Abstract
Class IA phosphoinositide 3-kinases (PI3Ks) signal downstream of tyrosine kinases and Ras and control a wide variety of biological responses. In mammals, these heterodimeric PI3Ks consist of a p110 catalytic subunit (p110α, p110β, or p110δ) bound to any of five distinct regulatory subunits (p85α, p85β, p55γ, p55α, and p50α, collectively referred to as “p85s”). The relative expression levels of p85 and p110 have been invoked to explain key features of PI3K signaling. For example, free (i.e., non-p110-bound) p85α has been proposed to negatively regulate PI3K signaling by competition with p85/p110 for recruitment to phosphotyrosine docking sites. Using affinity and ion exchange chromatography and quantitative mass spectrometry, we demonstrate that the p85 and p110 subunits are present in equimolar amounts in mammalian cell lines and tissues. No evidence for free p85 or p110 subunits could be obtained. Cell lines contain 10,000–15,000 p85/p110 complexes per cell, with p110β and p110δ being the most prevalent catalytic subunits in nonleukocytes and leukocytes, respectively. These results argue against a role of free p85 in PI3K signaling and provide insights into the nonredundant functions of the different class IA PI3K isoforms.
Keywords: quantitative mass spectrometry, signaling, protein stability, gene knockout, lipid kinase
Phosphoinositide 3-kinases (PI3Ks) generate lipid second messengers that serve as membrane docking sites for a variety of downstream effector proteins such as protein kinases, regulators of small GTPases, and scaffolding proteins (1, 2). The class IA PI3Ks are heterodimers consisting of a p110 catalytic subunit and a smaller regulatory subunit with Src-homology 2 (SH2) domains. Mammals have three catalytic subunits (p110α, p110β, p110δ) and five regulatory subunits (p85α, p85β, p55γ, p55α, p50α) (1, 2). Under experimental conditions, each p110 isoform can bind any p85 isoform with no apparent preference (see among others, refs. 3 and 4). p85s have a dual effect on the p110 subunits because they stabilize the thermally labile p110s but also conformationally inhibit their catalytic activity (5). Upon cellular stimulation, SH2 domain-mediated recruitment of p85/p110 complexes to Tyr phosphorylated (pY) membrane-proximal proteins serves dual functions: It positions p110 in proximity with its substrates, and the engagement of the p85 SH2 domains relieves p85-mediated inhibition of p110, thus increasing enzymatic activity of p110 (6, 7).
Experiments using gene-targeted mice and p110 isoform-selective inhibitors have uncovered nonredundant physiological functions of the p110 isoforms. These functions include insulin signaling in metabolic tissues (p110α; refs. 8 and 9), integrin signaling in platelets (p110β; ref. 10) and signaling through a variety of receptors in leukocytes (p110δ; refs. 11–15). The mechanisms of this nonredundant signaling are not fully understood. Indeed, p110 isoforms have high homology in their primary sequence, interact nonselectively with the different p85s, and have the same lipid substrate preference (3). However, the capacity of the different p110 isoforms to bind Ras might differ among catalytic subunits (refs. 16–18 and reviewed in ref 19). Another key parameter may be the differential tissue distribution of p110 isoforms (3, 20), which may result in differences in the absolute levels of expression of the distinct p110s. Given that competitive recruitment of p110 (and p85) isoforms to receptors and adaptor proteins may be intrinsic to the PI3K signaling system, it is clear that the absolute amount of PI3K isoforms could contribute to their relative functional importance in different cell types. Knowledge of the absolute protein amounts of class IA PI3K isoforms is thus important to gain insight into their apparent nonredundant biological roles.
Quantitative assessment of p85 to p110 ratios is also important to formally test the well-publicized concept that p85 can exist without p110 in cells and can compete with heterodimeric p85/p110 complexes for pY binding sites, thereby decreasing PI3K signaling (21, 22). This hypothesis was put forward to explain the remarkable observation that four different lines of mice with targeted deletions of p85α or p85β exhibit enhanced PI3K signaling in insulin-responsive cells and tissues (23–26). Forced overexpression of p85 in skeletal muscle cells was further found to decrease PI3K-dependent phosphorylation of the Akt/PKB kinase, a key downstream target of PI3K (27). Immunoblotting and immunodepletion studies also provided evidence for a 30% excess of p85 over p110 in mouse embryo fibroblasts (28) and liver cells (29). According to the “free p85” model, the expression of non-p110-bound p85 is preferentially reduced in p85 knockout (KO) cells, allowing enhanced access of heterodimeric p85/p110 to pY binding sites, resulting in increased PI3K signaling and hypersensitivity in response to insulin (23–26, 30).
Methods for the quantification of proteins in cells, including absolute quantification MS, have recently been developed (31–33). The latter is based on the use of an isotopically labeled internal standard (IS) peptide that is spiked at known concentration in protein mixtures. This IS peptide has the same sequence as a section of the protein to be quantified; however, one or more amino acids are enriched in stable heavy isotopes of carbon, nitrogen, and/or hydrogen, which causes a shift in the MS spectrum without affecting the chemical properties of the peptide. After controlled proteolysis (usually with trypsin), MS analysis of both the endogenous and IS peptides allows for accurate and precise quantification of cellular proteins in absolute units (e.g., molecules per cell).
In this report, we used quantitative MS in conjunction with affinity and ion exchange chromatography to determine the absolute protein amount of each class IA PI3K isoform in mouse cell lines and tissues. Our data show that p85s and p110s are present in equimolar amounts and form obligate heterodimers. These findings have implications for the understanding of the nonredundant function and regulation of class IA PI3K isoforms, and necessitate a revision of the free p85 hypothesis.
Results
p85 and p110 Are Present in Equimolar Amounts in Mouse Cell Lines and Exist as Obligate Heterodimers.
Three different approaches were used to assess the relative expression and molecular association between p85 and p110 subunits in the murine WEHI-231 B lymphocyte and NIH 3T3 fibroblast cell lines. WEHI-231 cells were chosen as a representative leukocyte cell line with high p110δ expression, whereas NIH 3T3 cells are similar to mouse embryo fibroblasts, a cell type previously used to explore relative p85/p110 protein expression levels (28).
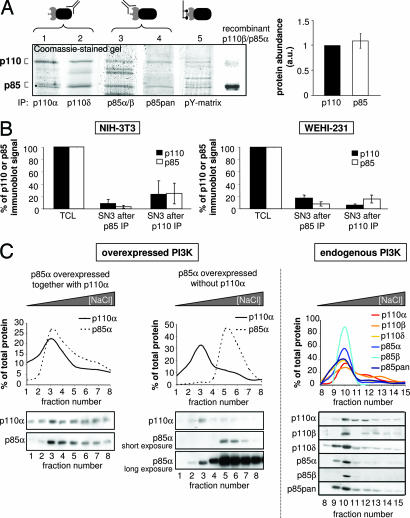
First, immunoprecipitates of p110 (Fig. 1A, lanes 1 and 2) or p85 isoforms (Fig. 1A, lanes 3 and 4), were separated by SDS/PAGE, followed by staining with colloidal Coomassie blue and measurement of optical densities of the p85 and p110 protein bands. Abs used in these experiments, several of which were made in-house, have been extensively validated in our laboratory (data not shown). PI3Ks were also enriched by using a matrix (Fig. 1A, lane 5) consisting of immobilized peptides that contain a pY present in a YxxM motif known to preferentially bind the SH2 domains of p85s (34) (this matrix is further referred to as a pY matrix and is described in detail in Materials and Methods). In eight independent experiments, no excess of p85 over p110, and vice versa, was found in WEHI-231 or NIH 3T3 cells (Fig. 1A Right).
Fig. 1.
Analysis of the ratio of p85 to p110 PI3K subunits in PI3K complexes. (A) PI3K subunits were immunoprecipitated from WEHI-231 and NIH 3T3 cell lysates by using isoform-specific Abs to p110 or p85 or were isolated by affinity purification by using a pY matrix (made up of immobilized peptides containing a phosphorylated Tyr residue in a YxxM motif that binds p85s), resolved by SDS/PAGE, and detected by colloidal Coomassie blue staining. Recombinant p85α/p110β was loaded as a control for protein separation. (Left) Representative SDS/PAGE gel. (Right) Average of the optical densities at 110 kDa and 85 kDa from eight independent SDS/PAGE gels, similar to the one shown in A Left. (B) Analysis of the presence of p85 and p110 in NIH 3T3 and WEHI-231 cell extracts after three rounds of immunodepletion with Abs to p110 or p85. Sequential IPs, using a mixture of p110 or p85 Abs, were performed on TCL. Samples of preabsorbed TCL and of SN after three rounds of depletion (SN3) were resolved by SDS/PAGE, followed by detection of class IA PI3K proteins by immunoblotting (using Abs to p110α, p110β, p110δ, p85α, and p85β) and quantification of optical densities. The graph represents the averages from two independent experiments. (C) Elution profiles of p110 and p85 after anion exchange chromatography of TCL. Cell extracts of HEK-293T cells, transiently transfected with p85α in the presence (Left) or absence (Center) of overexpressed p110α, were resolved by anion exchange chromatography, followed by immunoblotting for p85α and p110α in the different elution fractions. Immunoblotting signals were quantified to generate elution profiles. (Right) Endogenous PI3K isoforms from WEHI-231 cells were analyzed by anion exchange chromatography and immunoblotting as described above. A representative experiment of two is shown.
We next carried out immunodepletion experiments on total cell lysate (TCL) of NIH 3T3 or WEHI-231 cells by using Ab mixtures against all class IA p110 isoforms (p110α, p110β, p110δ) or against p85 isoforms (p85α, p85β). Preextraction TCL and supernatant (SN) remaining after three rounds of depletion (referred to as SN3) were analyzed for expression of all class IA PI3K isoforms by immunoblotting (using Abs distinct from those used for immunodepletion whenever possible), followed by quantification of the immunoblot signal (Fig. 1B). We calculated that a 30% excess of free p85 over p110 in cells (as suggested to be the case in mouse embryo fibroblasts; ref. 28) would lead to the detection in SN3 of three times more p85 than p110 if 90% of the p110 subunits were depleted from the TCL after three rounds of immunoprecipitation (IP) with p110 Abs [supporting information (SI) Fig. 5]. Such excess was not observed (Fig. 1B), arguing against the existence of free class IA PI3K isoforms in NIH 3T3 and WEHI-231 cells.
We next tested whether p110-bound p85 behaved differently than p110-free p85 upon fractionation of TCL by anion exchange chromatography. p85α was transiently overexpressed in human HEK-293T cells with or without p110α, and its elution profile was assessed under both conditions. Separation by anion exchange is based on overall charge (i.e., on pI), and a later elution of free p85 (i.e., at higher NaCl concentration) would be consistent with its lower pI compared with p110α (theoretical pI of 5.8 and 6.9, respectively; note that the pI of the p85α/p110α heterodimer is likely to be closer to that of p110 due to the larger size of the latter). As can be seen from Fig. 1C Left, p85α coeluted with p110α upon coexpression. In contrast, p85α overexpressed without p110α was found to elute at higher ionic strength than endogenous p110α (Fig. 1C Center) and endogenous p85α (Fig. 1C Center, long exposure), consistent with p85 being more acidic than the p85-p110 dimer. Thus, free and p110-bound p85s have different retention times on anion exchange HPLC. TCL of WEHI-231 cells was next subjected to this type of fractionation, and the presence of endogenous p110 and p85 isoforms in the different fractions was assessed by immunoblotting. As can be seen from Fig. 1C Right, p85α/β and all catalytic subunits tightly coeluted in the same HPLC fractions, providing further evidence against the existence of p110-free p85 in these cells under physiological conditions.
Absolute Quantification MS Reveals 10,000–15,000 p85/p110 Dimers per Cell, Without Excess of p85.
We next used quantitative MS (31) to determine the absolute levels of p85 and p110 subunits. The application of this approach to a specific set of proteins entails two stages. First, isotopically labeled protein-specific IS peptides are synthesized by using selection criteria that include amino acid composition (absence of Met, Cys, and Trp given their chemical reactivity; absence of Asp followed by Gly and N-terminal Gln or Asn due to their chemical instability), nonredundancy among other proteins, and coverage in different species. IS peptides used for specific class IA PI3K isoforms are shown in SI Table 1. These IS peptides were further characterized for retention time on reverse-phase columns, detection limit by MS, and fragmentation by MS/MS. In a second stage, the IS peptides were mixed with cell extracts and analyzed by MS to quantify endogenous amounts of PI3K proteins by comparing integrated peak areas of known amounts of IS peptides with those of the respective endogenous peptides. To test the performance of the quantitative MS method, we compared the integrated peak areas of known amounts of labeled IS peptides when run in the presence of increasing amounts of endogenous PI3K peptides (by trypsinization of PI3K immunoprecipitated from increasing amounts of cell lysates) and found that this approach can quantify PI3K subunits with good precision (≈10% variation, partial data shown in SI Fig. 6A and B; see SI Text for details). We further tested whether this MS method could detect small quantitative differences between PI3K subunits. Known amounts of recombinant p85α and p110δ proteins were quantified by MS in test samples made up of a constant amount of p110δ and increasing amounts of p85α. The observed precision and accuracy of PI3K isoform quantification were on average 11% and 7%, respectively, indicating that a potential excess of 30% p85 over p110 could be detected by this approach (SI Fig. 6C). For the purpose of this paper, we define precision as a measure of reproducibility, and we define accuracy as how close the calculated amount is to the real value.
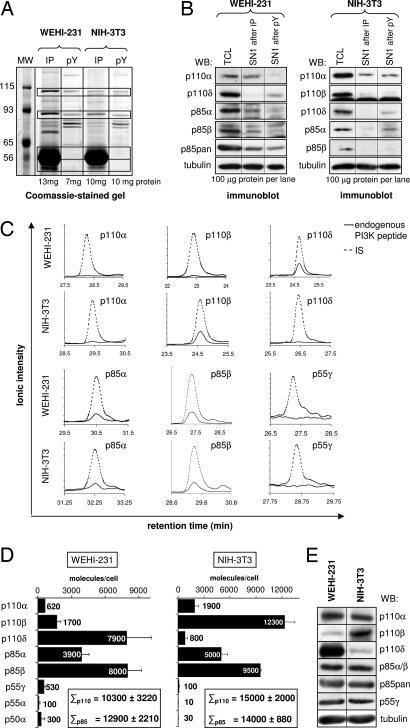
We next isolated PI3K complexes from WEHI-231 or NIH 3T3 lysates by using a mixture of PI3K Abs or the pY matrix described above, followed by SDS/PAGE separation and colloidal Coomassie blue staining (Fig. 2A). Immunoblot analysis of the SN remaining after one round of PI3K IP/pull-down (SN1; Fig. 2B) revealed that PI3K subunits were effectively depleted by both affinity procedures. Fig. 2C shows examples of the extracted ion chromatograms of 2-pmol IS peptides (dashed lines) and endogenous PI3K peptides (solid lines) isolated from cells. The integrated peak areas of IS and endogenous peptide were used to calculate the absolute amount of each PI3K isoform per mg TCL (SI Fig. 7) and the number of molecules of each PI3K isoform per cell (Fig. 2D). This analysis revealed that WEHI-231 and NIH 3T3 cells expressed similar amounts of class IA PI3Ks, namely 10,000–15,000 p85/p110 dimers per cell. In each case, the total number of all p110 molecules matched that of the p85s, again arguing against the existence of an excess of p85 over p110 in the cell lines used. In WEHI-231 cells, the predominant p110 isoform was p110δ, in line with the previously documented high expression of p110δ in leukocytes (3, 20). In NIH 3T3 cells, p110β was most abundant. Surprisingly, the amount of p85β (8,000–9,000 molecules per cell) was more than double that of p85α (4,000–5,000 molecules per cell) in both cell lines, with the levels of p55γ, p55α, and p50α being 500 molecules or less per cell. The differences between WEHI-231 and NIH 3T3 in expression levels of each individual PI3K isoform detected by quantitative MS correlated well with the relative protein expression of the same class IA PI3K isoform in these cell lines as detected by immunoblotting of TCL (Fig. 2E).
Fig. 2.
Determination of the absolute amounts of class IA PI3K subunits in murine cell lines. (A) Class IA PI3Ks were isolated from the indicated cell lines by IP using a mix of p110 and p85 Abs or by absorption onto the pY matrix, followed by separation by SDS/PAGE and visualization by colloidal Coomassie blue staining. Horizontal boxes indicate the gel sections that were excised for the further analysis shown in C and D and in SI Fig. 7. The abundant, ≈56-kDa protein in lanes 2 and 4 is the heavy chain of the Ab used for IP. (B) Protein depletion was assessed by immunoblotting for class IA PI3K isoforms in TCL compared with SN1 after one round of IP or pY pull-down. (C) Representative extracted ion chromatograms of endogenous and IS (2-pmol) peptides of immunoprecipitated class IA PI3Ks. For each subunit, all endogenous and IS peptides listed in SI Table 1 were analyzed. (D) The number of class IA PI3K molecules per cell as determined by quantitative MS. The total of all catalytic and regulatory subunit protein amounts is shown. (E) Immunoblot analysis of class IA PI3K isoforms in 100-μg TCL of WEHI-231 and NIH 3T3 cells.
Correlation Between Class IA PI3K Isoform mRNA and Protein Levels.
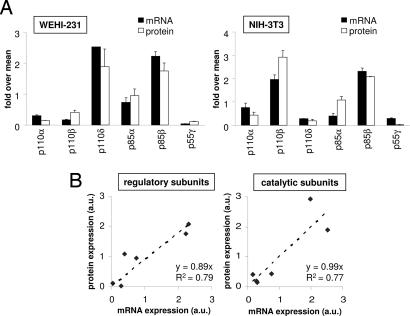
To investigate the relationship between protein and mRNA levels of class IA PI3Ks in WEHI-231 and NIH 3T3 cells, we next determined the amount of mRNA for each PI3K isoform by quantitative real-time RT-PCR. cDNA was prepared from total mRNA and amplified by real-time PCR by using primers specific for each class IA PI3K isoform. In each cell line, a reasonably good correlation between mRNA and protein levels was found for each class IA PI3K isoform (Fig. 3A). When these data from both cell lines were pooled, a linear correlation between mRNA and protein levels was found for both the regulatory subunits (Fig. 3B Left) and the catalytic subunits (Fig. 3B Right), indicating that transcriptional regulation is an important parameter in the regulation of the relative levels of p85 and p110.
Fig. 3.
Relationship between protein and mRNA levels of PI3K isoforms in WEHI-231 and NIH 3T3 cell lines. (A) Class IA PI3K mRNA and protein amounts in WEHI-231 and NIH 3T3 cells. Each mRNA or protein value was standardized to the mean of all catalytic and regulatory subunits, respectively. (B) Relationship between mRNA and protein expression of class IA PI3K subunits. Each mRNA and protein value was standardized to the mean of all catalytic and regulatory subunits, respectively, independent of the cell line used. Graphs were constructed by using the pooled data shown in A.
Absolute Amounts of Class IA PI3K Isoforms in Mouse Tissues.
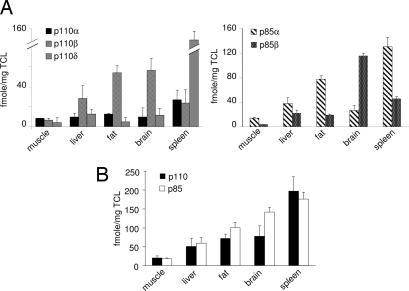
We next used quantitative MS to determine the amounts of class IA PI3K proteins in fresh murine tissues. PI3Ks were immunoprecipitated from homogenized tissues, resolved by SDS/PAGE, and stained by colloidal Coomassie blue (SI Fig. 8A). Immunoblotting of samples before and after one round of IP revealed effective depletion of most PI3K isoforms (SI Fig. 8B). As shown in Fig. 4A, the most abundant catalytic subunits were p110β (in liver, brain, and fat) or p110δ (in spleen), with a lower abundance of p110α in most tissues tested (Fig. 4A Left). p85 expression was more variable and, depending on the tissue, p85α or p85β was found to be the major isoform (Fig. 4A Right). Importantly, no excess of p85 over p110 subunits was found in all tissues investigated, apart from brain, which showed a 2-fold excess of p85 protein over p110 (Fig. 4A). This finding is surprising, given that no brain phenotypes have been reported in p85 KO mice. However, it is possible that the very rapid and selective degradation of p110δ over other p110 isoforms in post-mortem brain (A. Bilancio and B.V., unpublished observations) contributes to the apparent excess of p85 in extracts of this tissue.
Fig. 4.
Quantification of class IA PI3K isoforms in murine tissues. (A) Protein expression levels of class IA PI3K isoforms in mouse tissues. The value for each class IA PI3K isoform was determined by comparison of peak integrated areas of endogenous and IS peptides (data not shown). (B) Total amount of p110 and p85 subunits in different mouse tissues.
Discussion
In this study, we document that the p85 regulatory and p110 catalytic subunits of class IA PI3Ks are present in equimolar amounts in mammalian cell lines and tissues and are assembled in obligate heterodimers. In other words, all available p85 is bound to p110, and no evidence for free p85 or free p110 could be obtained. We also report an estimation of the absolute amounts of class IA PI3K catalytic and regulatory proteins. In established cell lines, 10,000–15,000 p85/p110 dimers were present per cell. In tissues, total p85/p110 levels were more variable, but were still within a 10-fold range, with the lowest and highest total amount of p85/p110 in muscle and spleen, respectively (Fig. 4B). These values may be underestimated, because with the current level of technology, we cannot account for losses that may occur during affinity purification. In the future, this problem will be solved by using a new generation of mass spectrometers that have a sensitivity that will permit quantifying endogenous proteins expressed at low levels without the need for affinity purification. Nevertheless, by using several different affinity techniques before MS quantification, it was observed that the values we obtained were self-consistent irrespective of the affinity technique used. We thus believe that the ratios of p85/p110 will not change when using more sensitive mass spectrometry, albeit the absolute amounts may do so to a small extent, should the affinity purification step be excluded from the experimental workflow.
A surprising finding was the high levels of p85β compared with p85α, especially in cell lines (Fig. 2D). Indeed, p85α was generally believed to be more abundant than p85β, a conclusion that was largely based on the more pronounced phenotypic impact of p85α gene KO in mice than that of p85β KO. Indeed, whereas p85α KO mice die perinatally (24), p85β KO mice are viable (25) and have less severe metabolic and immunological phenotypes (24, 35, 36). Previous analysis of PI3K protein expression by immunoblotting and silver staining (28) suggested that p85β contributes only 10% of total regulatory subunits in mouse embryo fibroblasts (which are similar to the NIH 3T3 cells used in our study). We found that the levels of p85β were within the same range as those of p85α, and in some cases (such as in brain, WEHI-231, and NIH 3T3 cells) even exceeded those of p85α. The reasons for this discrepancy between our results and these of Ueki et al. (28) are unclear at the moment but may be related to differences in PI3K Ab reagents.
In line with the previously documented enrichment of p110δ in leukocytes (3, 20), p110δ levels were high in spleen and in WEHI-231 B lymphoma cells. The data available to date suggest that the relative importance of the p110δ isoform in leukocytes exceeds that of the other class IA p110 isoforms (refs. 11, 14, and 15 and our unpublished results), suggesting that the expression level of a p110 isoform can contribute to its relative contribution to signaling. It is, however, likely that other parameters, such as specific activity, may also have to be taken into account when dissecting causes for the nonredundant functions of class IA PI3Ks. Indeed, the specific activity of p110α appears to be higher than that of the other class IA PI3K isoforms (37, 38), and might “compensate” for the lower expression of p110α relative to the other PI3K isoforms (Figs. 2D and 4A).
Our quantitative MS analysis revealed that the p85 regulatory and p110 catalytic subunits of class IA PI3Ks are present in equimolar amounts in cell lines and tissues. Affinity purification (Fig. 1A) and immunodepletion experiments (Fig. 1B), as well as analytical ion exchange chromatography (Fig. 1C), further support the notion that all p85 subunits are p110-bound and that no free p85 or p110 subunits exist in cell lines and tissues. Our data are in line with previous work demonstrating that the unstable p110s are stabilized by binding to p85 subunits (5) and that free p85α is unstable in the absence of p110α (39, 40). It was further shown that the interaction of p85 with p110 is very strong and can withstand high concentrations of urea, salt, or detergent (34, 41). It is therefore rather unlikely that the p85-p110 association will be dynamically regulated in vivo. A reasonably good correlation was found between the expression levels of the mRNA and protein of each p85 and p110 subunit (Fig. 3), suggesting that transcriptional control of PI3K expression, together with “cross-stabilization” of p85 and p110 proteins (5, 39), are important mechanisms to achieve equimolarity between the class IA PI3K subunits.
Our findings argue against the free p85 hypothesis (22), which had been put forward to explain the increased insulin-induced PI3K signaling in p85α and p85β KO mice (23–26). This hypothesis invokes the existence of non-p110-bound (“free”) p85, which can compete with heterodimeric p85/p110 for pY binding sites, thereby dampening PI3K signaling. In p85 KO cells, the levels of free p85 were thought to be preferentially reduced, leading to enhanced access of lipid kinase-competent, heterodimeric p85/p110 complexes to pY binding sites and thus to increased PI3K signaling (22). In addition to the data presented here, other recent experimental evidence also argues against the model of free p85. Indeed, monomeric p85 is unstable: Pulse–chase experiments of cells transiently overexpressing p85α with or without p110α revealed that the half-life of free p85α is at least three times less than that of p85α bound to p110α (39). In line with these observations, loss of expression of p110α or p110β leads to a concomitant reduction in p85 levels (39, 40). Moreover, introduction of increasing amounts of free p85α in pan-p85α KO adipocytes failed to decrease insulin-stimulated PI3K activity in pY complexes but reduces Ser-473 phosphorylation of Akt (21). The latter observation indicates that p85α can decrease cellular PI3K lipid levels through mechanisms that are independent of its capacity to compete for pY docking sites. Alternative models to explain the increased PI3K signaling in p85 KO cells are therefore needed. As argued in refs. 30 and 42, loss-of-expression of p85 has the capacity to alter lipid phosphatase activity in cells and, in support of this, decreased activity of the 3-phosphoinositide phosphatase PTEN has recently been reported in p85α KO liver cells (43).
In summary, we have subjected the class IA PI3K system to a rigorous quantitative analysis. Our data do not support the free p85 hypothesis, which had become an established concept in the field. It will now be important to determine relative expression of the other players in this signaling system, for example the receptors and adaptor proteins to which the different PI3K isoforms can bind. It is anticipated that such quantitative assessment of PI3K signaling will contribute to a better understanding of the regulation of this important biological system.
Materials and Methods
Abs and Reagents.
Abs to class IA PI3Ks were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) (p110β; sc-602, raised against C-terminal peptide) or Upstate (Charlottesville, VA) (p85pan; catalog no. 06-195; raised against the full-length p85α protein) or were made in house: Abs to p110α (SK214/15; polyclonal Ab raised against C-terminal peptide; ref. 44), p110β (no. 2.1; polyclonal Ab raised against C-terminal peptide; ref. 44), p110δ (polyclonal Ab raised against C-terminal peptide; ref. 44), p85α (U10, monoclonal Ab raised against the p85α BH-domain; refs. 45 and 46), p85β (monoclonal Abs T12 and T15; ref. 45), p85α/p85β (U2, monoclonal Ab raised against N-terminal SH2 domain of p85α; cross-reacts in IP with p85β). Other sources of reagents were as follows: IS peptides (Sigma Genosys, The Woodlands, TX), recombinant p110δ/p85 (Upstate), and p85 (Jena Bioscience, Jena, Germany), colloidal Coomassie blue G-250 (GelCode Blue Stain Reagent; Pierce, Rockford, IL), cell culture reagents (Invitrogen, Carlsbad, CA), in-gel digestion and liquid chromotography (LC) solvents (Rathburn, Walkerburn, U.K.), and protein standards and other buffer components (Sigma, St. Louis, MO).
Cell Culture.
The WEHI-231 cell line was cultured in RPMI medium 1640 supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.05 mM β-mercaptoethanol. NIH 3T3 and HEK-293T were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. All cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Murine Tissue Extraction.
Murine tissue was extracted from 8- to 10-week-old C57BL/6 mice, cut into sections, and instantly snap-frozen in liquid nitrogen. Cells were extracted by manual homogenization of tissue in Triton X-100 lysis buffer (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 supplemented with protease and phosphatase inhibitors).
DNA Transfection.
HEK-293T cells were transfected with PI3K expression vectors by using calcium phosphate precipitation (catalog no. K2780-01; Invitrogen). The expression vector used was pSG5, in which expression of untagged bovine p85α or bovine p110α is driven by the SV40 early promoter.
Generation of pY-Affinity Matrix.
The pY matrix was generated by coupling pYVPMLG (Alta Bioscience, Birmingham, U.K.) to Actigel (primary amines coupled to agarose) beads (Sterogene, Carlsbad, CA). For protein enrichment and depletion, 100 μl of pY matrix was added to 1 mg of cell lysate, followed by incubation at 4°C for 2 h.
Immunodepletion.
Cells were lysed in Triton X-100 lysis buffer. One milligram of TCL was incubated for 2 h with Ab mixture either directed against p110 [3 μg of Abs in total; mix made up of 1 μg of Ab to each p110 isoform (p110α, p110β, p110δ)] or with Ab mixture directed against p85 [5 μg of Ab in total; mix made up of 1 μg of p85α Ab (U10), 1 μg of U2, 1 μg each of T12 and T15 p85β Abs, and 1 μg of p85pan Ab (catalog no. 06-195; Upstate)], followed by incubation with protein A or G Sepharose (Amersham Biosciences, Piscataway, NJ). The resulting SN1 after p110 or p85 depletion was subjected to two further rounds of IP similar to that described above, eventually giving rise to SN3.
Immunoprecipitation.
Per round of IP, 1 mg of TCL was incubated for 2–5 h with 16–24 μg of Ab mix [made up of 2–3 μg of Abs to each p110 subunit (p110α, p110β, p110δ), p85α (U10), p85β (T12 and T15), p85α/p85β (U2), and p85pan (catalog no. 06-195; Upstate)], followed by incubation with protein A or G Sepharose.
Anion Exchange Chromatography.
Cells were lysed as described above, and lysates were desalted by gel filtration (Econo-Pac 10DG column; Bio-Rad, Hercules, CA) using chromatography buffer A (20 mM Tris·HCl, pH 7.6, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, and 0.1% octyl glucoside). Buffer B was 0.5 M NaCl dissolved in buffer A. Anion exchange chromatography was performed using a Tricorn Mono Q 4.6/100 PE column (Amersham Biosciences) connected to an Integral 100Q HPLC system (Applied Biosystems, Foster City, CA). Separation of class IA PI3Ks by anion exchange chromatography was performed according to Shibasaki et al. (47) by applying gradient elution from 0–100% buffer B in 20 column volumes (balancing the gradient with buffer A).
In-Gel Digestion.
In-gel digestion was performed as described in ref. 48, except that peptides were not alkylated due to a Cys residue within the sequence of one IS peptide. 1–2 pmol of IS peptide was added to the gel pieces at the same time as trypsin. Extracted peptides were dried in a SpeedVac and resuspended in 0.1% (vol/vol) formic acid.
LC-MS.
LC-MS was performed as described in ref. 48. “Pseudo multiple reaction monitoring” experiments were performed by setting the Q1 to transmit the m/z of peptide molecular ions, which were fragmented in the collision cell. Fragment ions thus produced were sampled by TOF and their areas in extracted ion chromatograms used for quantification.
Data Analysis.
Raw MS and MS/MS data were converted into peak lists by using MassLynx version 4.0 (Waters, Milford, MA). The m/z peaks were smoothed (Savitzky–Golay method, two smoothings) and centered (80% peak height for centroids). Charge states were calculated by the software, and peaks were de-isotoped. Correctness of selected identifications was confirmed manually by assigning all of the fragment ions in MS/MS spectra to theoretical peptide fragmentations (Protein Prospector was used to obtain theoretical fragment ions). Quantitative data were obtained from protein-derived peptides by inputting their m/z values and retention times into the “Quantify Method” provided with the MassLynx software. This feature of the software automatically obtains extracted ion chromatograms for each of the input m/z values, which, together with knowledge of retention time, are the basis for peak area selection and integration.
Real-Time PCR.
Total RNA was extracted from cells by using the RNeasy Mini kit (catalog no. 74104; Qiagen, Valencia, CA). mRNA was subsequently reverse transcribed by using the SuperScript II Reverse Transcriptase (catalog no. 18064; Invitrogen). This cDNA was used as the template for real-time PCR amplification. TaqMan Universal PCR Master Mix, primers, and probes were obtained from Applied Biosystems (catalog no. 4364103). For each gene of interest, known amounts of plasmids for this gene were used to create a standard curve. Real-time PCR generated a series of CT values (the PCR cycle at which amplification of each target gene is first detected) for endogenous and plasmid-born cDNA, which allowed for the determination of mRNA copy numbers for each individual gene.
Acknowledgments
We thank Katja Björklöf, Julie Guillermet-Guibert, Lazaros Foukas, and Klaus Okkenhaug for critically reading the manuscript. Work in the B.V. Laboratory was supported by the Ludwig Institute for Cancer Research, with additional funds from the International Association for Cancer Research (to P.R.C. and B.V.), the Roche Research Foundation, Switzerland (B.G.), Overseas Research Scheme U.K. (B.G.), Janggen-Pöhn Stiftung, Switzerland (B.G.), Uarda-Frutiger Fonds, Switzerland (B.G.), and the Medical Research Council (G.N.).
Abbreviations
- a.u.
arbitrary unit
- IP
immunoprecipitation
- IS
internal standard
- KO
knockout
- LC
liquid chromatography
- PI3K
phosphoinositide 3-kinase
- pY
phosphotyrosine
- SH2
Src-homology 2
- SN
supernatant
- TCL
total cell lysate.
Footnotes
Conflict of interest statement: Bart Vanhaesebroeck is a consultant for PIramed (Slough, U.K.).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700373104/DC1.
References
- 1.Vanhaesebroeck B, Waterfield MD. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 3.Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, Zvelebil MJ, Higashi K, Volinia S, Downward J, Waterfield MD. Proc Natl Acad Sci USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry MJ, Yonezawa K, Kasuga M, Waterfield MD. EMBO J. 1994;13:511–521. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter CL, Auger KR, Chanudhuri M, Yoakim M, Schaffhausen B, Shoelson S, Cantley LC. J Biol Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 7.Rordorf-Nikolic T, Van Horn DJ, Chen D, White MF, Backer JM. J Biol Chem. 1995;270:3662–3666. doi: 10.1074/jbc.270.8.3662. [DOI] [PubMed] [Google Scholar]
- 8.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 9.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, et al. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 11.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 12.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, Humphries LA, Rawlings D, Reynolds H, Vigorito E, Turner M. J Exp Med. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Wang D, Ihle JN. Mol Cell Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali K, Bilancio A, Thomas M, Pearce W, Gilfillan AM, Tkaczyk C, Kuehn N, Gray A, Giddings J, Peskett E, et al. Nature. 2004;431:1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- 15.Bilancio A, Okkenhaug K, Camps M, Emery JL, Ruckle T, Rommel C, Vanhaesebroeck B. Blood. 2006;107:642–650. doi: 10.1182/blood-2005-07-3041. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Viciana P, Sabatier C, McCormick F. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurosu H, Katada T. J Biochem (Tokyo) 2001;130:73–78. doi: 10.1093/oxfordjournals.jbchem.a002964. [DOI] [PubMed] [Google Scholar]
- 18.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 19.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 20.Chantry D, Vojtek A, Kashishian A, Holtzman DA, Wood C, Gray PW, Cooper JA, Hoekstra MF. J Biol Chem. 1997;272:19236–19241. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- 21.Ueki K, Fruman DA, Yballe CM, Fasshauer M, Klein J, Asano T, Cantley LC, Kahn CR. J Biol Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- 22.Luo J, Cantley LC. Cell Cycle. 2005;4:1309–1312. doi: 10.4161/cc.4.10.2062. [DOI] [PubMed] [Google Scholar]
- 23.Terauchi Y, Tsuji Y, Satoh S, Minoura H, Murakami K, Okuno A, Inukai K, Asano T, Kaburagi Y, Ueki K, et al. Nat Genet. 1999;21:230–235. doi: 10.1038/6023. [DOI] [PubMed] [Google Scholar]
- 24.Fruman DA, Mauvais-Jarvis F, Pollard DA, Yballe CM, Brazil D, Bronson RT, Kahn CR, Cantley LC. Nat Genet. 2000;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- 25.Ueki K, Yballe CM, Brachmann SM, Vicent D, Watt JM, Kahn CR, Cantley LC. Proc Natl Acad Sci USA. 2002;99:419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Mauvais-Jarvis F, Bluher M, Fisher SJ, Jozsi A, Goodyear LJ, Ueki K, Kahn CR, Fruman DA, Hirshman MF, et al. Mol Cell Biol. 2004;24:320–329. doi: 10.1128/MCB.24.1.320-329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueki K, Algenstaedt P, Mauvais-Jarvis F, Kahn CR. Mol Cell Biol. 2000;20:8035–8046. doi: 10.1128/mcb.20.21.8035-8046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueki K, Fruman DA, Brachmann SM, Tseng YH, Cantley LC, Kahn CR. Mol Cell Biol. 2002;22:965–977. doi: 10.1128/MCB.22.3.965-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauvais-Jarvis F, Ueki K, Fruman DA, Hirshman MF, Sakamoto K, Goodyear LJ, Iannacone M, Accili D, Cantley LC, Kahn CR. J Clin Invest. 2002;109:141–149. doi: 10.1172/JCI13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhaesebroeck B, Ali K, Bilancio A, Geering B, Foukas LC. Trends Biochem Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick DS, Gerber SA, Gygi SP. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A. Anal Chem. 2003;75:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 34.Fry MJ, Panayotou G, Dhand R, Ruiz-Larrea F, Gout I, Nguyen O, Courtneidge SA, Waterfield MD. Biochem J. 1992;288:383–93. doi: 10.1042/bj2880383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fruman DA, Snapper SB, Yballe CM, Davidson L, Yu JY, Alt FW, Cantley LC. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 36.Deane JA, Trifilo MJ, Yballe CM, Choi S, Lane TE, Fruman DA. J Immunol. 2004;172:6615–6625. doi: 10.4049/jimmunol.172.11.6615. [DOI] [PubMed] [Google Scholar]
- 37.Beeton CA, Chance EM, Foukas LC, Shepherd PR. Biochem J 350 Pt. 2000;2:353–359. [PMC free article] [PubMed] [Google Scholar]
- 38.Meier TI, Cook JA, Thomas JE, Radding JA, Horn C, Lingaraj T, Smith MC. Protein Expr Purif. 2004;35:218–224. doi: 10.1016/j.pep.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Brachmann SM, Ueki K, Engelman JA, Kahn RC, Cantley LC. Mol Cell Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. Proc Natl Acad Sci USA. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kazlauskas A, Cooper JA. EMBO J. 1990;9:3279–3286. doi: 10.1002/j.1460-2075.1990.tb07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geering B, Cutillas P, Vanhaesebroeck B. Biochem Soc Trans. 2007;35:199–203. doi: 10.1042/BST0350199. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi CM, Tran TT, Kondo T, Luo J, Ueki K, Cantley LC, Kahn CR. Proc Natl Acad Sci USA. 2006;103:12093–12097. doi: 10.1073/pnas.0604628103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanhaesebroeck B, Jones GE, Allen WE, Zicha D, Hooshmand-Rad R, Sawyer C, Wells C, Waterfield MD, Ridley AJ. Nat Cell Biol. 1999;1:69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 45.Gout I, Dhand R, Panayotou G, Fry MJ, Hiles I, Otsu M, Waterfield MD. Biochem J. 1992;288((Pt 2)):395–405. doi: 10.1042/bj2880395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.End P, Gout I, Fry MJ, Panayotou G, Dhand R, Yonezawa K, Kasuga M, Waterfield MD. J Biol Chem. 1993;268:10066–10075. [PubMed] [Google Scholar]
- 47.Shibasaki F, Homma Y, Takenawa T. J Biol Chem. 1991;166:8108–8114. [PubMed] [Google Scholar]
- 48.Cutillas PR, Geering B, Waterfield MD, Vanhaesebroeck B. Mol Cell Proteomics. 2005;4:1038–1051. doi: 10.1074/mcp.M500078-MCP200. [DOI] [PubMed] [Google Scholar]