Abstract
Type III secretion systems (T3SS), found in several Gram-negative pathogens, are nanomachines involved in the transport of virulence effectors directly into the cytoplasm of target cells. T3SS are essentially composed of basal membrane-embedded ring-like structures and a hollow needle formed by a single polymerized protein. Within the bacterial cytoplasm, the T3SS needle protein requires two distinct chaperones for stabilization before its secretion, without which the entire T3SS is nonfunctional. The 2.0-Å x-ray crystal structure of the PscE-PscF55–85-PscG heterotrimeric complex from Pseudomonas aeruginosa reveals that the C terminus of the needle protein PscF is engulfed within the hydrophobic groove of the tetratricopeptide-like molecule PscG, indicating that the macromolecular scaffold necessary to stabilize the T3SS needle is totally distinct from chaperoned complexes between pilus- or flagellum-forming molecules. Disruption of specific PscG–PscF interactions leads to impairment of bacterial cytotoxicity toward macrophages, indicating that this essential heterotrimer, which possesses homologs in a wide variety of pathogens, is a unique attractive target for the development of novel antibacterials.
Keywords: bacterial pathogenicity, chaperones, x-ray crystallography
Type III secretion systems (T3SS) are protein-secretion machineries that have been linked to the transport of virulence effectors from the cytosol of a variety of Gram-negative pathogens directly into the host cell cytoplasm. Although many of the protein components of T3SS from different microorganisms share considerable structural and functional similarities, transported effectors are bacterium-dependent and play a variety of subversive roles that range from induction of apoptosis to the suppression of host defense mechanisms (1–3). As a consequence, pathogens that harbor T3SS cause a wide diversity of diseases, including plague (Yersinia pestis), typhoid fever (Salmonella typhi), and bacillary dysentery (Shigella dysenteriae). Pseudomonas aeruginosa, a major nosocomial bacterium whose pathogenesis highly depends on a functional T3SS (4, 5), is the causative agent of opportunistic infections in neutropenic and other immunocompromised individuals as well as in severe burn victims. In addition, P. aeruginosa is the primary cause of chronic infections in ventilator-assisted and cystic fibrosis patients and ultimately leads to loss of lung function and death in the latter group (6). The natural resistance of P. aeruginosa to a broad range of antibiotics, as well as its widespread presence in hospital settings worldwide, has proven to be a challenge necessitating the search for yet-unexplored antibacterial development targets.
The injectisome, a key component of the T3SS, is composed of membrane-embedded protein rings extended by a hollow needle formed by a single polymerized protein. It is widely accepted that the T3SS needle could be the conduit through which effectors and toxins are exported, although other paths for effector transport are still possible (2, 7–9). The T3SS needle protein is synthesized in the bacterial cytoplasm but polymerizes only after secretion to the bacterial surface, following a highly regulated sequence of events (8, 10–13). Thus, within the bacterial cytoplasm, the T3SS needle-forming protein must be prevented from undergoing self-assembly, while at the same time being primed for secretion and subsequent polymerization into a T3SS-effector-permissive conduit. T3SS needle-forming proteins have been shown to be highly helical in nature, and an elegant model of the T3SS needle from Shigella flexneri, which combines a 17-Å electron microscopy map onto which the crystal structure of the needle-forming monomer was modeled, suggests that self-assembly occurs through head-to-tail association of the monomers (14, 15).
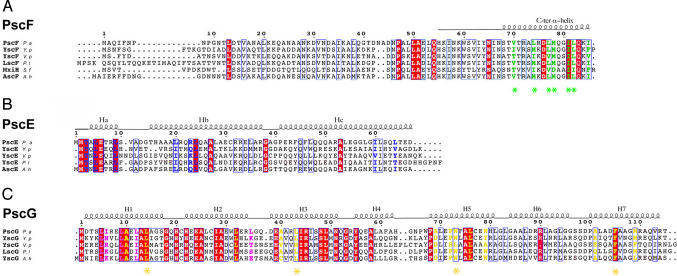
In P. aeruginosa, the T3SS needle is formed by polymerized PscF, an 85-residue protein that displays sequence similarity to T3SS needle-forming proteins from other major pathogens, such as YscF from Y. pestis (69%). PscF is found in the bacterial cytoplasm uniquely within a soluble heterotrimeric complex with two partners, PscE and PscG; clinical isolates mutated to lack either one of these two molecules are viable but noncytotoxic (16). Interestingly, homologs of PscE and PscG exist in a variety of pathogens (Fig. 1), suggesting that, unlike pili, flagella, or even other T3SS-associated filaments, the complexity of the T3SS needle formation mechanism requires that the needle protein be stabilized by two distinct partners. To define the mechanism and structural requirements for T3SS needle protein stabilization within the bacterial cytoplasm, we solved the structure of the PscE-PscF55–85-PscG complex from a clinical strain of P. aeruginosa (CHA) to 2.0-Å resolution. PscG and PscE fold in the form of a cupped hand that stabilizes the highly amphipathic C-terminal helix of PscF, whose hydrophobic face plays a key role in T3SS needle polymerization and stability. Strains in which the PscG–PscF recognition platform was mutated show dramatically decreased cytotoxicity profiles, revealing that the T3SS needle stabilization ternary complex is a previously undescribed, tractable macromolecular target for the development of new antibacterials applicable against a wide variety of pathogens.
Fig. 1.
Structure-based sequence alignment of T3SS needle proteins and their cytoplasmic partners from diverse pathogens. (A) PscF from P. aeruginosa (causative agent of nosocomial infections), YscF from Y. pestis (bubonic plague), YscF from Vibrio parahaemolyticus (gastrointestinal illness), LscF from Photorhabdus luminescens (insect pathogen), MxiH from S. flexneri (bacillary dysentery), and AscF from Aeromonas hydrophila (respiratory illness in humans, reptiles, and birds). Residues mutated in this study and shown to be involved in polymerization and needle stability are highlighted with stars, whereas those which interact with PscG are shown in green. (B) PscE homologs from bacteria described above share a major homology region within their N-terminal domains. PscE residues which interact with PscG are shown in blue. (C) PscG homologs share high conservation within residues which interact with PscE (purple) and PscF (yellow). Residues mutated to Ser in this study are indicated with stars.
Results and Discussion
A Complex Unique System to Stabilize the T3SS Needle Protein.
Crystals of the heterotrimeric complex (PscE1–67-PscF1–85-PscG1–115), in which all three proteins were expressed in full-length form and in tandem from the pscEFGHI section of the exsD-pscBCDEFGHIJKL P. aeruginosa operon in an Escherichia coli system (16), diffracted only to 7 Å. However, papain treatment of the heterotrimer yielded a stable complex in which PscE and PscG remained intact, but the first 54 residues of PscF were removed, suggesting that this region could be inherently flexible. The PscE-PscF55–85-PscG complex yielded crystals that diffracted to 2.0 Å, and structure solution was accomplished by performing a single anomalous dispersion experiment by using the scattering of 3 Ni+2 ions introduced into the crystallization solution.
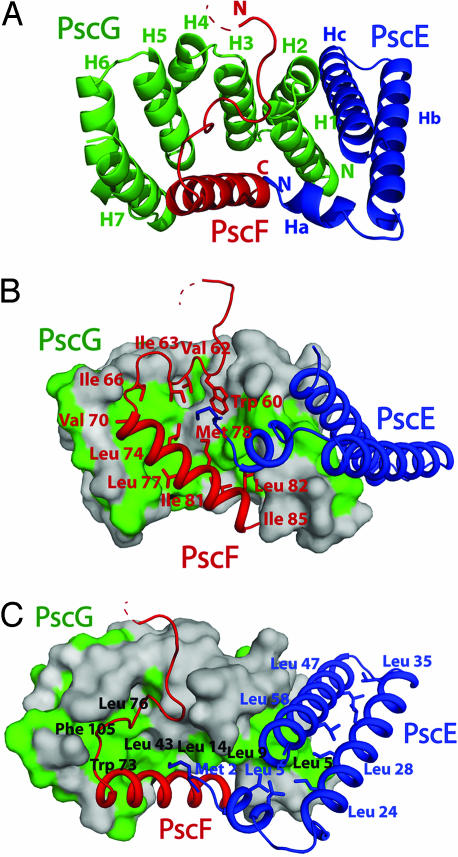
The structure of PscE-PscF55–85-PscG reveals that stabilization of the T3SS needle protein requires a macromolecular system that is totally distinct from what has been described for other fiber-forming molecules, such as the flagellum, the enteropathogenic E. coli (EPEC) T3S-associated filament or various pili (17–19); in addition, it does not display any similarity with other T3SS–chaperone complexes reported to date (20). PscG displays a seven-helical array harboring three helix–turn–helix motifs (H1-H6) that are highly reminiscent of a tetratricopeptide (TPR) domain [green in Fig. 2A and supporting information (SI) Fig. 5], although a TPR fold for PscG was not predicted from sequence analyses. The concave surface of PscG, formed by helices 1, 3, 5, and 7, displays a highly hydrophobic platform (green in Fig. 2 B and C), which “grasps” the C-terminal helix of PscF55–85 (red in Fig. 2). Each PscG helix provides between one and two residues to the interaction region, with Leu 14, Leu 43, Trp 73, and Phe 105 playing central roles (Fig. 2C; see below). PscG also interacts intimately with PscE (blue in Fig. 2) through a hydrophobic platform formed by the outside faces of H1 and H2 (Fig. 2A). In this sense, PscG represents an unusual TPR-like molecule, since both its convex and concave faces are used for partner molecule recognition.
Fig. 2.
Formation of the PscE- PscF55–85-PscG heterotrimer involves different platforms of hydrophobic interactions. (A) The seven-helical TPR-like structure of PscG (green) stabilizes PscF55–85 (red) through its concave side and interacts with PscE (blue) by using its convex face. (B) Hydrophobic residues that line PscF (red) are stabilized within the concave face of PscG's nonpolar palm (green). Note that PscF residues 60–67, which form an extended coil, are also bound within the palm. (C) A second hydrophobic platform, formed by the outside face of PscG helices H1 and H2, is responsible for recognition of PscE. The antiparallel nature of PscE helices Hb and Hc is also maintained through hydrophobic interactions.
The presence of an interaction platform between PscG and PscE reflects the fact that, both in vitro and in vivo, PscE serves as a stabilizing element for PscG (and vice versa). P. aeruginosa strains that lack chromosomally encoded pscE are incapable of producing PscG stably; the same occurs in pscG-deleted strains, which lack PscE and, in both cases, strains are noncytotoxic. In addition, PscE and PscG can be purified in vitro as a 1:1 complex that is more stable than isolated PscE or PscG (16). PscE folds into two antiparallel helices, Hb and Hc, that are preceded by a short independent helix (Ha). Hc and Hb are lined with hydrophobic residues within their common interacting region; all three helices associate onto PscG's hydrophobic surface (Fig. 2 B and C). Notably, the only PscE residue that interacts with PscF is Met 2, which precedes the highly conserved Ha helix, protruding deeply into a largely hydrophobic cleft formed by PscF residues Met 78, Trp 60, and Ile 63 (Fig. 2B). Thus, the fold of the PscE–PscG complex is reminiscent of a cupped hand, with the concave side of PscG representing the “palm.” Interestingly, the homologs of PscE and PscG in Y. pestis (YscE and YscG, respectively) have been shown to interact by two-hybrid screening (21), revealing the prevalence of this interaction amongst different bacterial species.
Needle Protein Amphipathicity Is Guaranteed Through TPR Recognition.
PscF55–85 is composed of two subdomains: an extended coil (PscF55–67), which is the continuation of the flexible N terminus that was removed by proteolysis before crystallization, and a C-terminal 17-residue, 25-Å-long α-helix (PscF68–85) (Figs. 2 and 3A). The extended coil interacts with the PscG palm through hydrophobic residues Trp 60, Val 62, Ile 63, and Ile 66 (Fig. 2B). The C-terminal, PscF68–85 helix is essential for polymerization of the full-length PscF, because a mutant lacking this region, PscF1–67, behaves as a monomer in gel filtration (SI Fig. 6) and chemical cross-linking experiments (not shown); the C-terminal region of the needle protein has also been shown to be essential for needle assembly in other T3S systems, as well as in the flagellar filament (15, 22, 23). In addition, PscF68–85 is highly amphipathic. The side of the helix that is stably embedded within the PscG palm is lined with hydrophobic residues (Figs. 2B and 3A), which are well conserved among a variety of bacterial species (Fig. 3D), forming a nonpolar platform that involves a total of 16 amino acids coming from both proteins. In contrast, the face that points to the outside of the PscE-PscG hand is highly hydrophilic. Thus, PscG's helix H7 interacts with PscF's Val 70 and Leu 77, H5 with Leu 74, Leu 77, Met 78 and Ile 81, H3 with Ile 81 and Ile 85, and H1 with Leu 82 and Ile 85.
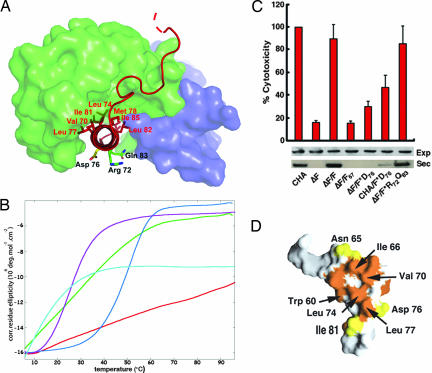
Fig. 3.
PscG grasps PscF's amphipathic tail. (A) Hydrophobic residues within the C-terminal helix of PscF (PscF68–85, in red) are shielded within PscG's concave groove, whereas hydrophilic residues point to solvent. PscG and PscE are represented as light green and blue surfaces, respectively. (B) Thermal denaturation curves obtained by circular dichroism measurements of wild-type PscF (red), PscF V70K/I77K (green), PscF L74K/I81K (purple), PscF M78K/L82K (blue), and PscF1–67 (light blue). Wild-type PscF is the only molecule that is robustly resistant to thermal denaturation. (C) Cytotoxicity profiles of P. aeruginosa strains carrying a pscF chromosomal deletion complemented by either wild-type (ΔF/F) or mutant pscF-expressing plasmids. Strains complemented by wild-type PscF or PscF R72A/Q83A are as cytotoxic as wild-type (CHA), whereas the one complemented with PscF D76A displays greatly reduced cytotoxicity. Supplying the wild-type strain with PscF D76A generated a dominant-negative effect. Below, Western blots revealing the cytoplasmic expression (exp) and secretion (sec) of T3SS effector molecule PopB in the same strains. Although all strains express PopB in the cytoplasm, only strains carrying either wild-type PscF or PscF R72A/Q83A are able to secrete PopB at levels comparable to the wild-type (CHA) strain. Error bars refer to standard deviation measurements for experiments performed in triplicate. (D) Mapping of PscF residues that are identical (yellow) and similar (orange) within different bacterial species from the point of view of PscG's concave side.
The stabilization of the C terminus of PscF within the grasp of an interaction platform generated by a TPR domain starkly deviates from structures of other fiber-forming molecules. The complex between a fragment of flagellin (FliC) and its chaperone FliS, reveals that, contrary to PscF, flagellin wraps itself around FliS in extended conformation and is highly exposed to solvent (17) (SI Fig. 7). Similarly, EspA, the building block of the EPEC T3SS-associated filament, also associates itself only laterally with its chaperone CesA through coiled coil interactions (SI Fig. 7; ref. 18). These distinct modes of binding point to the necessity of securing the T3SS needle protein within a stable cleft, which is provided by the scaffold provided by PscG and PscE.
Electron microscopy images of the bacterial flagellar filament, type III needle, and type IV pilus have revealed that the assembly of these structures involves packing of protein subunits within longitudinal helical arrays; in the case of the flagellum and type IV pilus, hydrophobic interactions have been shown to play key roles in the maintenance of the mechanical infrastructure (14, 23–25). These observations prompted us to investigate whether hydrophobic amino acids on the amphipathic C terminus of PscF could play a similar role in T3SS needle polymerization. To test this hypothesis, we mutated nonpolar residues aligned on the PscF69–85 helical axis to Lys (V70/I77; L74/I81; M78/L82; Fig. 3A) in the context of full-length PscF and tested purified proteins for their ability to self-assemble into stable polymers with size exclusion chromatography (SI Fig. 6) and melting temperature circular dichroism assays (Fig. 3B). Contrary to wild-type PscF, which when expressed on its own, polymerizes spontaneously and is a thermodynamically stable molecule (Fig. 3B and ref. 16), none of the mutant PscF molecules were able to form stable polymers. All mutants displayed melting transition temperatures of 50°C or lower, contrasting sharply with the robust behavior of wild-type PscF under the same experimental conditions (Fig. 3B). In addition, all three mutants eluted in gel filtration at positions which were intermediate between that of wild-type PscF (void volume) and PscF1–67, which was used as a negative polymerization control (SI Fig. 6) and formed heterogeneous oligomeric mixtures, as verified by negative staining electron microscopy (not shown). Thus, much as in the case of the type IV pilus and bacterial flagellum, hydrophobic residues aligned along the polymerization domain of the type III secretion needle-forming protein are responsible for its structural integrity and provide key interactions that ensure its robust nature.
The striking amphipathic character of PscF69–85 also brings into question the role of its hydrophilic face, which is notably decorated with residues Asp 76, Arg 72, and Gln 83 (the two latter residues not being aligned on the helical axis with Asp 76). To investigate their function, we mutated the three residues into Ala, introduced the mutated plasmids into the P. aeruginosa CHA strain carrying an endogenous deletion of PscF (ΔF), and tested expression and secretion capabilities of the resulting strains, as well as their cytotoxicity toward macrophages. A P. aeruginosa ΔF strain carrying double mutant PscF R72A/Q83A was able to express and secrete T3SS translocator PopB, intoxicating target cells (Fig. 3C). However, a ΔF strain carrying PscF D76A, although able to express PopB at wild-type levels, was unable to secrete effectors at any detectable level; in addition, it displayed a dramatic decrease in cytotoxicity. Interestingly, when a wild-type CHA strain, which carries native PscF, was complemented with the plasmid expressing PscF D76A, PopB secretion was also diminished, and cytotoxicity was ≈50% of that displayed by the uncomplemented CHA strain (Fig. 3C, lane 6). The dominant-negative effect displayed by PscF D76A on the P. aeruginosa isolate could be a consequence of its incorporation into polymerizing needles concomitantly with wild-type PscF molecules, thus disrupting T3SS needle functionality, although other possibilities cannot be excluded. Notably, mutations in the analogous residues in YscF (Asp 77) and MxiH (Asp 73; Fig. 1) also yielded strains incapable of secreting T3SS effectors or infecting target cells upon induction (22, 26). A model of the MxiH T3SS needle generated by the fitting of the crystal structure of MxiH monomers onto a 17-Å electron microscopy needle reconstruction suggests this residue could be located at the interface between subunits within the polymerized needle (14), which would explain its ability to disrupt normal needle function in strains carrying a mixture of viable and nonviable copies of the needle protein.
A Protein Interaction Region as a Template for Rational Drug Design.
The importance of maintenance of needle protein functionality for bacterial cytotoxicity, as well as the ubiquitous presence and sequence similarity between PscF, PscG, and PscE homologs in a variety of pathogens (Fig. 1), suggest that this ternary complex can be explored as a novel target for the development of new antibacterials. To identify the region in the complex that displays the most susceptibility to disruption, thus being interesting for exploration through targeting by a small molecule, we investigated the importance of both the N terminus of PscE (from Met 2 to the C terminus of helix Ha) and the concave surface of PscG (the palm), regions that are conserved among a variety of bacterial species (Figs. 1 and 4C), in T3SS functionality and bacterial cytotoxicity.
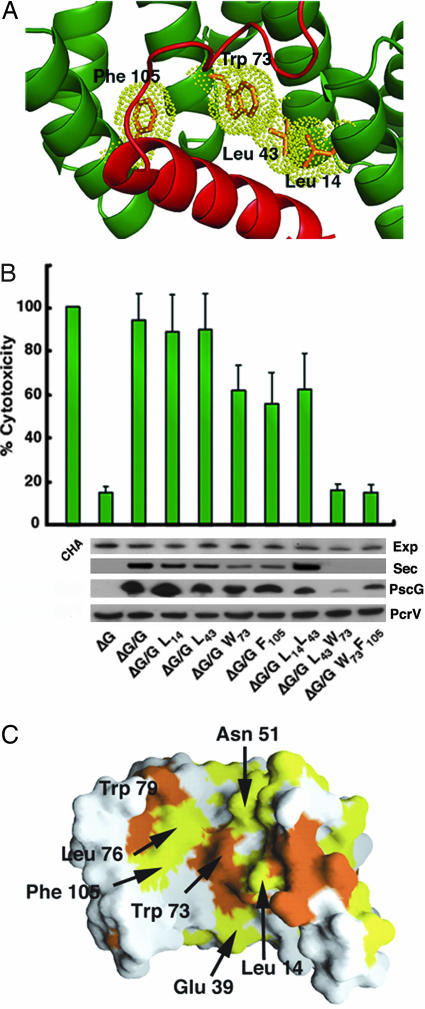
Fig. 4.
The PscG–PscF interaction platform is essential for T3SS viability. (A) Closeup of the PscG–PscF interaction region, showing the four hydrophobic residues that were mutated to Ser in this study. (B) Analysis of the effect of PscG mutations on cytotoxicity of the CHA P. aeruginosa isolate, from which pscG was chromosomally deleted. Strains expressing mutants PscG L43S/W73S and W73S/F105S can express (Exp), but not secrete (Sec), T3SS effector PopB and display only background cytotoxicity. A lower amount of PscG is detected in the cytoplasm of the double-mutant strains, whereas the levels of PcrV, an unrelated T3SS effector, are maintained. (C) Mapping of PscG residues that are identical (yellow) and similar (orange) within different bacterial species, revealing a highly conserved interface.
Four clones harboring mutations or deletions on the N terminus of PscE (PscE M2A, PscE M2H, PscE3–67, and PscE12–67, the latter of which lacks the entire Ha helix) were used to complement a P. aeruginosa CHA isolate carrying a pscE deletion (ΔE); interestingly, all four strains were fully cytotoxic (SI Fig. 8A). In addition, PscE12–67-PscF-PscG could still be purified as a 1:1:1 complex and displayed only a mild decrease in thermostability in circular dichroism temperature scanning assays when compared with full-length PscE-PscF-PscG (SI Fig. 8B). Hence, our results show that the first 11 residues of PscE, albeit the interaction of Met 2 with PscF and the high conservation of residues that interact with PscG, play at most a minor role in T3SS functionality.
Within the PscG palm, four central residues in the PscF-interacting region of PscG (Leu 14, Leu 43, Trp 73, and Phe 105; Fig. 4A) were mutated into Ser, and mutant plasmids were used to complement a P. aeruginosa CHA isolate in which pscG was deleted (ΔG) (Fig. 4B). In addition, mutant ternary complexes were also expressed and purified in E. coli and submitted to thermal denaturation assays (SI Fig. 9). Expression, secretion, and cytotoxicity assays revealed that strains expressing PscG W73S or PscG F105S were ≈50% less toxic toward macrophages than the ΔG strain complemented with wild-type PscG. This result was in accordance with those from thermal denaturation tests, which showed ternary complexes carrying these single mutations to be less stable than wild type (SI Fig. 9). Notably, double mutants PscG L43S/W73S and PscG W73S/F105S displayed cytotoxicity profiles that were comparable to the background level displayed by the uncomplemented ΔG strain (Fig. 4B, last two lanes). In vitro, the ternary complex carrying the L43S/W73S mutations on PscG could not be isolated; after purification, only a 1:1 PscE-PscG complex could be identified (not shown). In addition, Western blots performed with anti-PscG antibodies revealed that the double mutant strains harbor less PscG than other more cytotoxic strains (Fig. 4B), which is most likely due to of the instability of PscG in the absence of PscF. These results corroborate data from previous in vivo studies, which showed that the presence and stability of each member of the ternary complex is essential for preservation of the complex as a whole (16). PscG residues Leu 43, Trp 73, and Phe 105 lie on helices H5 and H7 within the concave face of PscG's TPR region, and interact with PscF's Leu 74 and Ile 81, which were shown to play key roles in polymerization and provide thermodynamic stability (see above). Consequently, the disruption of this 770-Å2 essential interface strongly affects TTSS functionality.
The sequence similarity observed within PscF, PscG, and PscE homologs suggests that different T3SS-carrying pathogens possess a similar heterotrimeric scaffold that regulates needle protein stabilization before secretion. In addition, the highly conserved PscG–PscF interface (Figs. 3D and 4C), which harbors residues essential for bacterial cytotoxicity, is a potential template for the design of inhibitors capable of disrupting PscF–PscG interactions. This approach could allow for the development of inhibitors that block T3SS viability without killing the bacterium, making the appearance of drug-resistant strains less likely. Thus, the PscF-PscE-PscG heterotrimeric complex presented here represents a yet-unexplored target for antibiotherapy development.
Experimental Procedures
Sample Preparation, Crystallization, and Data Collection.
The full-length PscE1–67-PscF1–85-PscG1–115 complex was purified as a 1:1:1 complex as described (16), but high-quality crystals were not obtained. After treating the complex with a 1:1,000 papain:protein ratio for 4 h, three stable bands (PscE1–67, PscF55–85, and PscG1–115) were identified by mass spectrometry and N-terminal sequencing. The DNA sequence corresponding to the first 54 residues of PscF was then removed from the pscEFG sequence by performing Slicing Overlap Extension PCR. The final PCR product was introduced into pET-15b (Novagen) and transformed into E. coli BL21 (DE3). The new heterotrimeric complex, PscE-PscF55–85-PscG, was purified by using the same protocol as for the full-length form (16), with the exception that the histidine tag was removed with thrombin at a 1:1,000 ratio before gel filtration in 25 mM Hepes, pH 6.7/0.1 M NaCl/1 mM EDTA/10 mM DTT. Crystals were obtained in hanging drops in 100 mM Tris·HCl, pH 7.4/0.8 M LiSO4/10 mM NiCl2 at room temperature and were cryoprotected by gradual incubation in increasing amounts of glycerol to a final concentration of 25%. Data sets were collected at the European Synchrotron Radiation Facility beamline ID14-EH2 at 100 K. Crystals belong to space group P6222 and contain one heterotrimer per asymmetric unit. All data were processed by using DENZO and SCALEPACK (27).
Structure Determination.
Experimental phases were calculated by using the anomalous signal from three Ni+2 ions, obtained by collecting data from a single crystal that diffracted to 2.2 Å. The Ni+2 sites were located by SHELXD (28) by using the AutoSHARP protocol (29). An interpretable electron density map was obtained after solvent flattening, and the PscE-PscF55–85-PscG model was automatically traced by using ARP/wARP (30). Molecular replacement with AmoRe (31) was performed with this model to phase reflections from a second crystal that diffracted to 2.0 Å. Refinement with CNS (32), which included energy minimization, temperature factor refinement, and simulated annealing steps, was intercalated with iterative cycles of manual model building in COOT (www.ysbl.york.ac.uk/∼emsley/coot). Data collection, phasing, and refinement statistics are included in SI Table 1.
Circular Dichroism.
Thermodynamic stability assays were performed on a Jasco (Easton, MD) J-810 spectropolarimeter with a 1-mm cell in 20 mM sodium phosphate, pH 7.2/0.1 M NaCl at protein concentrations of 0.2 mg/ml for all samples. The circular dichroism signal was recorded at 222 nm in a range of 4–96°C with scan rate of 1°C per min. Spectra were corrected against those of the buffer reference. The measured ellipticity was adjusted to the same starting value for all samples for a better visualization of the curves.
T3SS Effector Synthesis, Secretion, and Cytotoxicity Assays.
PscF mutants V70K/I77K, L74K/I81K, and M78K/L82K were obtained by using the Quick Change mutagenesis kit (Stratagene, La Jolla, CA), cloned into pET22b (Novagen), transformed into E. coli BL21 (DE3), and purified as wild-type PscF (16). Other PscF and PscG mutants were created by using pIApG/PscF, pIApG/PscE, and pIApG/PscG as templates (16). Mutated plasmids were introduced into a cytotoxic P. aeruginosa cystic fibrosis isolate CHA carrying appropriate chromosomal deletions of pscE, pscF, or pscG (ΔE, ΔF, and ΔG, respectively). Mutant and control strains were grown on Pseudomonas Isolation Agar (Difco, Lawrence, KS) plates or in liquid Luria Broth (LB) at 37°C with agitation. Carbenicillin was used at 500 μg/ml for Pseudomonas Isolation Agar plates and 300 μg/ml in LB. For induction of type III secretion in vitro, P. aeruginosa overnight cultures were diluted to an optical density at 600 nm (A600) of 0.2 in LB containing 5 mM EGTA and 20 mM MgCl2. Incubation was prolonged for additional 3 h until the cultures reached A600 values of 1.0. The samples containing 30 μl of culture supernatants or total cells were directly analyzed by Western blotting by using anti-PopB, anti-PcrV, or anti-PscG antibodies. For cytotoxicity assays, the bacteria were cultivated to an OD600 of 1 and added to macrophage cell line J774 at a Multiplicity of Infection of 5. Cell death was assessed at 3 h postinfection by using a Cytotoxicity Detection Kit, LDH (Roche, Indianapolis, IN). All tests were performed in triplicate.
Acknowledgments
We thank Otto Dideberg for continuous support, Guy Schoehn and the CIBB electron microscopy platform for help with negative staining analyses, and laboratory members for helpful discussions. This work was supported by grants from the French Cystic Fibrosis Foundation (Vaincre la Mucoviscidose) and the Direction des Sciences du Vivant (DSV/ Commissariat à l'Energie Atomique). M.Q. is the recipient of a CFR fellowship (Commissariat à l'Energie Atomique), and S.P. is the recipient of a postdoctoral fellowship from the DSV/Commissariat à l'Energie Atomique. We thank D. Lemaire and B. Dublet for mass spectrometry analyses, as well as the ESRF ID14 staff for access to beamlines.
Abbreviations
- T3SS
type III secretion systems
- TPR
tetratricopeptide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates listed in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2UWJ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610098104/DC1.
References
- 1.Galán JE, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh P. Mol Biol Rev. 2004;68:771–795. doi: 10.1128/MMBR.68.4.771-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornelis G. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 4.Vance RE, Retsch A, Mekalanos JJ. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kornish JP. J Infect Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 6.Lyczak JB, Cannon CL, Pier GB. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 7.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J, Aizawa S-I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 8.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 10.Kubori T, Sukhan A, Aizawa S-I, Galán JE. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlovits TC, Kubori T, Lara-Tejero M, Thomas D, Unger VM, Galán JE. Nature. 2006;441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 12.Kimbrough TG, Miller SI. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Journet L, Agrain C, Broz P, Cornelis GR. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 14.Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. Proc Natl Acad Sci USA. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. J Mol Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, Pastor A, Elsen S, Dessen A, Attree I. J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 17.Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters H.K., III, Pokross M, Waugh DS. Nat Struct Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 18.Yip CK, Finlay BB, Strynadka NC. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 19.Sauer FG, Barnhart M, Choudhury D, Knight SD, Waksman G, Hultgren SJ. Curr Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 20.Yip CK, Strynadka NC. Trends Biochem Sci. 2006;31:223–230. doi: 10.1016/j.tibs.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Day JB, Guller J, Plano GV. Infect Immun. 2000;68:6466–6471. doi: 10.1128/iai.68.11.6466-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. J Biol Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- 23.Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 24.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Mol Cell. 2006;23:651–652. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Yonekura K, Maki-Yonekura S, Namba K. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 26.Torruellas J, Jackson MW, Pennock JW, Plano GV. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 29.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 30.Morris RJ, Perrakis A, Lamzin VS. Methods Enzymol. 2003;374:229–244. doi: 10.1016/S0076-6879(03)74011-7. [DOI] [PubMed] [Google Scholar]
- 31.Navaza J. Acta Crystallogr D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]