Abstract
Selenoproteins are an elite group of proteins containing a rare amino acid, selenocysteine (Sec), encoded by the codon, UGA. In eukaryotes, incorporation of Sec requires a Sec insertion sequence (SECIS) element, a stem–loop structure located in the 3′-untranslated regions of selenoprotein mRNAs. Here we report identification of a noncanonical form of SECIS element in Toxoplasma gondii and Neospora canine, single-celled apicomplexan parasites of humans and domestic animals. This SECIS has a GGGA sequence in the SBP2-binding site in place of AUGA previously considered invariant. Using a combination of computational and molecular techniques, we show that Toxoplasma and Neospora possess both canonical and noncanonical SECIS elements. The GGGA-type SECIS element supported Sec insertion in mammalian HEK 293 and NIH 3T3 cells and did so more efficiently than the natural mammalian SECIS elements tested. In addition, mammalian type I and type II SECIS elements mutated into the GGGA forms were functional but manifested decreased Sec insertion efficiency. We carried out computational searches for both AUGA and GGGA forms of SECIS elements in Toxoplasma and detected five selenoprotein genes, including one coding for a previously undescribed selenoprotein, designated SelQ, and two containing the GGGA form of the SECIS element. In contrast, the GGGA-type SECIS elements were not detected in mammals and nematodes. As a practical outcome of the study, we developed pSelExpress1, a vector for convenient expression of selenoproteins in mammalian cells. It contains an SBP2 gene and the most efficient tested SECIS element: an AUGA mutant of the GGGA-type Toxoplasma SelT structure.
Keywords: genome, RNA structure, selenocysteine, selenoprotein
Selenocysteine (Sec)-containing proteins (selenoproteins) are rare but widely distributed in all domains of life (1), including bacteria (2, 3), archaea (4), and eukaryotes (5–7). The human genome possesses 25 genes encoding such proteins (7). The class of selenoproteins is defined by the occurrence of Sec, the 21st amino acid encoded by the UGA codon. Selenoproteins use the high reactivity of Sec, which is located in catalytic centers and serves redox function analogous to the functions of redox-active Cys residues (8). In addition to the UGA codon, a cis-acting element is present within selenoprotein genes, which is also essential for recognition of UGA as the Sec codon. This element is a stem–loop structure, known as the Sec insertion sequence (SECIS) located in coding regions of bacterial and in 3′-UTRs of archaeal and eukaryotic selenoprotein genes (9, 10).
The principal features of the eukaryotic SECIS element include a segment containing four non-Watson–Crick base pairs UGAN…. NGAN (designated the quartet or core), an unpaired A preceding the quartet, and an unpaired AA or CC motif in the apical loop or bulge that is separated from the quartet by 11–12 nucleotides (11–14). Although having low sequence conservation, the secondary structure of eukaryotic SECIS elements is strictly conserved and thermodynamically stable (15, 16). Based on these observations, several algorithms have been developed and successfully applied in genomic searches to identify these stem–loop structures, and thereby selenoprotein genes, in nucleotide sequence databases (5, 7, 17–19).
It has been established that the quartet is involved in the interaction with SECIS-binding protein 2 (SBP2) (20, 21), which, in turn, is essential for the formation of a complex with the Sec-specific elongation factor, known as EFsec, and tRNA[Ser]Sec (22, 23). This protein–RNA complex functions by inserting Sec in response to UGA codons (24). Recently, additional components of the complex were reported, including the L30 protein that interacts with the SECIS element in vitro and in vivo and competes with SBP2 for SECIS binding (25). In addition, SECp43 and SLA have roles in selenoprotein biosynthesis through interaction with tRNA[Ser]Sec in a supramolecular complex (26, 27).
It has been found that the four consecutive non-Watson–Crick base pairs are part of a recurrent motif, the kink-turn (K-turn), and constitute the functional motif of the SECIS structure, which is recognized by SBP2. It has been proposed that the invariant U in the conserved AUGA sequence forms a non-Watson–Crick base pair with another U in 5′DI SECIS RNA (12, 28). Similarly, it is thought this U may form a base pair with the corresponding nucleotide independent of its identity. The analysis of crystal structures of RNA–protein complexes suggested the residue at this position binds in a pocket of RNA-binding proteins (29–31). It was also proposed that the SECIS core is a K-turn-like motif, which provides greater flexibility and allows switching between open and closed kinked conformations. In turn, this feature triggers a major conformational change in the SBP2-bound complex (25, 28).
In our study, we found that the U in the quartet, previously considered an invariant residue, is not conserved in some apicomplexan selenoprotein genes. Instead, a previously undescribed form of SECIS element was found that has a GGGA sequence in place of the conserved AUGA sequence. We characterized this structure in detail using a combination of computational and molecular approaches.
Results and Discussion
Identification of a Noncanonical Form of Eukaryotic SECIS Element.
A search for Toxoplasma selenoprotein genes was carried out by homology analyses involving all known selenoproteins as queries. This procedure identified homologs of four mammalian selenoproteins: SelW, SelK, SelS, and SelT [supporting information (SI) Figs. 6–9]. Their genes had predicted Sec residues encoded by UGA codons. Analysis of the 3′-UTRs in these selenoprotein genes revealed the presence of canonical SECIS elements in SelK and SelW genes (Fig. 1A). However, no suitable structure was found in the SelT 3′-UTR. The use of relaxed settings and the loose pattern of SECISearch did not yield candidate SECIS structures in the Toxoplasma SelT gene.
Fig. 1.
SECIS elements identified in Toxoplasma and Neospora. (A) Canonical (shown in white background) and the GGGA-type (shown in gray background) SECIS elements identified in T. gondii and N. caninum are shown. The SECIS core and the unpaired AA nucleotides in the apical loop are shown in bold. (B) Selenoprotein Q (SelQ). EST sequences (GenBank accession nos. CN615432.1 and CF268978.1) were used for sequence reconstruction. Locations of the initiator AUG codon, Sec-encoding UGA codon, stop signal, and the SECIS element are indicated.
The lack of a standard SECIS element in the SelT gene suggested the presence of a noncanonical structure. Manual analysis of the SelT 3′-UTR using MFOLD revealed a SECIS-like structure that satisfied all SECIS element requirements with one notable exception: the SBP2-binding site (e.g., SECIS core or quartet) had a GGGA sequence instead of AUGA (Fig. 1A). The U in the AUGA sequence is considered invariant and is present in all known eukaryotic SECIS elements. To examine whether the GGGA sequence in the SECIS core represented a sequencing error, we analyzed additional protozoan sequences. EST sequences of Neospora caninum, another apicomplexan parasite, revealed a SelW homolog containing a canonical SECIS element and a SelT homolog containing a GGGA-type SECIS element (Fig. 1A). The occurrence of the same noncanonical SECIS-like structure in two different organisms was a strong indication that this structure is the true SECIS element.
The New Type of SECIS Element Is Functional.
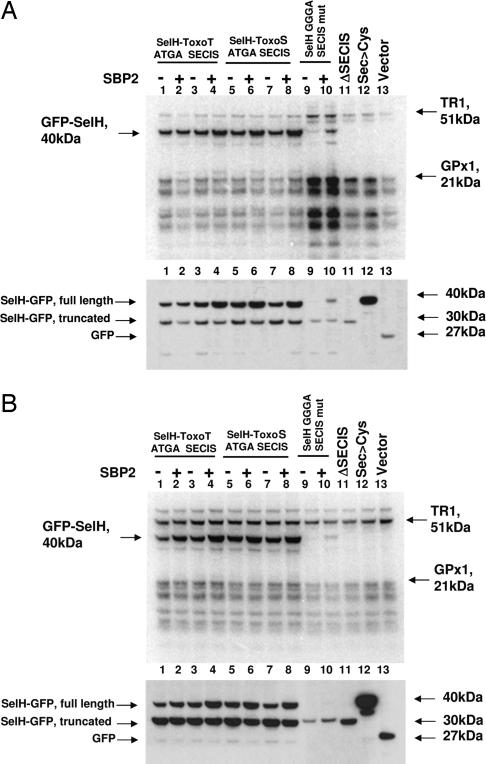
We prepared the constructs coding for GFP SelH fusion proteins, in which the natural SelH SECIS element was replaced with Toxoplasma SelT or SelS SECIS elements (Fig. 2A) and expressed these proteins in mammalian HEK 293 (Fig. 2B) and NIH 3T3 (Fig. 2C) cells. Expression of the fusion protein was predicted to result in an ≈40-kDa product (Fig. 2A). Indeed, metabolic labeling of the transfected cells with 75Se revealed a 40-kDa band (lanes 1–8, Fig. 2 B Upper and C Upper). This band was not present in cells transfected with the corresponding constructs lacking 3′UTRs (lanes 11, Fig. 2 B and C) or the constructs in which the Sec-encoding codons were mutated to cysteine codons (lanes 12, Fig. 2 B and C). We also examined whether SBP2 could influence expression levels of the expressed selenoprotein by cotransfecting with an SBP2 construct. In each case, SBP2 increased efficiency of Sec insertion (i.e., the 40-kDa selenoprotein band appeared to be more enriched). Thus, the new type of SECIS element is not only functional, but its function could be stimulated by mammalian SBP2. Moreover, when certain constructs were used, the GGGA form of SECIS element appeared to be more efficient than the natural mouse SelH element (e.g., compare lanes 1–4 and 9 and 10, Fig. 2 B and C).
Fig. 2.
The GGGA form Toxoplasma SECIS element supports Sec insertion in mammalian cells. (A) A scheme illustrating GFP-fusion constructs and cloning strategies used in the study, as described in detail in Materials and Methods. Predicted sizes of GFP-SelH fusion proteins are displayed at the top. Mouse SelH:Toxoplasma SECIS chimeras were generated by cloning the corresponding GGGA forms of Toxoplasma sequences immediately downstream of the SelH stop codon (into construct 2 in the scheme). Distances between stop codons and SECIS elements for WT SelH and Toxoplasma SelT and SelS SECIS elements are shown. Short versions of fusions were designated as “SECIS” and long as “3′UTR.” In B, HEK 293 cells and in C, NIH 3T3 cells were transfected with the constructs shown in A or cotransfected with the SBP2 construct as indicated; lanes 1 and 2 correspond to construct 3 in the scheme in A (GFP-mSelH-Toxoplasma SelT GGGA-type SECIS); lanes 3 and 4 correspond to construct 4 (GFP-mSelH-Toxoplasma SelT GGGA-type 3′UTR); lanes 5 and 6 correspond to construct 5 (GFP-mSelH-Toxoplasma SelS GGGA-type SECIS); lanes 7 and 8 correspond to construct 6 (GFP-mSelH-Toxoplasma SelS GGGA-type 3′UTR); lanes 9 and 10 correspond to construct 1 (GFP-mSelH); lane 11 corresponds to construct 2 (GFP-mSelHΔSECIS); lane 12 corresponds to GFP-mSelHSec>Cys; and lane 13 corresponds to GFP (control). Cells were labeled with 75Se. (Upper) Selenoprotein patterns on SDS/PAGE gels. Migration of major endogenous selenoproteins, thioredoxin reductase 1 (TR1) and glutathione peroxidase 1 (GPx1) is shown on the right. (Lower) Western blots of the same samples probed with GFP antibodies. The bands corresponding to GFP-SelH fusions are indicated on the left and their sizes on the right.
The New Type of SECIS Element Is Highly Efficient in Sec Insertion.
Another way to monitor efficiency of Sec insertion is to probe lysates of transfected cells in Western blot assays with anti-GFP antibodies to determine the ratio between full-length and truncated forms of the fusion protein (Fig. 2 B Lower and C Lower). The truncated form is generated by termination of protein synthesis at the UGA codon because of competition of Sec insertion and translation termination, whereas the full-length protein is made when UGA is read as the Sec codon, and translation continues until the true stop signal. The ratio of full-length and truncated forms of fusion proteins that resulted from transfections with various GFP-SelH chimeras differed in the cell lines used in the study. In HEK 293 cells, the full-length form was predominant, whereas in NIH 3T3 cells, the truncated form was generally more abundant, suggesting lower efficiency of Sec incorporation in NIH 3T3 cells under the conditions used in our study. Quantification of the ratio of full-length and truncated forms (SI Fig. 10) revealed that the abundance of the full-length protein expressed from the constructs carrying Toxoplasma SECIS elements was comparable to that containing a natural SelH SECIS element. In some cases (e.g., Toxoplasma SelT 3′UTR construct; see lane 4), the full-length protein was both the major selenoprotein in HEK 293 cells and significantly exceeded the corresponding truncated form of protein. Thus, the GGGA-type of SECIS element not only is functional but also is extremely efficient in Sec insertion in mammalian cells.
AUGA to GGGA Mutants of Mammalian SECIS Elements of Both Type I and II Are Functional.
To further characterize the GGGA form of the SECIS element, we tested whether mammalian SECIS elements are functional if they contain these structures. In this experiment, we used GFP-SelS (7) and GFP-SelM (13) constructs, in which we mutated the natural AUGA forms of SECIS elements to the GGGA forms (Fig. 3A), and these were transfected into HEK 293 (Fig. 3B) and NIH 3T3 (Fig. 3C) cells. We found the mutant forms were characterized by significantly decreased Sec insertion (compare lanes 1–2 to 3–4 for SelM and lanes 6–7 to 8–9 for SelS). We also mutated the mouse SelH SECIS to the GGGA form and transfected cells with this construct (compare lanes 9 and 10 in Fig. 3 B and C to lanes 9 and 10 in Fig. 4 A and B). Again, the mutant SECIS forms were less efficient in supporting Sec incorporation. Nevertheless, these structures were functional and stimulated by SBP2. The mammalian SelM SECIS element may also be viewed as noncanonical, because it has a CC sequence in the apical bulge that corresponds to the AA sequence in almost all other eukaryotic SECIS elements. Finally, SelH on one side and SelS and SelM on the other represent types I and II SECIS elements, respectively, which differ by the presence of an additional mini helix (32). It is clear that both of these SECIS types can use the GGGA form of SECIS element.
Fig. 3.
GGGA mutant forms of mammalian SECIS elements are functional. (A) Mammalian SECIS elements used in the study that represent three known types of eukaryotic SECIS elements. Mutations made in the structures are indicated. In B, HEK 293 cells and in C, NIH 3T3 cells were transfected with the following constructs: lane 1, GFP-mSelM (WT); lane 2, GFP-mSelM (WT) + SBP2; lane 3, GFP-SelM AT>GG SECIS mutant; lane 4, GFP-SelM AT>GG SECIS mutant + SBP2; lane 5, GFP (control); lane 6, GFP-mSelS (WT); lane 7, GFP-mSelS (WT) + SBP2; lane 8, GFP-mSelS AT>GG SECIS mutant; lane 9, GFP-mSelS AT>GG SECIS mutant + SBP2; and lane 10, GFP (control). Cells were labeled with 75Se. Migration of proteins expressed from the constructs and major endogenous selenoproteins are indicated.
Fig. 4.
The ATGA form of the GGGA-type Toxoplasma SECIS element is highly efficient in Sec insertion. In A, HEK 293 cells and in B, NIH 3T3 cells were transfected with the following constructs: lane 1, GFP-mSelH-Toxoplasma SelT GG>AT SECIS mutant; lane 2, GFP-mSelH-Toxoplasma SelT GG>AT SECIS mutant + SBP2; lane 3, GFP-mSelH-Toxoplasma SelT GG>AT 3′UTR mutant; lane 4, GFP-mSelH-Toxoplasma SelT GG>AT 3′UTR mutant + SBP2; lane 5, GFP-mSelH-Toxoplasma SelS GG>AT SECIS mutant; lane 6, GFP-mSelH-Toxoplasma SelS GG>AT SECIS mutant + SBP2; lane 7, GFP-mSelH-Toxoplasma SelS GG>AT 3′UTR mutant; lane 8, GFP-mSelH-Toxoplasma SelS GG>AT 3′UTR mutant + SBP2; lane 9, GFP-mSelH (WT); lane 10, GFP-mSelH (WT) + SBP2; lane 11, GFP-mSelHΔSECIS; lane 12, GFP-mSelH Sec>Cys; and lane 13, GFP (control). Upper represent selenoprotein patterns based on metabolic labeling of cells with 75Se. Lower shows Western blots developed with anti-GFP antibodies.
Computational Searches for Canonical and Noncanonical SECIS Elements in Apicomplexa.
Demonstration of the function of the GGGA-type SECIS elements raised the possibility that such structures might occur in various organisms, and that they might have been missed in the previous genomic searches. To address this possibility, we carried out computational analyses of genomic sequences for both canonical and noncanonical SECIS elements. In apicomplexans that contain both AUGA and GGGA forms of SECIS elements, we identified five selenoproteins in Toxoplasma gondii and three in N. caninum. SelT, SelW, and SelS-like proteins occurred in both organisms and corresponded to known selenoproteins. SelK was detected only in Toxoplasma. However, the complete genome is available for T. gondii but not for N. caninum, which likely has additional selenoprotein genes.
In addition, we identified a previously undescribed selenoprotein gene, which we designate here as SelQ (Fig. 1B). This protein was detected in T. gondii, but its homologs were not detected in other organisms. In the SelQ sequence, Sec was a C-terminal penultimate residue, and it was followed with a C-terminal cysteine. This Sec location is similar to that in thioredoxin reductases, SelS and SelK, but none of these other proteins has a C-terminal cysteine.
GGGA-Type SECIS Elements Are Restricted to Apicomplexa.
We tested whether the GGGA-type SECIS elements occur in nematodes and mammals. In the nematode searches, we analyzed Caenorhabditis elegans and Caenorhabditis briggsae genomes (SI Fig. 11). A similar procedure using the AUGA pattern detected a single selenoprotein gene in these organisms that codes for thioredoxin reductase, but the GGGA pattern detected no selenoprotein genes. Similarly, the search of human and mouse genomes identified known selenoprotein genes, but SECIS elements of the GGGA-type were not detected (SI Fig. 12). Finally, we scanned the entire National Center for Biotechnology Information EST database for GGGA-type SECIS elements (SI Fig. 13), but this search did not identify selenoprotein genes besides the Toxoplasma and Neospora selenoproteins described above. Thus, it appears that the GGGA-type SECIS elements are restricted to apicomplexa.
The GGGA-Type Toxoplasma SECIS Element Is Highly Efficient.
As discussed above, the Toxoplasma SelT and SelS SECIS elements were found to be highly efficient in Sec insertion in mammalian cells; in addition, comparison of AUGA and GGGA forms of mammalian SECIS elements revealed that the AUGA forms were more efficient. Thus, we hypothesized that the AUGA form of the Toxoplasma SelT and SelS elements could be even more efficient in Sec insertion. To functionally characterize such forms of Toxoplasma SelT and SelS SECIS elements, we transfected HEK 293 (Fig. 4A) and NIH 3T3 (Fig. 4B) cells with various GFP-SelH constructs and metabolically labeled these cells with 75Se. The expected 40-kDa selenoprotein band was detected (lanes 1–10, Fig. 4 A Upper and B Upper). Once again, for all constructs, cotransfection with SBP2 increased Sec insertion (analyzed by abundance of the 75Se-labeled form and the ratio of full-length and truncated forms; Fig. 4 A Lower and B Lower). Quantification of the bands (SI Fig. 14) revealed that the most efficient Sec insertion occurred in the case of the construct containing the GGGA to ATGA mutant of Toxoplasma SelT SECIS element (lanes 1–4, Fig. 4 A and B).
Development of a Vector for Overexpression of Selenoproteins in Mammalian Cells.
Expression of Sec-containing proteins in mammalian cells, especially at high levels, is challenging. We took advantage of the efficient Sec insertion by the ATGA form of the Toxoplasma SelT SECIS element, as well as of the dependence of this process on SPB2, to develop a vector for overexpression of selenoproteins in mammalian cells. For this purpose, we used pBudCE4.1 vector, designed for simultaneous expression of two genes. This vector contains the human CMV immediate-early promoter and the human elongation factor 1α-subunit (EF-1α) promoter for high level, constitutive expression of recombinant proteins. We cloned a C-terminal functional domain of rat SBP2 for expression under the EF-1α promoter and the AUGA form of Toxoplasma SelT SECIS element into the second cloning site for expression of a selenoprotein under the CMV promoter (Fig. 5A). The resulting expression vector was designated as pSelExpress1. To test this vector for selenoprotein expression, we cloned the mouse GPx1 ORF containing an N-terminal His-tag into pSelExpress1 and separately into the corresponding vector lacking the SBP2 gene. HEK 293 cells were transfected with these constructs and the cells labeled with 75Se. We further enriched the recombinant GPx1 from the transfected cells on an affinity column. The abundance of the 24-kDa GPx1 band increased in the order GPx1-pBud-Toxoplasma SECIS<GPx1-pBud-Toxoplasma SECIS +SBP2<GPx1-pSelExpress1. We also probed the samples with anti-GPx1 antibodies (Fig. 5B Middle), which showed a similar pattern. Because GPx1 is a homotetramer, the native GPx1 form copurified with that expressed from pSelExpress1.
Fig. 5.
Vector for high expression of selenoproteins in mammalian cells. (A) Vector map of pSelExpress1. The GGGA>ATGA mutant of Toxoplasma SelT SECIS element is preceded by a multiple cloning site (MCS) and by the human CMV immediate-early promoter. The C-terminal portion of rat SBP2 is under the control of the human elongation factor 1α-subunit promoter. Other major features of the vector backbone are indicated. (B) Expression and enrichment of recombinant His-tagged GPx1 on metal-affinity resin. HEK 293 cells were transfected with GPx1-pBudCE4.1 (lane 1), GPx1-pBudCE4.1 cotransfected with SBP2 (lane 3), and GPx1-pSelExpress1 (lane 5) or with pBudCE4.1 as control (lane 7). Cell lysates were prepared as described in Materials and Methods, and GPx1 was enriched from each sample on TALON resin. Proteins bound to the resin were loaded in lanes 2, 4, 6, and 8, as shown. Top shows metabolic labeling of cells with 75Se; Middle, Western blot with anti-GPx1 antibodies; and Bottom, protein staining with Amido Black. Because GPx1 is a tetramer, the His-tagged GPx1 expressed from pSelExpress1 binds the endogenous GPx1 (21-kDa band), which is then also enriched on TALON resin (see lower bands in lanes 2, 4, and 6, but not in 8).
Selenoproteins are notoriously difficult targets for recombinant expression. The bacterial Sec insertion system is different from that in eukaryotes, in that the bacterial SECIS is present in the coding region downstream of the Sec codon, whereas the eukaryotic SECIS is in the 3′-UTR. Therefore, expression of recombinant proteins in E. coli requires modification of the coding regions of selenoproteins in the vicinity of their active sites. In addition, efficiency of Sec insertion into recombinant proteins is low, because the major products are often the truncated forms of selenoproteins. To overcome this problem, several methods have been proposed (33–36), but none is fully satisfactory. Furthermore, some selenoproteins can be expressed only in eukaryotes because of unique posttranslational modifications. However, the major problem remains the low yield and high cost of recombinant selenoprotein expression. Therefore, the development of the vector for selenoprotein overexpression in mammalian cells has an important practical outcome, and the use of this vector may stimulate research in molecular biology and biochemistry of selenoproteins.
In conclusion, we identified a noncanonical form of eukaryotic SECIS element and a previously undescribed selenoprotein, SelQ. The SECIS does not conserve the nucleotide previously considered invariant in the SBP2 binding site, yet it is fully functional. This SECIS was detected in only two aplicomplexan parasites, Toxoplasma and Neospora, but it supports Sec insertion in mammals. We used the new SECIS form to develop a tool for efficient expression of recombinant selenoproteins in mammalian cells.
Materials and Methods
Search for Canonical Toxoplasma SECIS Elements.
A stand-alone version of SECISearch with the default pattern was used (7). The search procedure included: (i) Analysis of primary nucleotide sequence and secondary structures. We used PatScan to search the target database for the candidates satisfying the NUGA_AA_GA pattern. (ii) Estimation of the free energy. RNAfold from the Vienna RNA package (www.tbi.univie.ac.at/∼ivo/RNA) was used to calculate the free energies for whole structures and separately for their upper stem-loops. The threshold value was −12.6 kcal/mol for the whole structure and −3.7 kcal/mol for the upper stem-loop. (iii) Protein identification (analysis of location of SECIS elements and identification of ORFs). Candidate structures located on the complementary strand were filtered out. (iv) ORF analysis. This final step consisted of sequence analyses of predicted ORFs and identification of candidate Sec-encoding UGA codons.
Search for Noncanonical Toxoplasma SECIS Elements.
The search for noncanonical SECIS elements was carried out as described above for canonical SECIS elements, except that NTGA was replaced by NGGA in the primary sequence. Although no noncanonical SECIS elements other than the GGGA-type structures were discovered by homology searches involving known selenoproteins, the search settings were relaxed to allow any nucleotide preceding GGA (or UGA) for better sensitivity.
Cloning Strategies.
GFP-fusion constructs developed and used in the study are shown in the scheme in Fig. 2A. SelH cDNA containing the in-frame TGA codon but lacking the entire 3′UTR was amplified and cloned into pEFGP-C3 (Clontech, Mountain View, CA), and all subsequent constructs containing Toxoplasma SECIS elements were developed by using this GFP-SelHΔ3′UTR fusion construct (Fig. 2A, construct 2). Toxoplasma SelT and SelS SECIS elements (130-bp region, constructs 3 and 5, respectively) or the sequences beginning with the corresponding stop codons and containing SECIS elements (≈300-bp region, constructs 4 and 6) were amplified and cloned immediately downstream of the SelH stop codon. The rationale was as follows: the SelH SECIS is located very close to the stop codon (construct 1). Therefore, the constructs having the 130-bp sequences of Toxoplasma SECIS elements were regarded as corresponding to substitution of the mammalian SECIS element with the Toxoplasma structures, whereas the constructs containing the 300-bp sequences of Toxoplasma SelT 3′UTR or the 350-bp sequence of Toxoplasma SelS 3′UTRs were substitutions that introduced the corresponding 3′UTRs. We further mutated the GG bases in the SECIS quartet in both Toxoplasma SelT and SelS to AT (i.e., Toxoplasma GG to AT mutants). Likewise, the AT bases in GFP-mSelHwt, GFP-mSelSwt, and GFP-mSelMwt (Fig. 4A) fusions were mutated to the GG (i.e., mouse AT to GG mutants) by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA).
The vector for expression of selenoproteins in mammalian cells was developed on the basis of pBudCE4.1 (Invitrogen, Carlsbad, CA). First, the C-terminal domain of rat SBP2 was cloned into the first cloning site for expression under the EF1α promoter. Subsequently, the Toxoplasma SelT AT>GG SECIS was cloned into the second multiple cloning site. Finally, mouse GPx1 sequence containing an in-frame TGA codon, but lacking a 3′UTR, was amplified and cloned into the vector. As a control, we used the construct wherein mGPx1-Toxoplasma SelT SECIS AT>GG mutant was cloned into pBudCE4.1 that did not have the rat SBP2 sequence. To quantify the ratio of full-length and truncated forms, we used Scion Image 4.0 (Scion, Frederick, MD) for image processing and analysis.
Cell Culture, Transfection, and Metabolic Labeling.
Mouse NIH 3T3 and human HEK 293 cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 units/ml streptomycin. Cells were seeded in six-well plates and transfected as follows: NIH 3T3 cells using Lipofectamin and Plus reagent (Invitrogen) according to the manufacturer's protocol and HEK 293 using the calcium-phosphate method in OPTI-MEM (Invitrogen), or cotransfected in a ratio of 2:1 with the rat SBP2 expression construct that was the generous gift of Paul Copeland and Donna Driscoll, Cleveland Clinic Foundation, Cleveland, OH. In 12–24 h after transfection, the medium was replaced with DMEM supplemented with 75Se (specific activity 1,000 Ci/mmol; 1 Ci = 37 GBq), and the cells were incubated for an additional 12–24 h.
Acknowledgments
We thank Drs. Donna Driscoll and Paul Copeland for the gift of the rat SBP2 expression construct and Dr. David Sibley (Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, MO) for providing Toxoplasma and Neospora EST clones. This work was supported by National Institutes of Health Grant GM061603 (to V.N.G.). The use of 75Se was supported by Department of Energy Grant DE-FG07–02ID14380.
Abbreviations
- Sec
selenocysteine
- SECIS
Sec insertion sequence.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610683104/DC1.
References
- 1.Hatfield DL, Gladyshev VN. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock A, Rother M, Leibundgut, Ban N. In: Selenium, Its Molecular Biology and Role in Human Health. Hatfield DL, Berry MJ, Gladyshev VN, editors. New York: Springer; 2006. pp. 9–29. [Google Scholar]
- 3.Stadtman TC. Annu Rev Biochem. 2002;71:1–16. doi: 10.1146/annurev.biochem.71.083101.134224. [DOI] [PubMed] [Google Scholar]
- 4.Rother M, Resch A, Wilting R, Bock A. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- 5.Lescure A, Gautheret D, Carbon P, Krol A. J Biol Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 6.Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 8.Johansson L, Gafvelin G, Arner ES. Biochim Biophys Acta. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 10.Low SC, Berry MJ. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 11.Berry MJ, Martin GW, 3rd, Low SC. Biomed Environ Sci. 1997;10:182–189. [PubMed] [Google Scholar]
- 12.Walczak R, Westhof E, Carbon P, Krol A. RNA. 1996;2:367–379. [PMC free article] [PubMed] [Google Scholar]
- 13.Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN. Mol Cell Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walczak R, Carbon P, Krol A. RNA. 1998;4:74–84. [PMC free article] [PubMed] [Google Scholar]
- 15.Martin GW, 3rd, Harney JW, Berry MJ. RNA. 1996;2:171–182. [PMC free article] [PubMed] [Google Scholar]
- 16.Martin GW, 3rd, Harney JW, Berry MJ. RNA. 1998;4:65–73. [PMC free article] [PubMed] [Google Scholar]
- 17.Kryukov GV, Kryukov VM, Gladyshev VN. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 18.Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Fomenko DE, Gladyshev VN. Genome Biol. 2005;6:R37. doi: 10.1186/gb-2005-6-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Low SC, Grundner-Culemann E, Harney JW, Berry MJ. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atkins JF, Gesteland RF. Nature. 2000;407:463–465. doi: 10.1038/35035189. [DOI] [PubMed] [Google Scholar]
- 25.Chavatte L, Brown BA, Driscoll DM. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 26.Xu XM, Mix H, Carlson BA, Grabowski PJ, Gladyshev VN, Berry MJ, Hatfield DL. J Biol Chem. 2005;280:41568–41575. doi: 10.1074/jbc.M506696200. [DOI] [PubMed] [Google Scholar]
- 27.Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ. Mol Cell Biol. 2006;26:2337–2346. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allmang C, Krol A. In: Selenium, Its Molecular Biology and Role in Human Health. Hatfield DL, Berry MJ, Gladyshev VN, editors. New York: Springer; 2006. pp. 51–63. [Google Scholar]
- 29.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 30.Chao JA, Williamson JR. Structure (London) 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Moore T, Zhang Y, Fenley MO, Li H. Structure (London) 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 32.Grundner-Culemann E, Martin GW, 3rd, Harney JW, Berry MJ. RNA. 1999;5:625–635. doi: 10.1017/s1355838299981542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckenroth B, Harris K, Turanov AA, Gladyshev VN, Raines RT, Hondal RJ. Biochemistry. 2006;45:5158–5170. doi: 10.1021/bi0517887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su D, Li Y, Gladyshev VN. Nucleic Acids Res. 2005;33:2486–2492. doi: 10.1093/nar/gki547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 36.Rengby O, Arner E. Appl Environ Microbiol. 2007;73:432–441. doi: 10.1128/AEM.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]