nkx2.5 (SI Fig. 8). Thus, in zebrafish the heart does not form from the most anterior lateral plate mesoderm but, rather, at an intermediate anteroposterior level.
Early Activation of Wnt/β-Catenin Signaling Promotes Cardiogenesis in ES Cells.
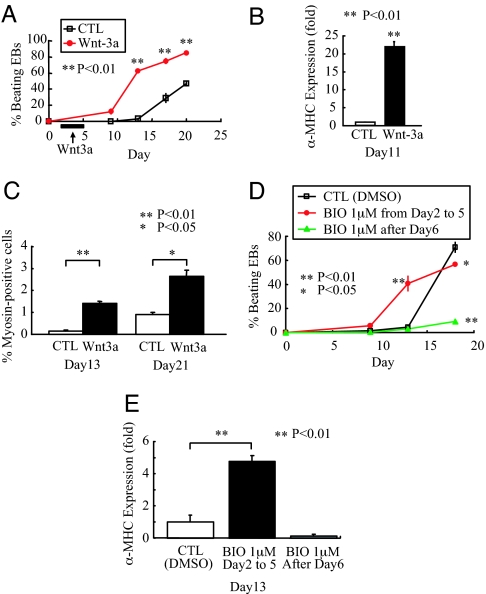
Based on the potent ability of Wnt/β-catenin signaling to regulate cardiac differentiation in zebrafish embryos, we next examined whether this pathway induces cardiogenesis in mouse ES cells. Mouse ES cells were differentiated as three-dimensional EBs, where they received 100 ng/ml recombinant mouse Wnt-3A every other day from days 2 to 5. Wnt-3A dramatically increased the fraction of EBs with beating activity at day 13 (Wnt-3A: 62.2 ± 2.2%, control: 3.3 ± 1.7%, P < 0.01), at day 17 (Wnt-3A: 74.8 ± 3.7%, control: 29.1 ± 6.2%, P < 0.01), and at day 20 (Wnt-3A: 85.1 ± 1.1%, control: 46.8 ± 3.0%, P < 0.01) (Fig. 3A). Although these results are most representative, we should note that, in some experiments, the control cultures exhibited a “catch up” phase of cardiac differentiation, when no significant differences in beating activity were observed at the last time point. To provide a second index of cardiac differentiation, we also determined the expression levels of cardiac-specific α-myosin heavy chain (α-MHC) mRNA in these groups. At day 11, α-MHC expression in EBs treated with Wnt-3A was increased 22-fold compared with control (P < 0.01; Fig. 3B). Finally, we determined the fraction of the cultures expressing sarcomeric MHC by using flow cytometry. Treatment with Wnt-3A increased the percentage of cardiomyocytes from 0.14 ± 0.04% to 1.4 ± 0.09% at day 13 (P < 0.01) and from 0.91 ± 0.08% to 2.64 ± 0.29% at day 21 (P < 0.05) (Fig. 3C).
Fig. 3.
Biphasic effect of Wnt/β-catenin signaling on cardiac differentiation in mouse ES cells. (A) Addition of Wnt-3A from day 2 to day 5 dramatically accelerated the onset of beating activity in EBs. ∗∗, P < 0.01 compared with control (CTL). Representative of three independent experiments, each performed in triplicate. (B) α-MHC expression at day 11 was examined by quantitative RT-PCR, normalized to GAPDH, and expressed as a ratio to control EBs. Wnt-3A treatment caused a 22-fold increase in α-MHC expression. ∗∗, P < 0.01 compared with control. Representative of three independent experiments, each performed in triplicate. (C) Analysis of sarcomeric MHC by flow cytometry demonstrates that early treatment with Wnt-3A enhances cardiomyocyte content at days 13 and 21 by 10- and 3-fold, respectively. (D and E) Analysis of beating activity (D) and α-MHC expression at day 13 (E) showed that the glycogen synthase kinase-3β inhibitor BIO has a biphasic role on cardiogenesis, enhancing cardiogenesis when given early and inhibiting cardiogenesis when given late. Note that in this experiment, control cultures exhibited a “catch-up” phase of accelerated differentiation that slightly surpassed that of day 18 cultures receiving early BIO treatment. A similar catch-up phase was occasionally observed by using recombinant Wnt-3A, although in most experiments, both early and late cardiogenesis was increased.
To define the time period when EBs were receptive to induction of cardiogenesis via Wnt/β-catenin signaling, experiments were repeated in which EBs received Wnt-3A from days 7 to 10. This later treatment did not result in a significant difference in beating activity compared with untreated EBs (data not shown). We hypothesized that the lack of an effect resulted from poor penetration of Wnt-3A into the core of the EB, where most cardiogenesis occurs. To address this issue, we used the small-molecule inhibitor of glycogen synthase kinase-3β, 6-bromoindirubicin-3′-oxime (BIO), which can readily penetrate an EB and results in activation of β-catenin signaling. These experiments showed a profound biphasic effect on cardiac differentiation (Fig. 3 D and E). Although addition of BIO at days 2 and 4 resulted in an ≈10-fold enhancement of beating activity by day 13, addition after day 6 markedly inhibited cardiogenesis, resulting in a 10-fold reduction in beating activity at the latest time point (day 18) (Fig. 3D). Analyses of α-MHC expression confirmed that BIO stimulated cardiogenesis early and inhibited it later in ES cell differentiation (Fig. 3E). Thus, the biphasic effect of Wnt/β-catenin signaling was observed in both the in vitro mouse ES cell system and in the in vivo zebrafish model.
Wnt3a Induces Mesoderm and an Inhibitory Feedback Loop in ES Cells.
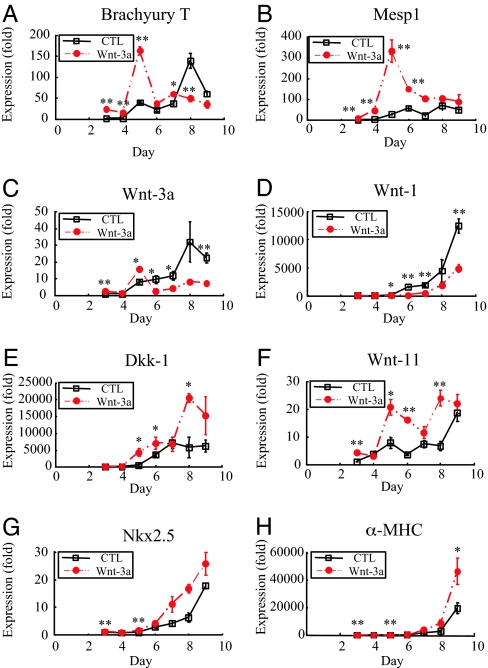
To understand possible mechanisms underlying cardiac induction by Wnt-3A, we studied regulation of mesodermal and Wnt-related genes during the early phases of EB differentiation by quantitative RT-PCR. Brachyury T, a mesoderm-specific transcription factor (21, 22), and the transcription factor Mesp1 are enriched in precardiac mesoderm (23, 24). Brachyury T expression was markedly accelerated by treatment with Wnt-3a, showing a peak of expression at day 5, rather than at day 8, in controls (Fig. 4A). Similarly, Wnt-3A-treated cultures showed an ≈320-fold induction of Mesp1, peaking at 5 days, which was greater and more rapid induction than the ≈65-fold induction observed at 8 days in controls (Fig. 4B). Analysis of additional mesoderm markers at day 13 indicated that Wnt-3A increased expression of the endothelial marker vascular endothelial cadherin by 3-fold and the pan-leukocyte marker CD45 by 7-fold (data not shown). Thus, in contrast to the zebrafish, early activation of Wnt/β-catenin signaling in mouse ES cells appears sufficient to induce mesoderm.
Fig. 4.
Effects of Wnt-3A treatment on expression of markers for mesoderm, the Wnt pathway, and cardiomyocyte differentiation. Differentiating EBs received Wnt-3A from day 2 to day 5. Triplicate cultures were harvested for RNA isolation at the indicated times and analyzed by quantitative RT-PCR for the indicated transcripts. Values were normalized to GAPDH, and control (CTL) values were arbitrarily set to 1.0 on day 3. ∗, P < 0.05 vs. control; ∗∗, P < 0.01 vs. control. (A and B) Wnt-3A accelerated expression of the pan-mesoderm marker Brachyury T by 3 days, indicating enhanced mesoderm induction. Wnt-3A markedly up-regulated expression of the precardiac mesoderm marker Mesp1, peaking at day 5 of differentiation. (C–E) Wnt-3A suppressed expression of Wnt-3A and Wnt-1 while inducing expression of Dkk-1, an inhibitor of Wnt/β-catenin signaling. This demonstrates a negative feedback loop for canonical Wnt signaling in differentiating ES cells. (F) Expression of Wnt-11 was up-regulated at multiple time points by treatment with Wnt-3A. Note the substantial increase in Wnt-11 in untreated control EBs over time (although delayed relative to Wnt-3A-treated). (G and H) Expression of the cardiac homeodomain transcription factor Nkx2.5 and contractile protein α-MHC was markedly increased by Wnt-3A by day 9.
We next examined expression of Wnt-related genes in the EBs treated with Wnt-3A (Figs. 4 C–F). Control EBs showed progressive increases in Wnt-1 and Wnt-3A beginning on day 5. Transcript levels of the Wnt/β-catenin inhibitor Dkk-1 increased by day 6 in controls and plateaued from day 7 to 9. Expression of Wnt-11 [implicated as a pro-cardiogenesis factor (15) (Fig. 5)] increased from day 3 to 5, and increased again sharply from day 8 to 9 in controls. Treatment with Wnt-3A markedly reduced expression of Wnt-1 (Fig. 4D) and Wnt-3A (Fig. 4C). In contrast, expression of Dkk-1 (Fig. 4E) and Wnt-11 (Fig. 4F) both increased after treatment with Wnt-3a. This is interesting, because in some contexts, Wnt-11, like Dkk, can inhibit Wnt/β-catenin signaling (25). These findings indicate that treatment with Wnt-3A induces a negative-feedback loop in differentiating ES cells, suppressing transcripts associated with β-catenin pathway activation and increasing transcripts associated with pathway inhibition. Finally, and consistent with our observations in zebrafish, Wnt-3A accelerated expression of the cardiac transcription factor Nkx2.5 and α-MHC in differentiating EBs, consistent with their earlier cardiac differentiation (Fig. 4 G and H).
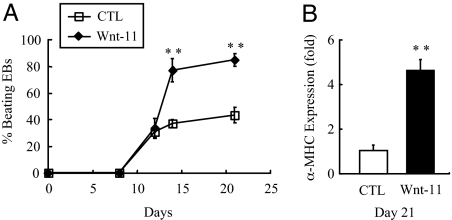
Fig. 5.
Overexpression of Wnt-11 enhances late cardiac differentiation in mouse ES cells. Mouse R1 ES cells were lentivirally transduced with a constitutively expressing human Wnt-11 construct plus EGFP or a GFP-only virus (control); the cells were then differentiated as EBs. (A) Although both groups initiated beating activity at the same time, overexpression of Wnt-11 significantly increased the extent of beating at days 14 and 21 (n = 3 per time point). ∗∗, P < 0.01 compared with control. These data are representative of six independent experiments, each performed in triplicate. In two of the six experiments, poor overall cardiac differentiation was noted (<10% of control EBs beating at 21 days), and Wnt-11 treatment did not improve differentiation in these cases. (B) α-MHC expression in EBs overexpressing human Wnt-11 was increased at day 21 (n = 3). ∗∗, P < 0.01 compared with control EBs expressing only EGFP. Data represent three experiments, each performed in triplicate.
Wnt-11 Overexpression Enhances Cardiogenesis.
The increase of Wnt-11 expression in the later phases of differentiation suggested this Wnt might also control cardiac differentiation from mesodermal cells. To test this, we overexpressed human Wnt-11 or EGFP as a control in ES cells and differentiated them as above. In contrast to treatment with Wnt-3A, continuous overexpression of Wnt-11 did not accelerate the onset of beating activity in EBs, but instead increased the final extent of cardiogenesis by 3- to 4-fold vs. controls (Fig. 5A). At day 21, α-MHC expression in the Wnt-11 group was increased 4.6-fold compared with controls (P < 0.01; Fig. 5B). These data are consistent with the notion that different Wnts, and perhaps multiple Wnt signaling pathways, have distinct roles in cardiac differentiation of ES cells.
Discussion
The changing response to Wnt/β-catenin signaling over time explains some of the apparent discrepancies in the literature regarding Wnts and cardiac differentiation (3–5, 10). Our data show that, depending on the time of activation, Wnt/β-catenin-dependent pathways have positive or negative effects on cardiogenesis in both the mouse ES cell system and the zebrafish model. We also show that Wnt-3A induces an inhibitory feedback loop in ES cells that results in the suppression of transcripts implicated in β-catenin pathway activation and the augmentation of transcripts associated with pathway inhibition, such as Dkk-1 and Wnt-11. Thus, one unanticipated effect of early Wnt/β-catenin stimulation appears to be suppression of the activity of the same pathway after mesoderm forms in differentiating ES cell cultures. This later suppression may also contribute to enhanced cardiogenesis. The gene expression time course prompted us to test whether overexpression of Wnt-11, also reported to be pro-cardiogenic (14, 15, 26), would enhance cardiac differentiation in our system. We found significant induction of cardiac differentiation by Wnt-11 (Fig. 5), but unlike Wnt-3A, Wnt-11 did not accelerate cardiac differentiation but, instead, enhanced its final extent. Additional studies will be required to determine whether Wnt-11 acts through antagonism of β-catenin signaling or through other pathways such as calcium signaling and the planar cell-polarity pathway (27).
Analysis of mRNA expression indicated that increased Wnt/β-catenin signaling enhanced mesoderm formation in the mouse ES cell system. These data are consistent with experiments by Lindsley et al. (13) and Gadue et al. (28), who found enhanced expression of mesoderm markers following early treatment of differentiating mouse ES cells with Wnt3A. In contrast, in zebrafish embryos, overexpression of Wnt8 did not increase total mesoderm differentiation, although blocking endogenous Wnt signaling by overexpression of Dkk-1 reduced mesoderm differentiation. These data indicate that β-catenin-dependent Wnt ligands are rate-limiting for mesoderm differentiation in EBs, whereas in the embryo, Wnt ligands are necessary but appear to be present above limiting concentrations.
Inhibition of cardiogenesis by Wnt/β-catenin signaling during and after gastrulation suggests that cells normally destined to become cardiomyocytes are redirected toward an alternate mesoderm fate. Kattman et al. (29) reported that mouse ES cells give rise to two waves of progenitors marked by expression of the VEGF receptor Flk-1. The first wave of Flk-1+ cells function like hemangioblasts, giving rise to hematopoietic cells and endothelium. The second wave gives rise to cardiomyocytes, endothelium, and smooth muscle cells. Naito et al. (8) reported that late addition of Wnt-3A to differentiating mouse ES cells resulted in expansion of hematopoietic and vascular fates. It is possible that activation of Wnt/β-catenin signaling after the formation of mesoderm redirects the Flk-1+ cardiovascular progenitors toward the hemangioblastic phenotype.
In summary, we demonstrated that Wnt/β-catenin signaling has a biphasic role in controlling differentiation of cardiomyocytes in vivo and in a scalable in vitro model that does not require hanging drops. At early stages of zebrafish development, Wnt/β-catenin signaling promotes cardiogenesis, whereas at later stages, Wnt signaling helps to define the proper size of the heart-forming field. Armed with this knowledge, it is now possible to significantly increase cardiac differentiation in ES cell cultures by manipulating Wnt signaling in a defined temporal sequence. This offers an example of how lessons from embryonic development can be applied to stem cell biology. Many other factors, such as TGF-β1 (30), bone morphogenetic proteins (30–34) or their endogenous antagonists (35), and fibroblast growth factors (31, 36, 37), have been implicated in cardiac induction of embryos and pluripotent cells. Further investigations are needed to clarify the interaction between Wnt signaling and these factors to optimally promote ES cell cardiac differentiation. Our findings establish a model of Wnt signaling in control of cardiac differentiation that may ultimately be useful in directing differentiation of stem cells into cardiomyocytes for cardiac repair applications.
Methods
Heat-Shock-Inducible Transgenic Fish Lines.
Establishment and characterization of the hsWnt8GFP transgenic line (16) and establishment and characterization of the hsDkk1GFP (18) have been published previously. For most experiments, homozygous carriers of the transgene were crossed to WT fish, resulting in clutches of embryos that were all heterozygous for the transgene, obviating the need to identify transgenic embryos. In some experiments, heterozygous carriers of the hsDkk1GFP transgene were crossed to WT fish, and transgenic embryos were sorted from WT embryos by GFP fluorescence 1 h after heat shock. As controls, embryos derived from incrosses of WT fish were used. Heat shocks were performed by adding embryo water preheated to 40°C to the embryos and incubating them in an air incubator at 37°C for 30 min, after which time they were moved to a 28.5°C incubator.
Whole-Mount in Situ Hybridization.
Two-color mRNA in situ hybridization was performed as described by Jowett and Lettice (38) with modifications according to Hauptmann and Gerster (39), and a combination of INT [2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl-tetrazolium chloride] and BCIP [5-bromo-4-chloro-3-indolyl-phosphate], both at 175 μg/ml, was used as alkaline phosphatase substrate in the second color reaction, producing a red-brownish color.
ES Cell Culture and Differentiation.
Details of mouse ES cell culture can be found in SI Methods. Briefly, undifferentiated R1 mouse ES cells were maintained in feeder cell-free culture by using leukemia inhibitory factor supplementation. EBs were formed by mass culture in ultra-low attachment plates and transferred to adherent growth conditions after 7 days. The number of spontaneously beating EBs was counted by using a DMIL microscope (Leica, Deerfield, IL) fitted with a grid system.
Lentiviral Transduction and Quantitative RT-PCR.
See SI Methods for details.
Statistical Analysis.
Results are expressed as means ± SEM. Groups were compared by ANOVA, followed by Fisher's partial least-squares difference test, with P < 0.05 considered as significant.
Supplementary Material
Acknowledgments
We thank Mr. Jamie Fugate for assistance with ES cell culture experiments. This work was supported in part by National Institutes of Health Grants HL61553, HL03174, HL64387, HL84642 (all to C.E.M.), GM69983 (to C.E.M. and R.T.M.), and GM073887 (to R.T.M.); the Cell Science Research Foundation (S.U.); and The Mochida Memorial Foundation for Medical and Pharmaceutical Research (S.U.). R.T.M. is an Investigator and G.W. is an Associate of the Howard Hughes Medical Institute.
Abbreviations
- BIO
6-bromoindirubicin-3′-oxime
- EB
embryoid body
- hpf
hours postfertilization
- MHC
myosin heavy chain.
Footnotes
The authors declare no conflict of interest.
See Commentary on 9549.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702859104/DC1.
References
- 1.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 2.Laflamme MA, Murry CE. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 3.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Genes Dev. 2001;15:316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider VA, Mercola M. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tzahor E, Lassar AB. Genes Dev. 2001;15:255–260. doi: 10.1101/gad.871501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manisastry SM, Han M, Linask KK. Dev Dyn. 2006;235:2160–2174. doi: 10.1002/dvdy.20878. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita JK, Takano M, Hiraoka-Kanie M, Shimazu C, Peishi Y, Yanagi K, Nakano A, Inoue E, Kita F, Nishikawa S. FASEB J. 2005;19:1534–1536. doi: 10.1096/fj.04-3540fje. [DOI] [PubMed] [Google Scholar]
- 8.Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Proc Natl Acad Sci USA. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T, Sano M, Songyang Z, Schneider MD. Proc Natl Acad Sci USA. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- 12.Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 13.Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Development (Cambridge, UK) 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg CA, Eisenberg LM. Dev Dyn. 1999;216:45–58. doi: 10.1002/(SICI)1097-0177(199909)216:1<45::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Pandur P, Lasche M, Eisenberg LM, Kuhl M. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 16.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. Curr Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 17.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 18.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Development (Cambridge, UK) 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 19.Christian JL, Moon RT. Genes Dev. 1993;7:13–28. doi: 10.1101/gad.7.1.13. [DOI] [PubMed] [Google Scholar]
- 20.Erter CE, Wilm TP, Basler N, Wright CV, Solnica-Krezel L. Development (Cambridge, UK) 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson DG, Peters G, Dickson C, McMahon AP. EMBO J. 1988;7:691–695. doi: 10.1002/j.1460-2075.1988.tb02864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson DG, Bhatt S, Herrmann BG. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 23.Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. Development (Cambridge, UK) 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- 24.Saga Y, Kitajima S, Miyagawa-Tomita S. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- 25.Torres MA, Yang-Snyder JA, Purcell SM, DeMarais AA, McGrew LL, Moon RT. J Cell Biol. 1996;133:1123–1137. doi: 10.1083/jcb.133.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terami H, Hidaka K, Katsumata T, Iio A, Morisaki T. Biochem Biophys Res Commun. 2004;325:968–975. doi: 10.1016/j.bbrc.2004.10.103. [DOI] [PubMed] [Google Scholar]
- 27.Kohn AD, Moon RT. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 28.Gadue P, Huber TL, Paddison PJ, Keller GM. Proc Natl Acad Sci USA. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kattman SJ, Huber TL, Keller GM. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, Puceat M. FASEB J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- 31.Kawai T, Takahashi T, Esaki M, Ushikoshi H, Nagano S, Fujiwara H, Kosai K. Circ J. 2004;68:691–702. doi: 10.1253/circj.68.691. [DOI] [PubMed] [Google Scholar]
- 32.Schlange T, Andree B, Arnold HH, Brand T. Mech Dev. 2000;91:259–270. doi: 10.1016/s0925-4773(99)00311-1. [DOI] [PubMed] [Google Scholar]
- 33.Schultheiss TM, Lassar AB. Cold Spring Harbor Symp Quant Biol. 1997;62:413–419. [PubMed] [Google Scholar]
- 34.Jamali M, Karamboulas C, Rogerson PJ, Skerjanc IS. FEBS Lett. 2001;509:126–130. doi: 10.1016/s0014-5793(01)03151-9. [DOI] [PubMed] [Google Scholar]
- 35.Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, et al. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- 36.Dell'Era P, Ronca R, Coco L, Nicoli S, Metra M, Presta M. Circ Res. 2003;93:414–420. doi: 10.1161/01.RES.0000089460.12061.E1. [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Sasse J, McAllister D, Lough J. Dev Dyn. 1996;207:429–438. doi: 10.1002/(SICI)1097-0177(199612)207:4<429::AID-AJA7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 38.Jowett T, Lettice L. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 39.Hauptmann G, Gerster T. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.