Abstract
Protein kinases are generally recognized as attractive drug targets to treat a variety of human diseases. Recent analysis of the Plasmodium falciparum kinome identified several kinases that are entirely unique to Plasmodium species. The specific functions and targets of most of these enzymes remain largely unknown. Here, we have identified a P. falciparum kinase (PfPK9/PF13_0085 ORF) that does not cluster with any of the typical eukaryotic protein kinases. PfPK9 protein expression was induced ≈18 h after red blood cell infection, and was mainly localized to the parasitophorous vacuolar membrane as well as the cytosol. Recombinant PfPK9 autophosphorylated in vitro and specifically phosphorylated the exogenous substrate histone H1, indicating that it is catalytically active. Phosphopeptide mapping studies showed that autophosphorylation occurred at three residues: T082, T265, and T269. We identified a P. falciparum homolog of the E2 ubiquitin-conjugating enzyme 13 (UBC13) as an endogenous substrate for PfPK9. PfPK9 phosphorylated UBC13 at S106, a highly conserved residue among eukaryotic E2s, and suppressed its ubiquitin-conjugating activity. Our findings not only describe a previously uncharacterized Plasmodium kinase and its likely in vivo target, but also suggest that modulation of UBC13 activity by phosphorylation may be a common regulatory mechanism in eukaryotes.
Keywords: ubiquitination
Malaria continues to be a major public health problem in the developing world and is responsible for ≈3 million deaths yearly. This disease is caused by a unicellular protozoan of the Plasmodium genus, in which Plasmodium falciparum is responsible for the most lethal form of malaria. P. falciparum has developed resistance to most antimalarial chemotherapeutics, thereby limiting our options for effective disease control (1). However, new chemotherapeutic strategies may be discovered through the investigation of molecular mechanisms required for parasite growth and development. The blood stage is responsible for the clinical manifestations of the disease and lasts 48 h. During this period, the parasite is metabolically very active and undergoes significant morphological changes. These processes probably involve the interaction of various signaling molecules within the parasite and with the host, and may represent points of therapeutic intervention.
Many eukaryotic intracellular processes employ modulation of protein phosphorylation by protein kinases and phosphatases as a form of regulation. Several studies have investigated parasite protein phosphorylation during various stages of the blood stage life cycle. During its intraerythrocytic development, P. falciparum was found to induce phosphorylation of numerous proteins both within the parasite and the host red cell (2, 3). Protein kinases are now widely recognized as valuable drug targets for the treatment of many human diseases (4). There is evidence that inhibition of kinase activity can have antimalarial actions. Both invasion and development of the parasite in red blood cells (RBCs) are inhibited by kinase inhibitors (5). Several groups have demonstrated potent antimalarial effects of cell cyclin-dependent kinase inhibitors in cell-based models of infection (6–9). However, we believe that drugs specifically targeting PfPKs with no human homologs are likely to be more successful because they are less likely to inhibit members of the human kinome, thereby avoiding potential toxicity issues.
Two independent analyses of the P. falciparum kinome based on genome sequence data (10) revealed either 85 or 99 genes encoding for proteins containing a kinase domain (11, 12). Although most of the Plasmodium kinases can be categorized into well established eukaryotic protein kinase families, several are completely unique to Plasmodium species, showing no significant sequence similarity outside the conserved kinase signature motif. In the past few years, multiple groups have characterized several Plasmodium kinases in a comprehensive manner (13–18). However, many of these studies have either examined kinases involved in sexual stage development or orthologs of higher eukaryotes. Although assigning putative roles to truly novel Plasmodium kinases can be a challenging task, their unique nature may play a significant role in identifying Plasmodium-specific signaling pathways for chemotherapeutic intervention. Investigating the regulation of unique Plasmodium-specific kinases and identifying cellular targets may unravel their functions and signaling roles.
To achieve the above objective, we characterized the P. falciparum protein kinase PfPK9. Although PfPK9 contains all of the kinase subdomains required for catalytic activity, it cannot be classified into established eukaryotic protein kinase families. It is located at the base of the calcium-dependent protein kinase (CDPK) family and the Protein kinase A/G/C family (12) but does not cluster with either of these groups [supporting information (SI) Fig. 6]. We show that PfPK9 is expressed in the later stages of the asexual life cycle and localized to the parasitophorous vacuolar membrane (PVM) as well as the cytosol. Moreover, we demonstrate that PfPK9 targets a P. falciparum ubiquitin-conjugating enzyme as an intracellular target. Ubiquitin-conjugating enzymes, known as E2 enzymes, catalyze a key second step in the ubiquitination cascade. These enzymes are defined by their ability to accept ubiquitin thioester from the ubiquitin-activating enzyme (E1) to form an E2-ubiquitin thioester. Recent studies in both yeast and mammalian systems have implicated phosphorylation as a regulatory mechanism for some E2s, such as UBC2, UBC3, and UBC6e (19, 20). Although ubiquitination is involved in modifying protein function and levels in eukaryotes, little is known about Plasmodium E2s. In this report, we identified PfUBC13, a putative P. falciparum E2, as a downstream target of PfPK9. UBC13, unlike other E2s, is engaged in nonproteolytic pathways. Here, we also demonstrate that phosphorylation of UBC13 negatively regulates its activity.
Results
Identification, Molecular Cloning, and Primary Structure of PfPK9 Gene.
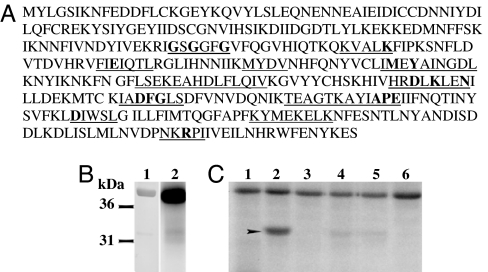
Upon availability of the P. falciparum database, we searched for proteins that contained all of the invariant residues of a kinase catalytic domain but exhibited no similarity to any of the known eukaryotic protein kinases. One of the putative ORFs identified was PF13_0085. The gene prediction algorithm GeneFinder on PlasmoDb predicted PF13_0085 to contain a 1.1-kb region with no introns, encoding a 367-aa protein (Fig. 1A). We confirmed its gene expression by synthesizing cDNA using oligo(dT)s in a reverse transcription reaction from P. falciparum mRNA, followed by amplification of the coding region with gene-specific primers. According to the genome-wide analysis of the P. falciparum kinome (11), PF13_0085 fails to associate strongly with any established eukaryotic protein kinase family. The closest related eukaryotic kinase is SNF-1 kinase, which shows a 36% identity in the kinase domain and an 11% identity overall. PF13_0085 possesses all of the 11 conserved kinase subdomains, including the invariant residues (Fig. 1A).
Fig. 1.
PF13_0085 (PfPK9) ORF, expression, and catalytic activity (A) The amino acid sequence of PfPK9 contains the catalytic domain, which includes all of the 11 conserved kinase subdomains (underlined). All invariant residues are indicated in bold. (B) Silver-stained gel of purified kinase (lane 1). The kinase exhibits the capacity to autophosphorylate (lane 2) as shown by autoradiography. (C) Activity of the kinase was tested against a battery of nonspecific substrates: myelin basic protein (lane1), histone H1 (lane 2), histone 2A (lane 3), histone 3 (lane 4), casein (lane 5), and kinase alone (lane 6). Histone H1 is robustly phosphorylated by the kinase.
Recombinant PfPK9 Is Catalytically Active.
The purified recombinant PfPK9 was catalytically active and autophosphorylated (Fig. 1B). In addition, we tested kinase activity against multiple protein substrates (Fig. 1C). PfPK9 phosphorylated histone H1 (lane2), whereas no activity was detected against myelin basic protein (lane 1), histone H2A (lane 3), and α- or β-casein (lanes 4 and 5).
PfPK9 Activity Is Positively Regulated by Autophosphorylation on Three Threonine Residues.
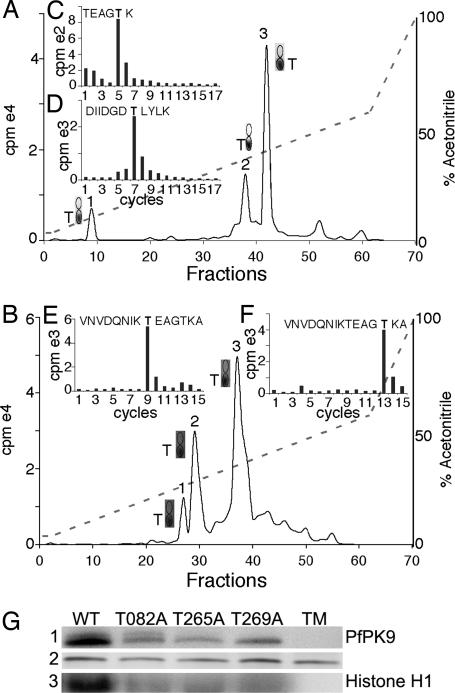
Many eukaryotic protein kinases are known to be activated by phosphorylation on important regulatory sites either by an upstream kinase or by itself. To investigate whether PfPK9 activity is likely to be regulated in vivo, we identified its autophosphorylation sites and determined their significance using a combination of phosphoamino acid analysis (PAA) and Edman cycling. PfPK9 was incubated with [γ-32P]ATP in vitro, separated on an SDS/PAGE gel, and digested with trypsin. Subsequently, the phosphotryptic peptides were resolved by reverse-phase HPLC, and radioactivity was measured for the collected fractions. Three distinct peaks were recovered (Fig. 2A) that were further subjected to both PAA and Edman degradation. PAA identified all of the peaks to be fragments containing phosphothreonines (Fig. 2A). To specifically identify the phosphorylated threonine residues, the phosphopeptides were cross-linked to Immobilon-P and subjected to sequential Edman degradation, and the released radioactivity at each cycle was measured. By using the cleaved radioactive analysis of phosphopeptides (CRP) algorithm, it was possible to identify two out of three phosphorylation sites unambiguously (21). Edman cycling of tryptic peptide 1 and 2 resulted in 32P release in cycles 5 and 7, respectively (Fig. 2 C and D). The CRP algorithm identified potential phosphorylation sites as T269 for peak 1 and T082, T147, or T158 for peak 2. To narrow the possible phosphorylation sites, an Asp-N digest was performed on the phosphorylated protein that after HPLC separation resulted in three radioactive peaks. Peak 2 processed by Edman gave a cycle 2 release, confirming T082 as the phosphorylation site (data not shown). The phospho-site on tryptic peptide 3 did not appear to be directly amenable to Edman degradation. Therefore, we digested autophosphorylated PfPK9 with chymotrypsin, resulting in the identification of three radioactive peptides (Fig. 2B). PAA confirmed that all were phosphothreonine sites (Fig. 2B). Edman cycling resulted in a cycle 9 and cycle 13 heat release for peaks 2 and 3, respectively (Fig. 2 E and F). CRP analysis confirmed the phosphorylation site as T265 (peak 2) and also reconfirmed T269 (peak 3). Peptides in peak 1 were not amenable to Edman degradation. Thus, using a combination of proteases namely trypsin, chymotrypsin, and Asp-N, we identified T082, T265, and T269 as the three autophosphorylation sites.
Fig. 2.
Identification and functional analysis of autophosphorylation sites on PfPK9. (A) [γ-32P]ATP-labeled PfPK9 was trypsin-digested and run on reverse-phase HPLC resulting in three radioactive peaks that were all phosphorylated threonines. (C) Cycle 5 32P release of tryptic peptide 1 identified T269 as a phosphorylation site. (D) Tryptic peptide 2 released cycle 7 32P, which in combination with analysis by Asp-N protease activity (data not shown) identified the phosho-T082 site. (B) [γ-32P]ATP-labeled PfPK9 chymotrypsin-digested and run on reverse-phase HPLC also resulted in three radioactive peaks corresponding to threonines. (E) Chymotryptic peptide 2 resulted in cycle 9 32P release and identified T265 as a phosphorylation site. (F) Chymotryptic peptide 3 generated a cycle 13 32P release confirming T269 as a phosphorylation site. T082, T265, and T269 were identified as the three autophosphorylation sites. (G) Mutation of the threonine sites highly reduced the autophosphorylation activity compared with the wild type (1) and abolished catalytic activity toward histone H1 (3). The triple phospho-mutant (TM) exhibits no autophosphorylation. Silver-stained protein gel of the wild-type and mutant kinases (2).
To further investigate the role of each identified phosphothreonine residue in PfPK9 activity, T-A mutations were introduced. All three mutants exhibit significantly reduced autophosphorylation in comparison to the wild-type enzyme. In the triple mutant, autophosphorylation was entirely eliminated (Fig. 2G). Catalytic activity toward histone H1 in all four phospho-mutants was also completely abolished (Fig. 2G). These findings indicate that autophosphorylation of T082, T265, and T269 positively modulates PfPK9 catalytic activity.
PfPK9 Is Expressed After Early Ring Stage in the Erythrocytic Life Cycle.
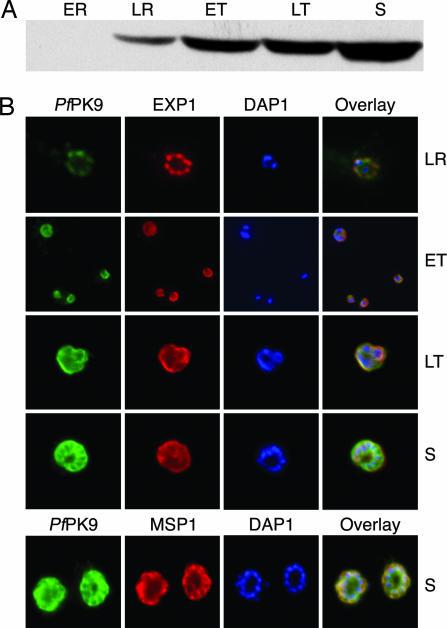
To examine its expression pattern and localization during the parasite's blood stage life cycle, we created a polyclonal antibody against full-length PfPK9. Samples were taken at different time points after RBC invasion and processed for Western blotting and immunofluorescence. PfPK9 expression was induced at the late ring stage and steadily increased up to the schizont stage (Fig. 3A). Immunofluorescence studies showed a punctuate staining in the late ring stage where PfPK9 colocalized frequently, but not exclusively, with exported protein 1 (EXP1) (Fig. 3B). EXP1 is a marker for the PVM, which forms the host–parasite interface. Membrane staining continued through early and late trophozoite stages, but additionally diffused cytoplasmic expression was also observed. Interestingly, by the schizont stage, PfPK9 did not colocalize with EXP1 anymore but colocalized significantly with the merozoite surface 1 (MSP1) protein (Fig. 3B). MSP1 is a marker for the plasma membrane a structure that exists just within the PVM. To rule out the possibility of the antibody cross-reacting with two different proteins at different life cycle stages, we preincubated the polyclonal PfPK9 antibody with recombinant PfPK9 protein before incubating with the fixed infected RBC. This treatment abolished the staining observed with the PfPK9 antibody, confirming that the immunofluorescence signal is not a result of antibody cross-reaction but is due to specific binding to the PfPK9 protein (SI Fig. 7). The observed colocalization analysis suggests that PfPK9 is initially expressed in the PVM, followed by additional scattered cytoplasmic expression, and is eventually localized in the parasite's plasma membrane.
Fig. 3.
PfPK9 expression and localization across the intraerythrocytic life cycle. (A) Immunoblot of sample extracts collected from various time points of the 48-h cycle. Equal number of parasites were collected at early ring (ER), late ring (LR), early trophozoite (ET), late trophozoite (LT), and schizont (S) stages. Protein expression steadily increases during life-cycle progression. (B) Immunofluorescence using polyclonal anti-PfPK9 antibody reveals a punctuate staining at late ring stage during which PfPK9 (green) appears to colocalize with exported protein 1 (red) in several spots. In addition to parasitophorous membrane staining (using EXP-1 as PVM marker), diffused cytoplasmic staining is observed during both early and late trophozoites. During the schizont stage, the expression pattern emerges to be like merozoite surface protein 1 (red), a plasma membrane protein.
Identification of P. falciparum Target Substrates Phosphorylated by PfPK9.
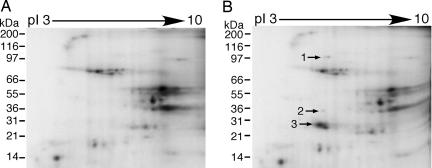
To identify endogenous substrates of PfPK9, we used a KESTREL (kinase substrate tracking and elucidation) approach (22). P. falciparum protein extract was first separated by anion exchange chromatography. The resulting fractions were used in kinase assays to identify potential endogenous PfPK9 substrates. Fractions 8 and 30 showed discrete phosphorylation events on addition of PfPK9 (SI Fig. 8B). Although separation by anion exchange chromatography before 1D electrophoresis readily separates proteins, unambiguous and reliable identification of proteins is complicated by comigration of several proteins in an apparently single band on the gel. To address this issue, 2D gel electrophoresis was used. Fractions 8 and 30 from seven identical anion exchange chromatography runs were subjected to in vitro kinase assays with PfPK9 and subsequently resolved by 2D gel electrophoresis (Fig. 4). Addition of wild-type PfPK9 increased the phosphorylation of three spots on the gel (compare Fig. 4 A and B). After the autoradiographed gel images from samples were superimposed on the corresponding silver-stained gels (SI Fig. 8 C and D), the three phosphorylated protein spots were excised, trypsin-digested, and analyzed by mass spectrometry. Spot 1 was below the threshold of detection of the instrument. Spot 2 was identified as recombinant PfPK9 protein, whereas Spot 3 was unambiguously identified as the PFE1350c ORF. A total of 80% sequence coverage by peptide mass fingerprint followed by 30% coverage by three peptides (SI Fig. 9) was attained by MS/MS spectra.
Fig. 4.
Identification of P. falciparum UBC13 homolog as a PfPK9 substrate. Fractions 8 and 30 from seven anion exchange chromatography runs were pooled together, and kinase assays were performed and resolved by 2DE. Autoradiography revealed increased phosphorylation in three spots upon addition of wild-type PfPK9 (B) compared with control (A). The proteins corresponding to the three spots were identified by mass spectrometry as PfPK9 (2) and PFE1350c (3) ORF.
PFE1350c Is a Homolog of Ubiquitin-Conjugating Enzyme 13 and a Specific Substrate of PfPK9.
The PFE1350c ORF was analyzed by using the BLASTP algorithm, which assigned the sequence to a ubiquitin-conjugating catalytic domain analogous to human UBC13 with an expectation score of 1e−51. ClustalW alignment with human UBC13 exhibited a 68% identity (SI Fig. 9C). Recombinant PfUBC13 was subjected to a kinase assay with PfPK9, and autoradiography of the reaction showed a clear phosphorylated band (Fig. 5A). To identify the phosphorylated residue, phosphorylated PfPK9 was excised from the gel and trypsin-digested, and the peptides resolved by reverse-phase HPLC. A single major peak of radioactivity was identified (Fig. 5B). Edman analysis of this peak resulted in a cycle 5 heat release (Fig. 5C), unambiguously assigning the phosphorylation site to S106.
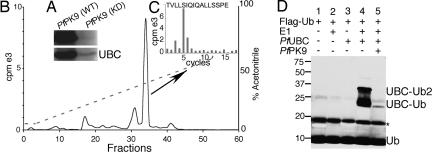
Fig. 5.
PfUBC13 is a PfPK9 substrate, and its phosphorylation suppresses the E2's ubiquitin-conjugating activity. (A) Recombinant PfUBC13, in the presence [γ-32P]ATP, was phosphorylated by wild-type PfPK9. (B) [γ-32P]ATP-labeled PfUBC13 was trypsin-digested, and radioactive peptides were resolved by HPLC, giving rise to a single peak. (C) Edman degradation produced a cycle 5 heat release, thereby unambiguously identifying the PfUBC13 phosphorylation site as Ser-106. (D) Thioester bond formation by phosphorylated PfUBC13 versus unphosphorylated form. Transfer of Flag-ubiquitin to PfUBC13 was analyzed by immunoblotting with anti-Flag monoclonal antibody. PfUBC13 preincubated with PfPK9 shows significantly reduced thioester bond formation (lane 5) compared with control (lane 4), which shows PfUBC13 conjugated to both mono- and diubiquitin. The asterisk indicates a nonspecific band.
PfPK9-Induced Phosphorylation of PfUBC13 Negatively Regulates Its Ubiquitin-Conjugating Activity.
Ubiquitin-conjugating enzymes mediate the transfer of the ubiquitin from the catalytic cysteine of a ubiquitin-activating enzyme (E1) onto a cysteine on itself through the formation of a thioester bond. PfPK9-induced phosphorylation of PfUBC13 may regulate its ubiquitin-conjugating activity. To test this possibility, we used a thioester assay to examine the transfer of ubiquitin from E1 to E2 (here PfUBC13). Because the PfUBC13 showed a high degree of identity with the human UBC13, human E1 was used to generate activated ubiquitin. E1 was first preincubated with Flag-ubiquitin and ATP to generate E1-ubiquitin thioester intermediates. In the presence of the E1 enzyme, PfUBC13 successfully formed mono- and diubiquitin thioester bonds (Fig. 5D, lane 4), indicating that it had potent ubiquitin-conjugating activity. In contrast, when PfUBC13 was phosphorylated by pretreatment with PfPK9, ubiquitin conjugation was highly suppressed (Fig. 5D, lane 5). These data demonstrate that PfPK9-induced phosphorylation is an important regulatory mechanism that determines the activity of PfUBC13.
Discussion
Although numerous phosphorylation events occur upon RBC invasion in the malarial parasite, Plasmodium, these signaling cascades have remained largely uncharacterized. Only recently, functions of certain parasite kinases have been elucidated, all of which have been in the P. berghei sexual stages. Because of its haploid nature, genetic manipulation of the parasite for functional studies on genes essential for the asexual stage is problematic. Until reliable conditional knockout strategies are developed for P. falciparum, we believe that biochemical characterization of unique Plasmodium-specific kinases and subsequent identification of their cellular targets will provide us valuable insights into the parasite's signaling pathways.
Here, we identified and characterized PfPK9, a highly conserved protein kinase among various Plasmodium species. PfPK9 exhibits 82% identity between the P. falciparum, P. vivax, and P. berghei forms. Like a number of well characterized kinases, PfPK9 activity is regulated by phosphorylation of residues within the catalytic subdomain VIII. Phosphorylation of T265 in subdomain VIII of PfPK9 (Fig. 1A) corresponds to that of T197 in protein kinase C-α, both of which are essential for catalytic activity of each kinase (23). Interestingly, a second threonine residue in the catalytic domain of PfPK9, T269, was also autophosphorylated (Fig. 1A). Although this residue is highly conserved in multiple protein kinases, we report functional phosphorylation at this site. Phosphorylation of both T265 and T269 was required for optimal catalytic activity (Fig. 2G). Finally, we identified a third phosphorylated threonine residue, T082. The T082 residue lies outside of the catalytic domain and is in fact part of a putative forkhead-associated domain (FHA) interaction motif. FHA, a phosphopeptide binding module, directly affects signal transduction events by mediating protein–protein interactions (24). Therefore, T082 phosphorylation could result in binding of PfPK9 to an FHA protein, thereby targeting the kinase to its substrates. Thus, autophosphorylation of multiple residues in PfPK9 may regulate kinase activity both temporally and spatially.
PfPK9 expression is induced in the late ring stage and continues to be expressed well into the schizont stage of the asexual life cycle where it exhibits maximal expression. The kinase initially colocalized with EXP-1, a PVM protein (Fig. 3B). In the intraerythrocytic stage, the PVM separates the parasite from the RBC cytosol and forms the host–parasite interface. Not only does the PVM contain small pores that allow transfer of low-molecular-weight solutes, it is also involved in macromolecular transport. Proteins localized to this membrane may also play a role in the sensing and transduction of signals from the host cytosol into the parasite. Two kinases are known to be secreted from the parasite cytosol: PfFEST localizes to the knobs under the RBC membrane (25), and calcium-dependent protein kinase 1 (CDPK1) is exported to the PVM (26). Although functional roles for these kinases have not been established, localization of these proteins at the two boundaries (RBC membrane and PVM) could facilitate coordination of phosphorylation based signaling events between host and parasite environments. Localization of PfPK9 to the PVM suggests that it functions as a receptor-kinase or mediator to transduce signal from the intraerythrocytic environment to the parasite cytosol. Interestingly, as the parasite matures into schizonts, there is a shift in the localization of PfPK9. In this stage, it localizes to the parasite's plasma membrane (Fig. 3B). This suggests that PfPK9 function is required in different cellular locations at different times during development. These findings provide evidence for spatial regulation by localization of PfPK9.
Identification of endogenous substrates can help to establish a potential physiological role for PfPK9 in the parasite. We adopted a KESTREL (kinase substrate tracking and elucidation) approach to identify PfPK9's downstream cellular targets. A putative PfUBC13 was unambiguously identified as a PfPK9 substrate (Fig. 5A). UBC13 is a highly conserved enzyme in eukaryotes and functions in a complex with ubiquitin E2 variant (UEV) protein. The UBC13-UEV heterodimer mediates assembly of K63-linked polyubiquitin chains on target proteins (27). Unlike the K46-linked polyubiquitin chains, which are a signal for 26S proteasome-mediated degradation, K63-linked chains mediate nonproteolytic pathways (28). UBC13's conjugating activity modulates various cellular processes, including DNA repair (29), mitotic progression (30), tumor suppressor p53 activity (31), and immune receptor signaling (32).
A few studies have demonstrated the regulation of E2 activity by phosphorylation. In yeast, cyclin-dependent kinase 1 and 2 phosphorylate UBC2 and increase its ubiquitin-conjugating activity 4-fold (20). Moreover, phosphorylated UBC6e has a reduced thioester-forming capacity in mammalian cell lines (19). Here, we show that PfPK9 phosphorylated PfUBC13 at residue S106 (Fig. 5C). S106 lies in a highly conserved region among E2s, and more importantly, it is a surface residue of helix 3, a region forming the binding site of the donor ubiquitin (33, 34). Phosphorylation of this residue could interfere with binding of ubiquitin to UBC13 and prevent thioester bond formation. Our results indicate that PfPK9 phosphorylation of PfUBC13 strongly suppressed its E2 activity. Consequently, PfPK9 may be an important regulator of the parasite's K63 ubiquitin-conjugating pathways. Because the S106 residue is conserved in a number of organisms including yeast and humans, it leads to the exciting possibility that phosphorylation-mediated modulation of UBC13 activity is a conserved regulatory mechanism in other species.
In summary, we showed that PfPK9 expression is developmentally controlled and its activity regulated by autophosphorylation of specific threonine residues. PfPK9 localization changed through the intraerythrocytic life cycle from the PVM to the plasma membrane, suggesting that it exerts its function at distinct cellular locations during development. Moreover, PfPK9-induced phosphorylation of PfUBC13 regulated its E2 activity, implying that PfPK9 can regulate a wide variety of cellular processes, such as DNA repair or cell cycling. Apart from the recent characterization of a deubiquitinating/deNeddylating enzyme (35), enzymes involved in ubiquitination have not been described in the malarial parasite. Not only have we characterized a P. falciparum kinase, but we also demonstrate the presence of an active parasite E2 enzyme and its regulation by phosphorylation.
Materials and Methods
P. falciparum Cultures.
P. falciparum 3D7 strain was cultured according to previously published methods (36).
Molecular Cloning of Wild-Type and Mutant PfPK9 cDNAs.
Total RNA was isolated from asynchronous P. falciparum 3D7 cultures, and reverse transcription was performed by using oligo(dT)s. The resulting cDNA was used as a template in PCRs to generate full-length PfPK9 (PF13_0085 ORF). The primer sets used for cloning PfPK9 gene were 5′-CGGGATCCATGTATTTGGGAAGTATAAAAAATTTCGAAG-3′ (forward) and 5′-GGAATTCTTATGACTCCTTATAATTTTCAAACCACC-3′ (reverse), and the amplified product was inserted into pGEX6P.1 vector. All site-directed mutagenesis was performed by using the QuikChange kit with the following primers: 5′-CGATAAAAGATATAATAGATGGTGATGCCTTATATTTGAAAGAAAAGAAAGAAG-3′ and the reverse complement (T082A), 5′-GATCAAAATATTAAAGCAGAAGCAGGAACAAAAGCATATATAGCACCAGAAATC-3′ and the reverse complement (T265A), and 5′-GATCAAAATATTAAGCAGAAGCAGGAGCAAAAGCATATATAGCACCAGAAATC-3′ and the reverse complement (T269A). PfUBC13 was cloned to the ORF PFE1350c by using the following primers: 5′-CGGGATCCATGTCAATACCTAGAAGAATAACAAAAGAAACACAAAACTTAGCG-3′ (forward) and 5′-GGAATTCTTATAAAACATTATTATTTGCATAAATTTTATTCCATTGTCTCGCAACATGTTCG-3′ (reverse), and the amplified product was inserted into the pGEX6P.1 vector.
Expression and Purification of Recombinant PfPK9 and PfUBC13.
All recombinant protein expression vectors containing the relevant genes were transformed into Rosetta-Lys strain. Cultures were grown in LB media containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol, induced with 0.1 mM IPTG at 18°C for 14 h. Bacterial lysates were incubated with glutathione-Sepharose resin, and GST-PfPK9 was finally cleaved with precession protease overnight.
Assay of PfPK9 Catalytic Activity.
Kinase assays were performed in 30-μl reactions in buffer containing 50 mM Hepes (pH 7.5), 10 mM MgCl2, 100 μM ATP, [γ-32P]ATP (specific activity of ≈7,500 cpm/pmol), and 1 μg of substrate [myelin basic protein, histone H1, histone H2A, casein (all bovine), or PfUBC13] incubated with 0.1 μg of PfPK9. The samples were resolved by SDS/PAGE, silver-stained, dried, and visualized by autoradiography.
Autophosphorylation and Identification of Phosphorylation Sites.
Recombinant PfPK9 was incubated in kinase reaction buffer as described above with 50 μM ATP and [γ-32P]ATP (10,000 cpm/nmol) for 30 min at 30°C. Autophosphorylated kinase was resolved by SDS/PAGE, visualized by silver staining, and in gel-digested with trypsin, chymotrypsin, or Asp-N (1.5 μg) overnight at 37°C. The protein digests were processed for phospho-site mapping as described (37).
Generation of Anti-PfPK9 Serum, Western Blotting, and Immunofluorescence.
PfPK9 antibodies were generated in rabbits by using purified recombinant PfPK9 (Proteintech, Chicago, IL). Parasites were isolated from infected erythrocytes by 0.1% saponin treatment. Extracts from specific parasite stages from sorbitol synchronized cultures were subjected to SDS/PAGE, transferred to nitrocellulose membrane, and probed with anti-PfPK9 antibody. Subsequently, the membrane was incubated with anti-rabbit IgG-HRP and finally developed with ECL substrate.
For immunofluorescence microscopy, infected erythrocytes were washed twice with PBS, fixed in 4% paraformaldehyde, and spotted on poly(lysine)-coated slides for 6 min. The cells were permeabilized with 0.1% Triton X-100 for 6 min, washed with PBS, and blocked for 1 h at room temperature. Polyclonal anti-PfPK9 (1:100 dilution) and monoclonal anti-EXP1 (1:1,000) and MSP1 (1:500) were added for 2 h at room temperature or overnight at 4°C. Anti-mouse IgG conjugated to rhodamine red and anti-rabbit IgG conjugated to FITC were added for 1 h at room temperature. Nuclei were stained with 1 μg/ml DAPI for 30 s. The slides were mounted in 50% (vol/vol) glycerol and 2.5% (wt/vol) triethylenediamine, and imaged by using a Zeiss Axio Imager wide-field epifluorescence microscope.
Substrate Identification and Validation.
Protein lysate from parasite pellet was fractionated by anion-exchange chromatography using a SMART Mono Q PC 1.6/5 column (Amersham Biosciences). Fifty-microliter fractions were collected over a 0–1 M NaCl gradient. Kinase assays were performed on 15 μl of the fractions in the presence of wild-type or kinase-dead forms of PfPK9 in 30-μl reactions as described previously. The samples were resolved by SDS/15% PAGE gel, silver-stained, dried, visualized by autoradiography, and analyzed for differences in phosphorylation (SI Fig. 8 A and B). Additionally, fractions 8 and 30 from seven anion exchange runs were pooled and concentrated in Ultrafree-0.5 centrifugal filter devices (Millipore). One hundred-microliter kinase reactions were performed with 1 μg of recombinant PfPK9. The reactions were eluted with 8 M urea and 1% Triton X-100 sample buffer and resolved by 2D electrophoresis (pH gradient 3–10). The gels were silver-stained, dried, laid on film, and analyzed for increased phosphorylation.
Silver-stained spots corresponding to increased phosphorylation were cut out and in gel-digested with trypsin, and the peptides were extracted by using previously developed methods (38). The mass spectra of the tryptic digests were generated on an ABI 4700 MALDI-TOF/TOF mass spectrometer (Applied Biosystems). The combined peptide mass fingerprint and MS/MS spectra were used to search an internal P. falciparum database (from PlasmoDb) with GPS Explorer software (Applied Biosystems).
Ubiquitin-Conjugating Assays.
Recombinant PfUBC13 was first phosphorylated by PfPK9 and then assayed for its ubiquitin-conjugating activity. Phosphorylation was initiated by incubating 30 mM PfUBC13 in a 30-μl reaction consisting of 20 mM Hepes (pH 7.5), 10 mM MgCl2, and 1 mM ATP in the presence of 0.1 μg of wild-type or kinase-dead PfPK9 for 30 min at 30°C. Two microliters of this solution containing PfUBC13 was removed and used for ubiquitination reactions. Purified 100 nM human E1 was preincubated with 1 mM Flag-ubiquitin in 20 mM Tris·HCl (pH 7.5), 10 mM MgCl2, and 1 mM ATP for 10 min. Two microliters of the PfUBC13 solution was added, and the reaction was continued for 20 min. The reaction was stopped with nonreducing Laemmli sample buffer and resolved by 10–20% gradient SDS/PAGE. The proteins were transferred to nitrocellulose membrane and probed with a 1:1,000 dilution of monoclonal anti-Flag antibody. Subsequently, the membrane was incubated with anti-mouse IgG-HRP and finally developed with ECL substrate.
Acknowledgments
We thank Steven Spoel and Laura Hagerty for critical reading of the manuscript, Elizabeth Snyder for help in the preparation of the manuscript, Dr. Jana McBride and the Malaria Research and Reference Reagent Resource Center for EXP1 and MSP1 antibodies, and the researchers involved in maintaining the PlasmoDB database for their contributions.
Abbreviations
- PAA
phosphoamino acid analysis
- PVM
parasitophorous vacuolar membrane
- RBC
red blood cell.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611601104/DC1.
References
- 1.Ridley RG. Nature. 2002;415:686–693. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- 2.Magowan C, Liang J, Yeung J, Takakuwa Y, Coppel RL, Mohandas N. Exp Parasitol. 1998;89:40–49. doi: 10.1006/expr.1998.4261. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Franklin RM, Kappes B. Mol Biochem Parasitol. 1994;66:329–343. doi: 10.1016/0166-6851(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P. Nat Rev Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 5.Dluzewski AR, Garcia CR. Experientia. 1996;52:621–623. doi: 10.1007/BF01969742. [DOI] [PubMed] [Google Scholar]
- 6.Geyer JA, Prigge ST, Waters NC. Biochim Biophys Acta. 2005;1754:160–170. doi: 10.1016/j.bbapap.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Harmse L, van Zyl R, Gray N, Schultz P, Leclerc S, Meijer L, Doerig C, Havlik I. Biochem Pharmacol. 2001;62:341–348. doi: 10.1016/s0006-2952(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 8.Waters NC, Geyer JA. Exp Opin Ther Targets. 2003;7:7–17. doi: 10.1517/14728222.7.1.7. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Z, Waters NC, Woodard CL, Li Z, Li PK. Bioorg Med Chem Lett. 2001;11:2875–2878. doi: 10.1016/s0960-894x(01)00578-9. [DOI] [PubMed] [Google Scholar]
- 10.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anamika , Srinivasan N, Krupa A. Proteins. 2005;58:180–189. doi: 10.1002/prot.20278. [DOI] [PubMed] [Google Scholar]
- 12.Ward P, Equinet L, Packer J, Doerig C. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. Cell. 2004;117:503–514. doi: 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 14.Khan SM, Franke-Fayard B, Mair GR, Lasonder E, Janse CJ, Mann M, Waters AP. Cell. 2005;121:675–687. doi: 10.1016/j.cell.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Vaid A, Syin C, Sharma P. J Biol Chem. 2004;279:24255–24264. doi: 10.1074/jbc.M312855200. [DOI] [PubMed] [Google Scholar]
- 16.Merckx A, Le Roch K, Nivez MP, Dorin D, Alano P, Gutierrez GJ, Nebreda AR, Goldring D, Whittle C, Patterson S, et al. J Biol Chem. 2003;278:39839–39850. doi: 10.1074/jbc.M301625200. [DOI] [PubMed] [Google Scholar]
- 17.Reininger L, Billker O, Tewari R, Mukhopadhyay A, Fennell C, Dorin-Semblat D, Doerig C, Goldring D, Harmse L, Ranford-Cartwright L, et al. J Biol Chem. 2005;280:31957–31964. doi: 10.1074/jbc.M504523200. [DOI] [PubMed] [Google Scholar]
- 18.Tewari R, Dorin D, Moon R, Doerig C, Billker O. Mol Microbiol. 2005;58:1253–1263. doi: 10.1111/j.1365-2958.2005.04793.x. [DOI] [PubMed] [Google Scholar]
- 19.Oh RS, Bai X, Rommens JM. J Biol Chem. 2006;281:21480–21490. doi: 10.1074/jbc.M601843200. [DOI] [PubMed] [Google Scholar]
- 20.Sarcevic B, Mawson A, Baker RT, Sutherland RL. EMBO J. 2002;21:2009–2018. doi: 10.1093/emboj/21.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackey AJ, Haystead TA, Pearson WR. Nucleic Acids Res. 2003;31:3859–3861. doi: 10.1093/nar/gkg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knebel A, Morrice N, Cohen P. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cazaubon SM, Parker PJ. J Biol Chem. 1993;268:17559–17563. [PubMed] [Google Scholar]
- 24.Hofmann K, Bucher P. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 25.Kun JF, Hibbs AR, Saul A, McColl DJ, Coppel RL, Anders RF. Mol Biochem Parasitol. 1997;85:41–51. doi: 10.1016/s0166-6851(96)02805-8. [DOI] [PubMed] [Google Scholar]
- 26.Moskes C, Burghaus PA, Wernli B, Sauder U, Durrenberger M, Kappes B. Mol Microbiol. 2004;54:676–691. doi: 10.1111/j.1365-2958.2004.04313.x. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann RM, Pickart CM. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 28.Pickart CM. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 29.Brusky J, Zhu Y, Xiao W. Curr Genet. 2000;37:168–174. doi: 10.1007/s002940050515. [DOI] [PubMed] [Google Scholar]
- 30.Bothos J, Summers MK, Venere M, Scolnick DM, Halazonetis TD. Oncogene. 2003;22:7101–7107. doi: 10.1038/sj.onc.1206831. [DOI] [PubMed] [Google Scholar]
- 31.Laine A, Topisirovic I, Zhai D, Reed JC, Borden KL, Ronai Z. Mol Cell Biol. 2006;26:8901–8913. doi: 10.1128/MCB.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, et al. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 33.Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Nat Struct Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 34.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 35.Artavanis-Tsakonas K, Misaghi S, Comeaux CA, Catic A, Spooner E, Duraisingh MT, Ploegh HL. Mol Microbiol. 2006;61:1187–1195. doi: 10.1111/j.1365-2958.2006.05307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trager W, Jensen JB. Bull WHO. 1977;55:363–365. [PMC free article] [PubMed] [Google Scholar]
- 37.Graves PR, Winkfield KM, Haystead TA. J Biol Chem. 2005;280:9363–9374. doi: 10.1074/jbc.M412538200. [DOI] [PubMed] [Google Scholar]
- 38.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]