Abstract
The architecture of cellular RNA polymerases (RNAPs) dictates that transcription can begin only after promoter DNA bends into a deep channel and the start site nucleotide (+1) binds in the active site located on the channel floor. Formation of this transcriptionally competent “open” complex (RPo) by Escherichia coli RNAP at the λPR promoter is greatly accelerated by DNA upstream of base pair −47 (with respect to +1). Here we report real-time hydroxyl radical (·OH) and potassium permanganate (KMnO4) footprints obtained under conditions selected for optimal characterization of the first kinetically significant intermediate (I1) in RPo formation. ·OH footprints reveal that the DNA backbone from −71 to −81 is engulfed by RNAP in I1 but not in RPo; downstream protection extends to approximately +20 in both complexes. KMnO4 footprinting detects solvent-accessible thymine bases in RPo, but not in I1. We conclude that upstream DNA wraps more extensively on RNAP in I1 than in RPo and that downstream DNA (−11 to +20) occupies the active-site channel in I1 but is not yet melted. Mapping of the footprinting data onto available x-ray structures provides a detailed model of a kinetic intermediate in bacterial transcription initiation and suggests how transient contacts with upstream DNA in I1 might rearrange the channel to favor entry of downstream duplex DNA.
Keywords: initiation, transcription, wrapping, kinetics
Specific transcription initiation by Escherichia coli RNA polymerase (RNAP: core subunit composition α2ββ′ω + σ70 = holoenzyme) at promoter sequences is determined by recognition of DNA (−10 and −35 hexamers) upstream of the start site (+1) by the specificity subunit σ70. Subsequent to binding, a series of large-scale conformational changes in both RNAP and promoter DNA create the initiation-competent open complex (RPo) (1). During these steps, the multisubunit bacterial RNAP acts as an intricate molecular machine and opens ≈14 bp of the DNA double helix. Defining the cascade of conformational changes that occur during initiation is essential to understand sequence- and factor-dependent regulation of the rate of transcription initiation and has important applications in chemical biology and in antibiotic design. However, the intermediates on this pathway are relatively unstable and short-lived and hence are difficult to trap unambiguously. To date, all structural information about complexes known to be on-pathway intermediates in RPo formation has come from chemical and enzymatic DNA footprinting methods.
Quantitative kinetic–mechanistic studies find that at least two kinetically significant intermediates, generically designated I1 and I2, precede formation of RPo by E. coli RNAP:
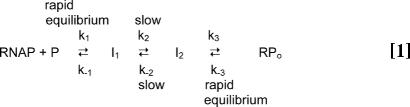 |
where the relatively slow interconversions between I1 and I2 are rate-limiting in both the forward and back directions (2, 3). In the mechanism shown in Eq. 1, I2 and RPo are characterized by their resistance to a short challenge with a polyanionic competitor such as heparin, which acts to sequester any free RNAP present during the challenge. (In contrast, after a 10 to 20 sec challenge with heparin, I1 complexes, which are in rapid equilibrium with free RNAP and promoter sequences, are eliminated from the population.) Given the high degree of conservation of bacterial RNAP and promoter DNA sequences, this mechanism is likely to describe the key steps in initiation in most prokaryotes. Moreover, conservation of many elements of sequence, structure, and/or function between bacterial and eukaryotic polymerase (pol II) subunits and transcription factors supports the inference that the bacterial intermediates may be homologs of initiation intermediates formed by pol II (4, 5).
Recently we (6) and Ross and Gourse (7) found that the presence of DNA upstream of the −35 promoter recognition hexamer greatly accelerates (up to ≈60-fold) the rate-determining isomerization step (conversion of I1 to I2). Strikingly, DNase I footprinting of I1 at the strong bacteriophage promoter λPR reveals that when nonspecific DNA upstream of base pair −47 is present, downstream DNA is protected to around +20, and thus bound in the active-site channel of RNAP. However, when DNA upstream of −47 is deleted, I1 is now less “advanced,” with downstream protection ending at +2 (template strand)/+7 (nontemplate strand) (6). How does DNA upstream of −47 alter downstream interactions in early transcription intermediates? Surprisingly, protection of DNA in I1 from DNase I cleavage extends upstream only to −52 (template) on “full-length” λPR, similar to what is observed in RPo (6). Because DNase I might displace weak upstream interactions in I1 (8, 9), other techniques are required to probe RNAP–DNA contacts in this transient intermediate.
Sclavi et al. (10) recently reported an elegant “real-time” footprinting study of RPo formation at the T7A1 promoter using x-ray-generated ·OH and rapid quench mixing. Unlike DNase I, ·OH is small and nonperturbing of weak interactions. It reacts rapidly and exhibits little if any sequence specificity, making it very useful for probing transient protein–DNA interactions (11). Additionally, the rapid rate of ·OH reaction with the DNA backbone means that fractional protection is approximately proportional to fractional occupancy for a short-lived, rapidly equilibrating intermediate like I1 (10). By following the appearance of protection of the DNA backbone as a function of time (milliseconds to seconds) after mixing at 37°C, this study (10) circumvented potential issues raised by trapping promoter complexes at low temperature (11) and possible displacement of weak upstream interactions by DNase I (8, 9).
In the absence of detailed kinetic–mechanistic information for this promoter, Sclavi et al. (10) used the footprinting data from base pair −60 to +20 to infer both the mechanism (e.g., sequence of intermediates and rate constants of their interconversions) and structure of intermediates preceding RPo. Evidence was obtained for three classes of intermediates (and two to three subclasses of the two early intermediates). The mechanistic analysis revealed that, in general, multiple complexes are populated at T7A1 at each time point in the kinetics. Intriguingly, the series of snapshots of open complex formation at T7A1 suggests that the backbone is protected to −70 (nontemplate)/−82 (template) at early times but to only approximately −60 (template/nontemplate) in RPo (see figure 6 in ref. 10). However, Sclavi et al. (10) did not interpret the far-upstream signal on either strand in their structural–mechanistic analysis.
What is the molecular basis of activation of transcription initiation by upstream DNA? Does the extent of upstream interactions change in RPo formation? Does placement of the downstream DNA (−11 to +20) in the active-site channel of RNAP in I1 require that regions of this DNA be single-stranded (i.e., melted)? Here we use kinetic data from nitrocellulose filter binding experiments (1) to select reaction conditions and times where a relatively homogeneous population of either I1 or RPo exists at the λPR promoter. By avoiding complications that might arise from a mixed population or from off-pathway complexes, these ·OH footprints obtained in real time provide unambiguous structural information regarding DNA backbone positions protected from cleavage in I1 and in RPo. Similarly, these conditions allow us to probe the extent of unstacking of thymine bases in I1 with KMnO4. The lack of KMnO4 reactivity of I1 demonstrates that downstream DNA enters the channel as a duplex and thus must be opened by RNAP in subsequent steps.
Results
Comparative Analysis of DNA Backbone Interactions in I1 and in RPo by ·OH Footprinting.
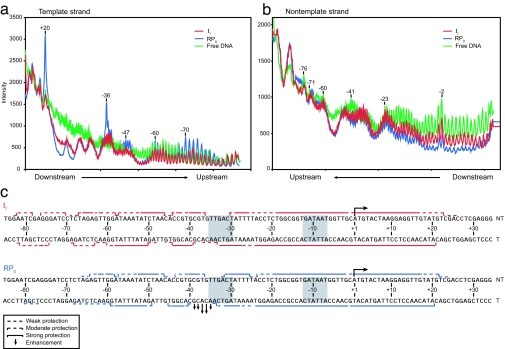
How does the presence of upstream DNA extend the protection of downstream DNA from +2 (nontemplate)/+7 (template) to approximately +20 in I1 (6)? Are direct contacts between RNAP and DNA upstream of base pair −47 responsible? Fig. 1 summarizes the ·OH footprints of I1 and of RPo at λPR obtained at 25 sec (>70% I1) and >2,500 sec (>95% RPo) after mixing, respectively (see Methods). Striking differences exist between these footprints in the upstream boundary, in the degree of protection of the far-upstream region of DNA (approximately −65 to −81) and in the degree of protection of the downstream DNA (approximately −17 to +16). Regions of promoter DNA from −82 (nontemplate)/−81 (template) to approximately +20 are protected in I1, whereas the upstream boundary of moderate DNA backbone protection in RPo ends at −64 (nontemplate) and −59 (template) [Fig. 1c; see supporting information (SI) Fig. 5 and SI Tables 1 and 2]. Very weak protection from −60 to −68 and from −77 to −80 on the template strand is observed in RPo; this protection pattern is consistent with previously reported footprints of RPo at λPR (8). Although the downstream boundaries of I1 and RPo are similar, the downstream region from −17 to +16 is much more strongly protected in RPo than in I1 (see Fig. 1 a and b). Periodic protection centered at −42 (template) and −54 (template) occurs in both I1 and RPo and presumably reflects binding of the C-terminal domains of the α subunit (αCTD) (12–14). As shown in Fig. 1, the amount of protection at these positions does not change when I1 isomerizes to RPo, indicating that the extent of occupancy of each of these sites by an αCTD is the same in I1 and RPo at λPR (see below).
Fig. 1.
Hydroxyl radical (·OH) footprints of RNAP–promoter complexes. Shown are representative scans of ·OH cleavage of RNAP–λPR promoter mixtures at 17°C at short (25 sec, >70% I1, <30% RPo; red) and long (>2,500 sec, <5% I1, >95% RPo; blue) times after mixing, compared with the cleavage of free DNA (green) on the template strand (a) and nontemplate strand (b). (c) The average protection in four to five independent experiments mapped onto the λPR sequence. (See SI Tables 1 and 2 for protection at each position observed in each independent experiment.) Numbers above the DNA cleavage products specifying fragment length in a and b and in between the template and nontemplate λPR sequences in c correspond to promoter position numbered with respect to the transcription start site (+1). (The ·OH enhancement observed at +20 on the template strand in RPo is not observed in any other ·OH footprints.)
To ensure that the small fraction of RPo (<30%) present under conditions used to footprint I1 does not significantly contribute to the observed footprint, heparin, an inert competitor for free RNAP, was added to the reaction 10 sec before the generation of ·OH. This challenge eliminates the population of the short-lived I1 complexes but not the long-lived RPo complexes (3). The ·OH footprints obtained after a 10 sec heparin challenge show only weak protection from approximately −18 to −13 (nontemplate) and from −5 to +7 (nontemplate) (see SI Methods and SI Fig. 6). Because these regions are within the most strongly protected parts of the ·OH footprint of RPo, we conclude that this residual protection results from the small subpopulation of RPo that forms before the heparin challenge (see Methods and SI Methods). Importantly, however, no protection exists outside of these regions, indicating that this small amount of RPo does not significantly contribute to the upstream protection pattern reported for I1 in Fig. 1c.
Comparison of the Extent of DNA Opening in RPo with I1 Using KMnO4 Footprinting.
We and others have previously invoked binding in the active site channel of RNAP to explain protection of downstream DNA (to approximately +20) in I1 (3, 9) and in RPo (10, 12, 15, 16). What is the state of the DNA when initially bound in this cleft? The narrow width of the channel (≈15 Å) in the crystal structures of free RNAP holoenzyme has prompted proposals that DNA must open before entering (16, 17). However, the acidic N-terminal domain (NTD) of σ70 (σ1.1), which binds in the channel in the free RNAP in solution (18, 19), is not observed in the available crystal structures. Placing σ1.1 in the channel may open the β/β′ jaws (20), which can “flex” as much as 50 Å apart (21). Such an “open” jaw conformation would permit dsDNA to enter before strand separation occurs.
To address whether entry of downstream DNA into the channel in I1 involves extensive DNA opening, KMnO4 was used to probe for unstacked, solvent-accessible thymine bases. Reactions were performed 20 sec after mixing at 17°C (>70% I1 and <30% RPo) and at >2,500 sec (>95% RPo; see Methods). At >2,500 sec, thymines at −11, −9, and −8 (template; Fig. 2) and at +2, −3, and −4 (nontemplate; SI Fig. 7) are strongly oxidized by the dose of KMnO4 used here. At 20 sec these thymines also exhibit a small amount of KMnO4 reactivity (Fig. 2 and SI Fig. 7). Because this KMnO4 reactivity remains in a sample pretreated with heparin (SI Table 3), we conclude that this signal originates primarily from the small subpopulation of RPo complexes that form in this short time (see Methods and SI Methods), and not to any significant extent from I1. [These signals from reactive thymines in the subpopulation of RPo are observable because base-paired and stacked thymines in dsDNA (e.g., free DNA) are not KMnO4-reactive.] We conclude from these results that promoter DNA in I1 is largely double-helical. Alternatively, open bases in I1 could be buried in interactions with polymerase and therefore not accessible to KMnO4. We do not favor this interpretation because it would require these presumably favorable interactions to exist only in this transient intermediate and to be disrupted to create the much more stable RPo complex.
Fig. 2.
Potassium permanganate (KMnO4) footprints of RNAP–promoter complexes. Shown are representative results from KMnO4 probing of complexes obtained early (20 sec, >70% I1, <30% RPo) and late (>2,500 sec, <5% I1, >95% RPo) in the time course of open complex formation at 17°C. G+A sequencing (40) and an uncut DNA control were performed. Positions of KMnO4-reactive thymines in RPo on the template strand are labeled. KMnO4 reactivity observed in the 20 sec lanes in the presence and absence of heparin is consistent with the small population of RPo complexes formed at this time after mixing. SI Table 3 quantifies the intensity of bands in this and two other independent experiments. See SI Fig. 7 and SI Table 3 for KMnO4 probing of the nontemplate strand.
Discussion
DNA Opening Occurs in the RNAP Active Site Channel in the Conversion of I1 to RPo.
Real-time ·OH and KMnO4 footprinting obviates the need for temperature downshifts or other shifts in solution conditions to stabilize or trap the first intermediate at the λPR promoter. At the short times after mixing examined here, I1 complexes are >70% and RPo complexes are <30% of promoter DNA. At this time point, downstream protection extends to +20, which can be accounted for only by placing DNA downstream from −11 into the active site channel of RNAP (Fig. 3) (3, 6, 15, 16). Together, the ·OH and KMnO4 data (figs. 1 and 2) and DNase I and KMnO4 footprints of I1 at 0°C (9) provide compelling evidence that opening of the DNA from −11 to +2 occurs only after it binds in the active site channel in I1. Cross-linking experiments on wild-type RNAP and on a mutant RNAP lacking the downstream lobe of β (E. coli βΔ186–433) have led to a similar “active” melting proposal (22).
Fig. 3.
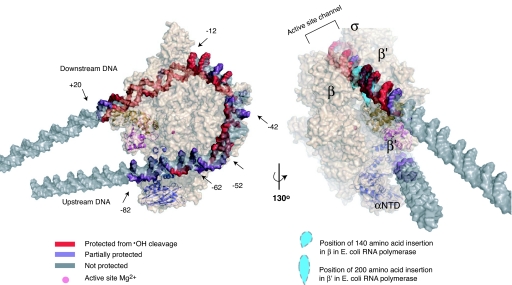
Model of the intermediate complex I1 preceding formation of the transcriptionally competent RPo. The conformation of RNAP (beige) is based on the x-ray crystal structure of the homologous T. thermophilus enzyme [Protein Data Bank ID code 1IW7 (16); T. thermophilus β′ residues 138–452, absent in E. coli (28), are not shown]. The modeled position of promoter DNA in I1 is based on DNase I (6), ·OH and KMnO4 footprinting and relevant crystal structures. The two views of I1 demonstrate the agreement between the model and the experimentally determined strong and moderate protection (red and purple, respectively) of the DNA backbone (gray) from ·OH cleavage (see Fig. 1). Strong protection seen at −12 to −19 likely involves the nonconserved region of E. coli σ70 not present in σA of T. thermophilus. Domains in E. coli subunits β and β′ (represented as blue teardrops, missing in T. thermophilus) are positioned at the sites of their insertion in the T. thermophilus sequence; these likely protect DNA downstream of +10 from ·OH cleavage. The upstream surface groove formed by β′ and the NTD of α, and the mobile downstream jaw (E. coli β′ residues 913-1262) and upstream DNA clamp (E. coli β′ residues 808–912), are highlighted in cartoon representations in dark blue, magenta, and gold, respectively. This figure was created using PyMol (41).
Alternative models of DNA opening during initiation invoke the passive capture of transiently open regions in the −10 hexamer by RNAP (16, 17, 23). These proposals are largely based on the conformational state of free RNAP captured in the crystal. Although atomic resolution structural data are necessary to establish molecular structure–function relationships, not all aspects of the current set of crystal structures need be relevant for hypotheses regarding RPo formation in solution. In particular, high-salt crystallization conditions (15) and/or crystal packing interactions may disfavor binding of σ1.1 in the channel, and this or other aspects of crystallization may induce the “narrow” conformation of flexible/mobile elements of the enzyme. The results reported here and the publication of a crystal structure of a dsRNA ligand bound in the structurally homologous cleft of the yeast RNAP (24) demonstrate that double-stranded nucleic acids can indeed access the channel, ruling out a general requirement for DNA opening to precede entry.
Recently, a subassembly of RNAP containing only regions 2 and 3 of σ70 and the NTD of β′ was shown to form a KMnO4-reactive complex with an A/T-rich promoter sequence (23). Does formation of this complex mirror the steps in RPo formation by RNAP? Unlike opening by the wild-type RNAP holoenzyme at λPR, formation of this open or distorted complex requires a highly negatively supercoiled DNA template. At this superhelix density, both experiments and DNA supercoiling thermodynamics indicate that the equilibrium fraction of open bases is significant at 37°C (1–5%, primarily A/T-rich regions). Because region 2 of σ70 (σ2) bound to the NTD of β′ selectively recognizes and binds the nontemplate strand of the −10 recognition hexamer (TATAAT) (25), possibly σ2-β′ recognizes and stabilizes this A/T-rich region as it transiently opens on negatively supercoiled DNA (but not on linear fragments). Kinetic data for this minimal system are required to define the mechanism of DNA opening at this supercoiled promoter and how it relates to opening by the wild-type RNAP machinery.
Structural Model of I1.
To explain why moderate to strong far-upstream protection is observed only in I1 and to explore how upstream DNA facilitates placement of downstream dsDNA in the active site channel, we modeled I1 using footprinting data (Figs. 1 and 2), patterns of DNase I hypersensitive cleavage sites (6), and relevant crystal structures (see Methods and SI Methods). In this model (Fig. 3), protection to position −81 (template)/−82 (nontemplate) in I1 is achieved by wrapping upstream DNA around the outside of RNAP. Protection to +20 arises from a sharp DNA bend (≈90°) at position −11/−12 that places downstream duplex DNA high in the channel (≈50 Å above the active site) (3). Whereas the channel protects downstream DNA from ·OH cleavage in I1, far greater protection is observed in RPo (Fig. 1 a and b). This difference presumably reflects formation of extensive contacts between RNAP and the DNA backbone that stabilize the open bubble (−11 to +2) in RPo.
A critical feature of the model (Fig. 3) is that the trajectory of upstream DNA in I1 is set by the interaction of the −35 hexamer with σ region 4 (σ4), the interaction of the proximal αCTD with the region centered at base pair −42, and the interaction of the distal αCTD with the region centered at base pair −54 (26). Together these interactions are predicted to bend promoter DNA from positions −30 to −57 by ≈90–100°, directing the flanking upstream DNA along a path along the outside of β′ that roughly parallels the active-site channel (Fig. 3).
The model predicts that DNA from positions −60 to −65 lies next to the C terminus of β′ and the ω subunit; the following helical turn of upstream DNA (from −71 to −81) falls in a surface groove formed by the interface between β′ and the NTD of the β′-associated α subunit. This surface groove contains pairs of conserved cationic (β′ R576, R610, K615; α H128) and anionic (β′ D571 and E616) residues (see SI Methods) positioned to be able to form intramolecular salt bridges in the absence of DNA, and hence modulate the affinity of RNAP for the upstream region of promoter DNA (27). Although the ·OH backbone protection data do not precisely define the phasing of the far-upstream DNA with respect to RNAP (see SI Methods), they unambiguously demonstrate that the “backside” of RNAP interacts extensively with this DNA in I1, but not in RPo.
Proposal: Upstream DNA Wrapping Triggers Conformational Changes in the RNAP Active Site Channel.
Results presented here and by Sclavi et al. (10) indicate that the extent of upstream DNA protection from ·OH cleavage is time-dependent. At early times after mixing, E. coli RNAP protects the DNA backbone at T7A1 (10) and λPR to approximately −80 but to only approximately −60 in RPo. At λPR, upstream protection from −71 to −81 spans the minor and the major groove for approximately one turn. This pattern is difficult to explain by transient interactions with the αCTDs [as deduced from cross-linking experiments on RPo formed at the lacUV5 promoter (12)] but is completely consistent with wrapping of upstream DNA on RNAP, as modeled here (Fig. 3). Moreover, the protection from −71 to −81 indicates that the DNA backbone is practically engulfed on all sides, which cannot be accounted for by binding in the surface groove alone.
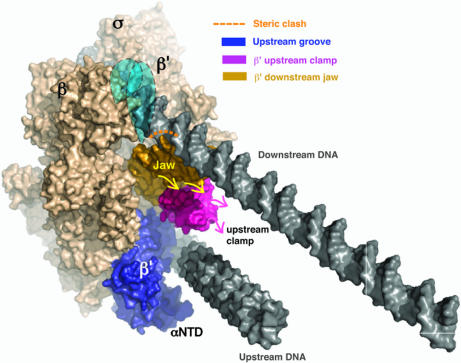
What domains of RNAP are responsible for the protection of far-upstream DNA in I1? One possible candidate is the conserved domain of β′ (E. coli residues 808–912; Thermus thermophilus residues 1106–1218) (Fig. 3), which lies directly above the surface groove in the T. thermophilus RNAP crystal structure (16). To explain the upstream protection data, we propose that this domain moves to contact this region of DNA when it wraps on RNAP in I1 (Fig. 4). This region, which we propose calling the “upstream DNA clamp,” is likely flexible in solution, as suggested by the prediction that it is partially disordered in free RNAP (1). Importantly, the upstream DNA clamp (magenta in Fig. 4) is N-terminal to and contacts another conserved domain called the “downstream jaw” (E. coli residues 913–1262; T. thermophilus residues 1219–1377) (gold in Fig. 4) or “downstream mobile clamp,” also predicted to be partially disordered in solution (1). In E. coli, the downstream jaw includes the so-called “trigger loop” and a flexibly tethered, independently folded ≈200-residue sequence insertion [E. coli residues 940–1139 (28), absent in T. thermophilus], shown as a blue shape in Fig. 4. The “plasticity” of the downstream jaw and the upstream DNA clamp is also suggested by the significant and possibly coupled changes in their position in various structures relative to the rest of RNAP (21). Consequently, we speculate that direct interactions between RNAP and upstream DNA (from −68 to −81) rearrange not only β′ residues 808–912 (magenta in Fig. 4), but also β′ residues 913–1262 (gold in Fig. 4). In Fig. 4, this motion is proposed to reposition the downstream jaw (and β′ insertion), thereby opening the downstream end of the channel and allowing the full entry of downstream DNA.
Fig. 4.
Surface representation of I1. The proposed upstream DNA binding groove (dark blue) and the steric clash between the β′ downstream jaw (gold; see text) and the downstream DNA that occurs in the conformational state of RNAP captured in the crystal structure are highlighted. To account for all of the ·OH protection observed from −71 to −82 (nontemplate), a subdomain of β′ (upstream clamp; magenta) is hypothesized to move to contact DNA in the upstream groove (dark blue). This rearrangement (magenta arrows) is proposed to be coupled to a movement in the adjacent domain forming the downstream jaw (yellow arrows; see text). These shifts would eliminate the steric clash seen in the model and allow full entry of downstream DNA into the cleft. Further descent of dsDNA into the active-site channel is proposed to be blocked by the presence of the NTD of σ (data not shown) (1). As a result, the transcription start site (+1) lies directly over but ≈50 Å above the active site. This figure was created by using PyMol (41).
Rearrangements in the β′ downstream jaw have recently been proposed to be required for RPo formation by the alternate specificity factor σ54 (29). For σ54, ATP-dependent activator proteins are hypothesized to remodel the downstream jaw by means of interactions with the NTD of σ54. Similarly, ATP hydrolysis by the transcription factor TFIIH has been proposed to drive large-scale protein rearrangements in the pol II preinitiation complex to allow DNA melting (30). This hypothesis contrasts with the generally accepted model that invokes ATP-driven conformational changes in the helicase domains of TFIIH to open DNA (4). Because (i) promoter opening still occurs when the helicase activity of TFIIH is substantially reduced by mutation (30) and (ii) both cross-linking (5) and footprinting (4) place TFIIH at the downstream end of the pol II channel, we propose that rearrangements in domains at this end of the channel remove impediments to entry of dsDNA and thus are critical to RPo formation by both bacterial and eukaryotic RNAP. In this model, by coupling promoter-mediated upstream DNA interactions to rearrangements in the downstream jaw, RNAP effectively distinguishes between promoter and nonpromoter DNA and bars nonpromoter DNA from accessing the active site located on the floor of the channel.
Role of DNA Bending and Upstream Wrapping in Transcription Activation.
In the model of I1 shown in Fig. 3, wrapping of upstream DNA on the backside of RNAP requires a series of optimally phased DNA bends. Although it is well established that many transcription activators/repressors (e.g., catabolite activator protein) bind upstream of promoter sequences and bend DNA, how DNA bending impacts the steps in transcription initiation is unclear. Nevertheless, many studies have demonstrated that altering the degree and/or phasing of upstream bending by introducing intrinsically bent DNA sequences (e.g., ref. 31) or by moving binding sites of activators (32) significantly increases (activates) or decreases (represses) the amount of transcription. Fig. 3 provides one possible explanation for these observations. In this model, σ4-αCTD–DNA interactions establish the trajectory/topology of upstream DNA beyond −60, thereby fundamentally determining whether far-upstream DNA contacts the CTD of β′. Any changes in the network of upstream DNA–protein and protein–protein interactions (Fig. 3) (e.g., introducing bent DNA via binding interactions with accessory proteins or mutating key residues in the protein–protein interfaces) will affect the path of upstream DNA and thus are predicted to alter the efficiency of initiation. Deleting upstream DNA beyond −65 is predicted to remove the contact with the downstream jaw. In this case, full entry of dsDNA into the channel would require coupled displacement of the downstream jaw, thus retarding formation of RPo (6, 7).
Far-Upstream DNA Unwraps in Converting I1 to RPo.
In forming RPo from I1, the upstream boundary shifts from −82 (nontemplate) to −64 (nontemplate) and aspects of the interactions of σ4 and the proximal αCTD with promoter DNA change: cleavage at −36 (template) and −37 (template) is now strongly enhanced relative to free DNA. The appearance of a DNA distortion in this region in RPo may reflect and/or result in an altered trajectory of DNA upstream of base pair −65 such that it is no longer directed toward the αNTD/β′ groove. The upstream protection remaining in RPo indicates that DNA from approximately −35 to −64 (nontemplate)/−59 (template) continues to wrap on the surface of RNAP [consistent with previous ·OH (8) and AFM (33, 34) studies of RPo at λPR]. Here Fig. 1 shows that the upstream DNA is even more extensively wrapped in I1.
Because the truncation of upstream DNA does not significantly change the dissociation rate constant at either λPR (6) or lacUV5 (7), we infer that unwrapping does not occur in the late steps that convert the transition state (I1–I2)‡ to I2 and RPo. Although we cannot rule out compensatory changes that mask unwrapping in later steps, we think it is most likely that the unwrapping of approximately −65 to −81 occurs in the conversion of I1 to (I1–I2)‡. This rate-limiting isomerization step exposes anionic surface, which may originate from the movement of σ1.1 out of the active site channel and/or unwrapping of upstream DNA (1). The ejection of σ1.1 may be driven by the entry of negatively charged DNA phosphates in the channel and facilitated by opening of the downstream end of the channel by upstream wrapping in I1 (see above). Exit of this single-stranded DNA mimic from the channel may disfavor the upstream wrap, either directly or indirectly. Unwrapping of upstream DNA is predicted to release the downstream jaw, making it available to fold on downstream DNA in RPo (1). These proposals for a series of coordinated motions in the polymerase machinery involved in DNA opening are currently being tested by kinetic studies on RNAP and promoter variants.
Methods
Buffers.
RNAP storage buffer (SB) is 50% glycerol, 10 mM Tris (pH 7.5 at 4°C), 100 mM NaCl2, 0.1 mM DTT, and 0.1 mM EDTA. DNase I storage buffer is 50% (vol/vol) glycerol, 10 mM Tris·HCl, and 5 mM MgCl2. The binding buffer (BB) for hydroxyl radical (·OH) and KMnO4 footprinting experiments is 12.5 mM Tris (adjusted to pH 8 at the temperature of the experiment), 3 mM MgCl2, 60 mM KCl, 0.6 M glycine betaine, and 100 μg/ml BSA. ·OH were generated by using the Fenton reaction [1.73 mM Fe(II), 2.3 mM EDTA, 0.98 mM Na ascorbate, and 0.08% H2O2]. ·OH stop buffer is 9.5 mM thiourea and 1.7 mM EDTA. Solvent-exposed thymines were probed with 4 mM KMnO4; strands at oxidized thymines were cleaved by adding 10% piperidine. KMnO4 stop buffer is 1 M 2-mercaptoethanol and 2.7 M ammonium acetate. TBE buffer and urea loading buffer have been described (8).
RNAP.
E. coli K12 RNAP σ70 holoenzyme containing the ω subunit was purified as previously described (35, 36). The promoter-binding activity of RNAP was assayed at the time of the footprinting experiments as described in (37) and was always 60–95%. All RNAP concentrations reported are for the active holoenzyme.
λPR Promoter DNA.
Full-length λPR promoter DNA fragments were isolated and labeled as described (6). DNA fragments end at −110 on nontemplate-labeled DNA and at −102 on template-labeled DNA; λPR DNA sequences extend from −60 to +20. This design eliminates λPRM by replacing the region upstream of −60 with nonspecific plasmid DNA sequences.
Determination of Fractions of Promoter DNA in I1 and RPo Complexes as a Function of Time After Mixing.
At 17°C, where the equilibrium constant K1 is a maximum and conversion of I1 to I2 is relatively slow, we previously found that the fraction of promoter DNA in I1 complexes (θI1) at 15 sec after mixing λPR promoter with 70 nM (excess) RNAP was 0.55 (6). To increase the fraction of I1 in the time interval (20–35 sec) of the footprinting experiments, we reduced the KCl and MgCl2 concentrations to those listed in Buffers and added 0.6 M glycine betaine. Based on observed dependences of K1 and k2 on these solute concentration variables (1) we calculate θI1 > 0.7 in the absence of heparin for the time interval and under the conditions of experiments used to footprint I1. See SI Methods.
·OH Footprinting.
A preliminary attempt to characterize I1 by ·OH footprinting was performed at 0°C at equilibrium (38) where the only complex predicted to be populated is I1 (9). Because the signal:noise ratio in these 0°C experiments was marginal, and to avoid the possibility of populating off-pathway complexes at 0°C (39), we identified solution conditions where I1 is >70% of promoter DNA (see above) at early times in the time course of RPo formation, and footprinted as follows. RNAP (70 nM in SB) and DNA (0.4–1 nM in BB) were mixed at 17°C. To characterize I1, RNAP and DNA were incubated for 15 sec, challenged with heparin (100 μg/ml final) in BB (or with the same volume of BB as a control) for 10 s, and subsequently probed with ·OH for 10 s starting 25 sec after mixing RNAP and DNA. RPo was characterized in an identical manner at >2,500 sec. ·OH cleavage was stopped after 10 sec by the addition of 6.5 μl of stop buffer and 100 μl of equilibrated phenol. Samples were phenol-extracted, ethanol-precipitated, and loaded onto an 8% sequencing gel as previously described (6).
Sixteen (nontemplate) and eight (template) sets of independent ·OH footprinting experiments were performed by using at least three independently labeled λPR fragments. Ten (five template and five nontemplate; numbered 1–5 in SI Tables 1 and 2) footprints of I1 and nine (four nontemplate and five template; numbered 1–4 and 1–5, respectively, in SI Tables 1 and 2) footprints of RPo were well aligned and included in the ·OH footprint analysis (see SI Methods for specific details of analysis and alignment criteria). RPo footprints reported here are consistent with footprints reported in ref. 8 (see SI Tables 1 and 2). In one footprinting experiment, the glycine betaine concentration was increased to 1 M; the resulting footprint was identical to the ones obtained when 0.6 M glycine betaine was used.
Heparin is a nonperturbing competitor for free RNAP under these reaction conditions. To footprint with ·OH in 10 sec, highly active RNAP holoenzyme (60–95% active) was used to reduce the glycerol concentration (from SB) to 2%.
KMnO4 Footprinting.
KMnO4 probing was performed at 17°C as previously described (6). To characterize I1 at 17°C, RNAP and DNA were incubated for 10 sec, challenged with heparin in BB (100 μg/ml final) or the same volume of BB (as a control) for 10 sec, and subsequently probed with KMnO4 (at 4 mM KMnO4 final) for 10 sec starting 20 sec after mixing RNAP and DNA. At 20 sec, I1 is >70% of promoter DNA and RPo is <30% based on ·OH footprint analysis (see SI Methods) and kinetic calculations. RPo was characterized at >2,500 sec after mixing RNAP and DNA by using the same KMnO4 concentration. Three independent footprinting experiments of I1 and RPo (with or without heparin) were performed on each strand. (See SI Methods and SI Table 3 for further details.)
PhosphorImager Analysis.
Gel image data were obtained by using a PhosphorImager (Molecular Dynamics, Piscataway, NJ) and analyzed as described (8) using ImageQuant (versions 4.2 and 5.2; Molecular Dynamics). Quadrilateral contours were drawn around each uncut band, around DNA blocks upstream of −85 and/or downstream of +30, and at each 10–20 positions in between. Background values were subtracted from these blocks by using the “object average” background correction function in ImageQuant and corresponding positions in an empty lane as a reference point. To account for differences in loading and ·OH cleavage, ImageQuant was used to calculate pixel intensities within each block. The blocks of DNA upstream of approximately −85 (when DNA is labeled downstream) or DNA downstream of approximately +30 (when DNA is labeled upstream) in the free DNA lane (−RNAP) were divided by the intensities of the corresponding blocks in the +RNAP lanes. This ratio was used to normalize each footprint lane (+RNAP) to the free DNA lane (−RNAP). Normalizations were checked visually by overlaying traces (pixel intensity versus distance) to ensure that peaks outside of the “footprint” (+RNAP) match the cleavage pattern and intensity of the corresponding free DNA (−RNAP) block. After normalization, if traces outside of the footprint did not match, the footprint was discarded (see SI Methods for details on ·OH footprint analysis).
Modeling of I1.
All modeling was done by using InsightII (Accelrys) on an Octane workstation (Silicon Graphics). In the model of I1, T. thermophilus holoenzyme (Protein Data Bank ID code 1IW7) (16) provides the basis for the protein surfaces shown in Fig. 3. See SI Methods for details of the modeling and of the sequence alignments.
Acknowledgments
We thank the reviewers, Pieter deHaseth, and Wayne Kontur for their comments on the manuscript. This work is supported by National Institutes of Health Grants GM23467 (to C.A.D., M.T.R., and R.M.S.), GM38660 (to R.L.), and GM074901 (to C.A.B.); the Dr. James Chieh-Hsia Mao Distinguished Wisconsin Graduate Fellowship (to C.A.D.); and the Department of Biochemistry Graduate Student Fellowship (to C.A.D.).
Abbreviations
- RNAP
RNA polymerase
- RPo
open complex
- I1
first kinetically significant intermediate
- CTD
C-terminal domain
- NTD
N-terminal domain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609888104/DC1.
References
- 1.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT., Jr Biochemistry. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buc H, McClure WR. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 3.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Record MT., Jr J Mol Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 4.Miller G, Hahn S. Nat Struct Mol Biol. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forget D, Langelier MF, Therien C, Trinh V, Coulombe B. Mol Cell Biol. 2004;24:1122–1131. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis CA, Capp MW, Record MT, Jr, Saecker RM. Proc Natl Acad Sci USA. 2005;102:285–290. doi: 10.1073/pnas.0405779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross W, Gourse RL. Proc Natl Acad Sci USA. 2005;102:291–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig ML, Suh WC, Record MT., Jr Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 9.Craig ML, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Saecker RM, Record MT., Jr J Mol Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 10.Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, Heumann H. Proc Natl Acad Sci USA. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenowitz M, Erie DA, Chance MR. Proc Natl Acad Sci USA. 2005;102:4659–4660. doi: 10.1073/pnas.0501152102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 13.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 14.Ross W, Ernst A, Gourse RL. Genes Dev. 2001;15:491–506. doi: 10.1101/gad.870001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 16.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 17.Murakami KS, Masuda S, Darst SA. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 18.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 19.Nagai H, Shimamoto N. Genes Cells. 1997;2:725–734. doi: 10.1046/j.1365-2443.1997.1600357.x. [DOI] [PubMed] [Google Scholar]
- 20.Mooney RA, Landick R. Genes Dev. 2003;17:2839–2851. doi: 10.1101/gad.1142203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Proc Natl Acad Sci USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brodolin K, Zenkin N, Severinov K. J Mol Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 23.Young BA, Gruber TM, Gross CA. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- 24.Kettenberger H, Eisenfuhr A, Brueckner F, Theis M, Famulok M, Cramer P. Nat Struct Mol Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- 25.Young BA, Anthony LC, Gruber TM, Arthur TM, Heyduk E, Lu CZ, Sharp MM, Heyduk T, Burgess RR, Gross CA. Cell. 2001;105:935–944. doi: 10.1016/s0092-8674(01)00398-1. [DOI] [PubMed] [Google Scholar]
- 26.Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL. Genes Dev. 1999;13:2134–2147. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saecker RM, Record MT., Jr Curr Opin Struct Biol. 2002;12:311–319. doi: 10.1016/s0959-440x(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 28.Chlenov M, Masuda S, Murakami KS, Nikiforov V, Darst SA, Mustaev A. J Mol Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 29.Wigneshweraraj SR, Savalia D, Severinov K, Buck M. J Mol Biol. 2006;359:1182–1195. doi: 10.1016/j.jmb.2006.04.063. [DOI] [PubMed] [Google Scholar]
- 30.Lin YC, Choi WS, Gralla JD. Nat Struct Mol Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 31.Newlands JT, Josaitis CA, Ross W, Gourse RL. Nucleic Acids Res. 1992;20:719–726. doi: 10.1093/nar/20.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavigne M, Kolb A, Buc H. Biochemistry. 1992;31:9647–9656. doi: 10.1021/bi00155a018. [DOI] [PubMed] [Google Scholar]
- 33.Cellai S, Mangiarotti L, Vannini N, Naryshkin N, Kortkhonjia E, Ebright RH, Rivetti C. EMBO Rep. 2007;8:271–278. doi: 10.1038/sj.embor.7400888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivetti C, Guthold M, Bustamante C. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess RR, Jendrisak JJ. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez N, Wiggs J, Chamberlin MJ. Arch Biochem Biophys. 1977;182:404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- 37.Roe JH, Burgess RR, Record MT., Jr J Mol Biol. 1984;176:495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 38.Raffaelle M. Madison: University of Wisconsin; 2003. Ph.D. thesis. [Google Scholar]
- 39.Buckle M, Pemberton IK, Jacquet MA, Buc H. J Mol Biol. 1999;285:955–964. doi: 10.1006/jmbi.1998.2391. [DOI] [PubMed] [Google Scholar]
- 40.Maxam AM, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMol Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]