Abstract
The cytoskeletal protein, actin, has its structure and function regulated by cofilin. In the absence of an atomic resolution structure for the actin/cofilin complex, the mechanism of cofilin regulation is poorly understood. Theoretical studies based on the similarities of cofilin and gelsolin segment 1 proposed the cleft between subdomains 1 and 3 in actin as the cofilin binding site. We used radiolytic protein footprinting with mass spectrometry and molecular modeling to provide an atomic model of how cofilin binds to monomeric actin. Footprinting data suggest that cofilin binds to the cleft between subdomains 1 and 2 in actin and that cofilin induces further closure of the actin nucleotide cleft. Site-specific fluorescence data confirm these results. The model identifies key ionic and hydrophobic interactions at the binding interface, including hydrogen-bonding between His-87 of actin to Ser-89 of cofilin that may control the charge dependence of cofilin binding. This model and its implications fill an especially important niche in the actin field, owing to the fact that ongoing crystallization efforts of the actin/cofilin complex have so far failed. This 3D binary complex structure is derived from a combination of solution footprinting data and computational approaches and outlines a general method for determining the structure of such complexes.
Keywords: actin-cofilin binary complex, molecular docking, radiolytic protein footprinting
Actin is a globular protein (G-actin) that polymerizes to filaments (F-actin) to form the cytoskeleton, a 3D network inside a eukaryotic cell. It provides mechanical, morphological, and locomotive support to cells and mediates cell division, exocytosis, and endocystosis (1–3). As a result, many normal and abnormal cellular processes depend on the rapid remodeling of the actin cytoskeleton and are regulated by many actin binding proteins (ABPs) through a series of polymerization and depolymerization reactions (2). Cofilin is an important ABP (4) that severs the F-actin (5, 6) and inhibits nucleotide exchange in the G-actin (7). Severing the filaments by cofilin near the slow-growing “pointed end” promotes dissociation of monomers accelerating actin deploymerization, whereas severing near the fast-growing “barbed end” induces filament extension (5, 6). This multifunctional behavior of cofilin is crucial to the spatial and temporal regulation of the actin filament network within various biological contexts. The lack of atomic-resolution structural data on actin/cofilin complexes prevents a full understanding of the functional mechanisms of cofilin action. Although the cofilin side of the interface of G-actin/cofilin and F-actin/cofilin complexes have been defined by mutagenesis (8), radiolytic footprinting (9), and NMR (10) (Fig. 1A), similar details with respect to the actin side of the interfacial residues are lacking. Based on the structural homology between cofilin and gelsolin segment 1 (GS1) (11) and the competitive binding of cofilin against profilin or GS1 (12, 13), cofilin was suggested to bind to G-actin at the cleft between subdomains 1 and 3, consistent with the crystal structures of G-actin/GS1 and G-actin/profilin complexes (Fig. 1B) (14–16). However, this proposed cofilin binding site lacks definitive experimental support, and a detailed model of the interface is lacking.
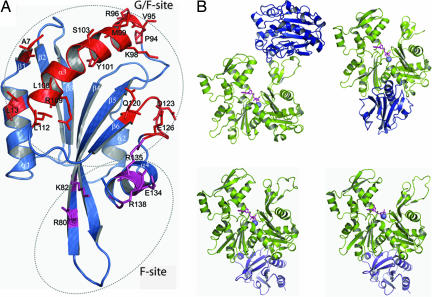
Fig. 1.
Structures of cofilin and actin/actin-binding protein complexes. (A) Structure of cofilin (Protein Data Bank ID code 1COF) showing the reported G/F-site (colored red) (8–10) and F-site (colored magenta) (8). The G/F-site participates in both G- and F-actin binding, whereas the F-site is required in addition to the G/F-site for F-actin binding. (B) Structures of actin/DNaseI (Protein Data Bank ID code 1ATN; Upper Left), actin/GS1 (Protein Data Bank ID code 1EQY; Upper Right), actin/profilin in the closed nucleotide cleft conformation (Protein Data Bank ID code 2BTF; Lower Left), and actin/profilin in the open nucleotide cleft conformation (Protein Data Bank ID code 1HLU; Lower Right).
We used radiolytic oxidative protein footprinting and mass spectrometry to probe the conformation of G-actin in the G-actin/cofilin binary complex in solution with resolution at the level of single side chains. The reactive residues buried at the interface of protein complexes or involved in the allosteric changes of the protein conformation are protected from oxidation, making it possible to detect changes in surface accessibility. This approach has been successfully applied to large proteins structures, including G- and F-actin structures, and has identified the binding interfaces for a number of protein complexes (17–28).
In the present study, actin residues involved in the binding interface and the allosteric changes in the G-actin/cofilin complex are identified by radiolytic protein footprinting coupled to mass spectrometry. Site-specific fluorescence experiments were conducted to confirm the results. These data are used to provide an atomic model of G-actin/cofilin complex by using comparative modeling and docking. The constructed model is used to understand the key hydrophobic and hydrophilic interactions in the actin/cofilin interface. These results illustrate the emerging power of structural mass spectrometry approaches in solution-based protein structural determination.
Results
Radiolytic Protein Footprinting Identifies Conformational Reorganization in Actin/Cofilin Complex.
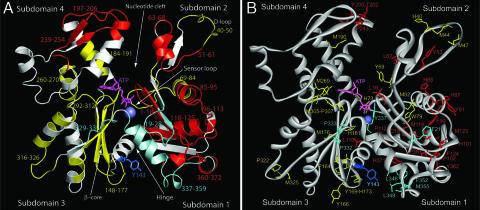
Protein samples were exposed to a white synchrotron x-ray beam for intervals from 0 to 200 ms. On these time scales, oxidative modifications dominate the chemistry compared with cross-linking events and cleavage (29), and effects on the global protein structure are minimal. Exposed samples were subjected to proteolysis followed by quantification of the peptides by electrospray ion source–mass spectrometry coupled to liquid chromatography. The oxidized probe sites within each peptide were confirmed by tandem mass spectrometry (20, 23). First-order rate constants of modification derived from dose–response curves (20, 23) for 20 peptides from trypsin and Glu-C digestion that cover 75% of actin sequence are listed in Table 1. These rate values are in accordance with the number, type (specific reactivity), and solvent exposure of the reactive amino acids in the respective peptides (20, 23–26). Fig. 2 depicts the various oxidized peptides and their reactive probes. Nine peptides showed substantial protection upon cofilin binding (≥4.0-fold; Fig. 2, red); five are located in subdomain 1, two in subdomain 2, and two in subdomain 4. Peptides that experience the level of protection seen for those located in subdomain 1 (20, 22–24, 26), particularly if they are clustered, provide strong candidates for identifying an interface. The seven peptides that are moderately protected (between 1.2- and 4.0-fold; Fig. 2, yellow) are distributed throughout the actin structure. The four peptides that exhibit no protection (between 0.8- and 1.2-fold; Fig. 2, cyan) are from subdomains 1 and 3. The tryptic peptide 119–147 showing no change in reactivity contains the probe Tyr-143 in addition to three other probes common with Glu-C-digested peptide 118–125, which is 4-fold protected. This observation indicated that Tyr-143 exhibits increased modification (essentially a negative protection; Fig. 2, blue) and hence increased solvent exposure upon complex formation. Thus, all of the actin peptides that are observed to show no change or increased modification are located in subdomains 1 and 3: three of four in subdomain 3. Taken together, these results suggest that the actin/cofilin interface consists of residues in subdomain 1 and does not contain residues from subdomain 3.
Table 1.
Modification rates for actin peptides in absence and presence of cofilin, averaged from duplicate experiments
| No. | Peptide | Protease | Oxidized residues | Subdomain | Modification rate, s−1 |
Degree of protection† | |
|---|---|---|---|---|---|---|---|
| Actin | Actin/cofilin | ||||||
| 1 | 1–18 | Trypsin | C10, L16 | 1 | 3.1 ± 0.2 | 0.7 ± 0.1 | 4.4 |
| 2 | 19–28 | F21 | 1 | 0.7 ± 0.1 | 0.8 ± 0.1 | 0.9 | |
| 3 | 40–50 | H40, M44, M47 | 2 | 12.9 ± 0.5 | 5.6 ± 0.6 | 2.3 | |
| 4 | 51–61 | Y53 | 2 | 0.9 ± 0.2 | 0 | High | |
| 5 | 63–68 | L67 | 2 | 0.4 ± 0.1 | 0.06 ± 0.03 | 6.7 | |
| 6 | 69–84 | Y69,H73,W79,M82 | 1+2 (70-) | 5.8 ± 0.6 | 2.3 ± 0.7 | 2.5 | |
| 7 | 85–95 | H87, H88, F90, Y91 | 1 | 2.7 ± 0.3 | 0.3 ± 0.1 | 9.0 | |
| 8 | 96–113 | H101, P102, L110, P112 | 1 | 1.8 ± 0.1 | 0.16 ± 0.05 | 11.3 | |
| 9 | 119–147 | M119, M123, F124, Y143 | 1+3 (145-) | 4.6 ± 0.1 | 4.0 ± 0.4 | 1.2 | |
| 10 | 148–177 | H161, P164, Y166, Y169, L171, P172, H173, M176 | 3 | 13.5 ± 0.5 | 6.3 ± 0.1 | 2.1 | |
| 11 | 184–191 | M190 | 4 | 4.4 ± 0.1 | 2.3 ± 0.2 | 1.9 | |
| 12 | 197–206 | F200-T202* | 4 | 0.7 ± 0.0 | 0 | High | |
| 13 | 239–254 | P243 | 4 | 2.5 ± 0.1 | 0.6 ± 0.1 | 4.2 | |
| 14 | 292–312 | M305-P307* | 3 | 3.7 ± 0.1 | 1.9 ± 0.4 | 1.9 | |
| 15 | 316–326 | P322, M325 | 3 | 4.9 ± 0.1 | 1.8 ± 0.0 | 2.7 | |
| 16 | 329–335 | P332, P333 | 3 | 1.5 ± 0.3 | 1.4 ± 0.2 | 1.1 | |
| 17 | 337–359 | Y337, L346, L349, F352, M355 | 1 (338-) +3 | 2.5 ± 0.3 | 3.1 ± 0.4 | 0.8 | |
| 18 | 360–372 | Y362, P367, H371 | 1 | 2.4 ± 0.2 | 0.5 ± 0.0 | 4.8 | |
| 19 | 118–125 | Glu-C | M119, M123, F124 | 1 | 2.0 ± 0.4 | 0.5 ± 0.1 | 4.0 |
| 20 | 260–270 | M269 | 4 | 2.0 ± 0.1 | 1.2 ± 0.2 | 1.7 | |
*Specific probe sites can not be determined definitely.
†Defined as ratio of modification rate of actin to actin/cofilin complex.
Fig. 2.
G-actin structure indicating the protection sites revealed by radiolytic footprinting. (A) Peptides that show substantial protection (4.0-fold and beyond decrease in the modification rate) are colored in red, moderate protection (between 1.2- and 4.0-fold) are in yellow, and nearly no protection is in cyan (0.8- to 1.2-fold). The residue Tyr-143 shown as a blue stick model shows negative protection (increased modification). (B) Modified amino acids of various protected peptides are shown as stick models.
Site-Specific Fluorescence Reveals Conformational Changes upon Cofilin Binding.
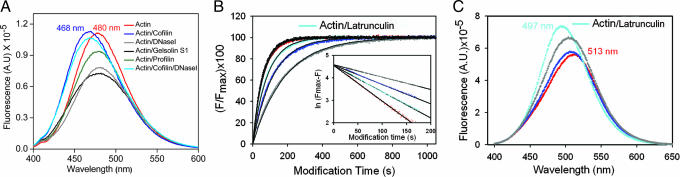
The fluorescence emission maximum, λem, is 480 nm for acrylodan-labeled Cys-374 of actin and its binary complexes with DNaseI, GS1, and profilin (Fig. 3A), indicative of unaffected solvent exposure of the label with binding of these ABPs. However, the emission maximum is blue-shifted (468 nm) for actin/cofilin complex (Fig. 3A), indicating that the label is protected from solvent, which could result from either cofilin binding to this region or an allosteric conformational change associated with cofilin binding to a distant site. For the ternary complex of actin/cofilin/DNaseI (30), the spectrum showed a similar λem as that of the binary actin/cofilin complex (468 nm; Fig. 3A) and is indicative of cofilin binding to the same site in both binary and ternary complexes, as the solvent exposure of the label is not affected by DNaseI. These data suggest that the mode of cofilin binding is distinct from that of profilin or GS1 or DNaseI (Fig. 1B).
Fig. 3.
Fluorescence of acrylodan-labeled actin and actin/actin binding factor complexes. (A) Fluorescence spectra of acrylodan-labeled Cys-374 in rabbit actin and actin/actin binding factor complexes as indicated. (B) Time course of fluorescence increase during acrylodan modification of Cys-203 in yeast actin (red) and in its binary complexes with cofilin (blue), DNaseI (gray), GS1 (black), and latrunculin (cyan). GS1 and profilin overlap, and the latter is not shown. (Inset) Plots of ln(F − Fmax) versus time, from which the rate constants are deduced. (C) Fluorescence spectra of the acrylodan-labeled Cys-203 yeast actin and actin/actin binding factor complexes. Color codes follow those from B. Spectra of actin, actin/GS1, and actin/profilin overlap, and the latter two are not shown.
Footprinting data reveal high protection afforded by cofilin to peptide 197–206 in subdomain 4 that is distant from the likely cofilin binding site (see Discussion and Fig. 2A). To probe for allosteric influence of cofilin in this region, fluorescence of acrylodan labeled to Cys-203 of the yeast actin mutant (Thr-203 replaced by cysteine and Cys-374 changed to alanine) was measured in the absence and presence of various actin binding factors (Fig. 3 B and C). As expected, Cys-203 was protected strongly from acrylodan modification by DNaseI (modification rate decreased from 15.8 × 10−3 s−1 to 5.5 × 10−3 s−1, 2.9-fold) or by latrunculin (11.6 × 10−3 s−1, 1.4-fold), which binds to the vicinity of this region (31), and no protection by GS1 or profilin, which bind to actin at sites distant from subdomain 4 (0.9- to 1.0-fold). In contrast, cofilin, which also binds to site(s) distant from subdomain 4, afforded significant protection to Cys-203 (8.6 × 10−3 s−1, 1.8-fold). A blue-shifted λem of actin with binding of cofilin (506 nm), DNaseI (504 nm), or latrunculin (497 nm) and nearly unchanged λem with binding of GS1 or proflin (513–514 nm; Fig. 3C) further confirms these results. These results are consistent with footprinting and show a prominent allosteric effect of cofilin on elements of subdomain 4 in actin.
Constructing the 3D Model of G-Actin/Cofilin.
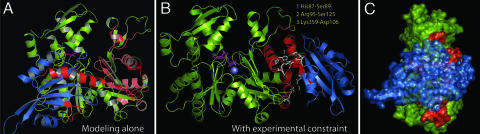
A model of G-actin in the putative closed cofilin-bound form was constructed with comparative modeling using SWISS-MODEL (32). Because our previous footprinting data (9) detected no conformational changes for cofilin with actin binding, we carried out rigid body docking of cofilin (Protein Data Bank ID code 1COF) (33) to the modeled G-actin structure by using the DOT program provided by ClusPro (see Methods) (34). The top-scored structure after energy filtering and clustering, without the use of experimental data (Fig. 4A), had neither the cofilin at the binding site identified by footprinting nor the cofilin's actin binding segments (G/F-site; Fig. 1A) at the interface. Next, we incorporated “attract” constraint (see Methods) in the docking step by using a few interfacial residues of G-actin from the footprinting data that are solvent-exposed and/or located at the surface including Trp-79, His-87, His-88, Tyr-91, Tyr-362, Pro-367, and His-371. The first ranked model obtained is shown in Fig. 4B and is consistent with the present footprinting data of the actin interfacial residues (see Discussion) and the reported (8–10) data on the cofilin interfacial residues (Fig. 1A and see Table 2). The same model was ranked second in the unsupervised docking, which suggests that this model has a high propensity for shape complementarity such as would be found in a native interface structure; the use of experimental data as attract constraints moved this model to top ranking.
Fig. 4.
Actin and cofilin docking and interaction analysis. (A) Top-scored G-actin/cofilin model from the docking method without the use of experimental constraints. Previously established G-actin binding segments of cofilin (G/F-site; see Fig. 1A) and the cofilin binding segments of actin as suggested by the current study are colored red. (B) Top-scored G-actin/cofilin model from the docking constrained with the footprinting data. Red-colored region marks the closely interacting segments. Hydrogen bonding/salt bridging at the interface are indicated as stick models. (C) Side view (rotated 90° to the left) of the surface rendered model shown in B.
Table 2.
Regions of contact at the binding surface
| Cofilin | Actin | H bond/salt bridge* |
|---|---|---|
| 1–5 (N terminus) | 359–363 (C terminus) | Lys-359–Asp-106 |
| 89–112 (α3 helix) | 48–52 (D-loop) | His-87–Ser-89 |
| 79–92 (cleft helix) | ||
| 124–128 (lower helix) | ||
| 359–363 (C terminus) | ||
| 120–126 (β6-α4 loop) | 93–100 (loop after cleft helix) | Arg-95–Ser-125 |
*Order of H bond/salt bridge is actin residue–cofilin residue.
Discussion
Cofilin Does Not Occupy Canonical ABP Binding Site.
Structural comparisons (11, 12, 35) and competitive binding studies (12, 13) suggest cofilin binds to the cleft formed between subdomains 1 and 3 of actin as preceded by the binding of GS1 and profilin. Contrary to this observation, the footprinting data do not detect significant protections within the cleft between subdomain 1 and 3 as a function of cofilin binding. On the other hand, footprinting reveals significant protections within subdomains 1 and 2. GS1 interacts with the hinge helices (Gly-137–Ser-145 and Arg-335–Ser-348) (Fig. 1B) located in the cleft between subdomains 1 and 3 in actin (14). Footprinting data on the G-actin/GS1 complex (25) revealed five peptides protected, including the hinge helices. Protected peptides were found only in subdomains 1 and 3, consistent with crystallographic data (14). Not only do we not see cofilin-dependent protections in the first hinge helix; its single probe residue Tyr-143 exhibits increased reactivity upon actin/cofilin complex formation, whereas the peptide 337–359 encompassing the second hinge helix showed no change in reactivity. Thus, there is no evidence for cofilin occupying the GS1 binding site.
Cofilin Occupies a Site in the Cleft Between Subdomains 1 and 2.
A cluster of protections mediated by complex formation was observed in the cleft between subdomains 1 and 2 (Fig. 2). The oxidation rate of Tyr-53 of peptide 51–61 (0.9 s−1, solvent-accessible surface area is 14 Å2), which is located in the cleft between subdomains 1 and 2, is reduced to effectively zero when cofilin binds (Table 1), indicating complete burial for this residue. Other peptides of subdomain 1, positioned adjacent to peptide 51–61, also show significant protections including: peptide 96–113 (11-fold protection), peptide 85–95 (9-fold protection), C-terminus peptide 360–372 (5-fold protection), N-terminus peptide 1–18, peptide 118–125 (4-fold protection), and peptide 69–84 (3-fold protection). Peptides 69–84 and 85–95 encompass the helix within the cleft located just below peptide 51–61, while peptide 118–125 belongs to another helix located below the cleft helix. Peptide 96–113 is a loop with a β-strand that connects both helices. The N and C termini are positioned next to these peptides. We suggest that this region is the center of the cofilin binding surface. Footprinting data for the actin/GS1 complex (25) revealed no change in reactivity for these peptides as a function of complex formation (protection range of 0.9- to 1.1-fold), further confirming that cofilin does not occupy the previously presumed binding site.
Cofilin Binding Increases the Closure of the Nucleotide Binding Cleft of G-Actin.
Protections for residues other than the putative cofilin binding surface are observed upon cofilin binding. Of particular interest are protections at residues Phe-200–Thr-202 (within peptide 197–206) and at residue Pro-243 (from peptide 239–254) from the top region of the nucleotide cleft in subdomain 4. In addition, Leu-67 (of peptide 63–68), Tyr-69 and His-73 (from peptide 69–84), and Met-190 (from peptide 184–191), all located within the cleft (Fig. 2B), are protected as a function of complex formation. Leu-16 of peptide 1–18 and His-40, Met-44, and Met-47 of the D-loop, all of which show significant protections upon cofilin binding, are located near the nucleotide cleft. These protections at the nucleotide cleft cannot be accommodated by cofilin binding as then the ternary complex of actin, cofilin, and DNaseI cannot be formed because of steric overlap (Fig. 1B). However, this constellation of protections observed at the nucleotide cleft region has been previously observed in footprinting experiments that compared the conformation of Ca-ATP–G-actin and Mg-ATP–G-actin (25). The footprinting data of Ca-ATP–G-actin form was entirely consistent with crystallographic data (31) showing a partially closed nucleotide cleft, whereas the footprinting data for Mg-ATP–G-actin indicated a more closed nucleotide cleft. Based on these data, we proposed a structural model with a highly closed cleft for Mg-ATP–G-actin (21). Our data indicate that cofilin induces a similar closure of the nucleotide cleft; overall, this result suggests a general mode of actin dynamic conformational change.
Fluorescence Confirms the Results of Footprinting.
Fluorescence spectral properties of actin in complex with cofilin are quite different from the analogous complexes with GS1 or profilin, in agreement with the proposed binding interface and cofilin's allosteric effect on the nucleotide cleft. Cofilin-induced changes in the 197–206 region on G-actin can be linked to the well established effects of cofilin on the nucleotide cleft in actin such as the inhibition of nucleotide exchange, an increase in the bound ε-ADP fluorescence (36), and a decrease in the accessibility of ε-ADP to collisional quenchers (36). The above effects are indicative of a further closure of the actin's nucleotide cleft by cofilin, consistent with the present data suggesting consequent conformational changes in subdomains 2 and 4 on actin.
Footprinting Results Explain Cofilin's Competition with GS1 or Profilin for Binding to Actin.
We observed cofilin's competition with GS1 and with profilin for binding to actin by fluorescence and by native gel electrophoresis [see supporting information (SI) Text]. Such competition data have been interpreted as evidence for the overlap of cofilin's binding site with these ABPs (12, 13). The observation that cofilin induces an allosteric structural change centered at the hinge that closes the large cleft (between subdomains 2 and 4) and opens the small cleft (between subdomains 1 and 3), clearly explains the competition data. Our previous footprinting data (25) indicated no conformational rearrangements of actin structure induced by GS1 binding. Further, profilin binding to G-actin is known to be accompanied by opening of the large cleft. Therefore, when GS1 or profilin is bound the domain motion toward cleft closure is blocked and cofilin's binding is inhibited.
3D Model of G-Actin/Cofilin Complex: Consistency with Footprinting Data.
The 3D model of the Ca-ATP-G-actin/cofilin complex, as constrained by footprinting data (Fig. 4 B and C), provides a valid framework for understanding the functional interactions of cofilin and actin. Residues from the footprinting probe set seen in Table 1, which are relatively exposed (≥30 Å2) at the cofilin binding surface of free actin that experience burial upon cofilin binding (≥ 25% reduction of solvent-accessible surface area), include: Trp-79 (53/55 to 33 Å2, 38–40% burial), His-87 (89/85 to 3.4 Å2, 96% burial), His-88 (33/25 to 17 Å2, 32–48% burial), Tyr-91 (72/86 to 1.4 Å2, 98% burial), Pro-102 (36 to 22 Å2, 40% burial), and His-371 (28/30 to 21 Å2, 25–30% burial). Moderately buried probe residues (<30 Å2 and ≥10 Å2) that are further buried with cofilin binding are Tyr-53 (14 to 4.6 Å2, 67% burial), His-101 (20/28 Å2 to 8 Å2, 60–71% burial), and Tyr-362 (13/10 to 7 Å2, 30–46% burial). Deeply buried probe residues (<10 Å2) from this region are Met-119 (0.5 to 0 Å2), Met-123 (0 Å2), and Phe-124 (4/0 to 0 Å2). These residues are likely to exhibit dynamic behavior in free actin, resulting in occasional oxidation events, but are fully inaccessible in the complex.
3D Model of G-Actin/Cofilin Complex: Molecular Details of the Interface and Charge Sensitivity.
The reported G-actin binding segments of cofilin (G/F-site; Fig. 1A) (8–10) are seen clearly making specific contacts with actin in the model (Fig. 4B) and are detailed in Table 2. The actin binding motif in cofilin, namely the long α3 helix, interacts with the lower segment of D-loop (residues 48–52), cleft helix (residues 79–92), residues 124–128 of the lower cleft helix, and residues 359–363 of the C terminus. The β6–α4 loop of cofilin closely interacts with residues 93–100, the loop after the cleft helix in actin. Although residues 1–5 of N terminus of cofilin are missing in the crystal structure of cofilin and in the model, the location and the directionality of this region of the N terminus is close to the actin C terminus and is consistent with the involvement of segment 1–5 in binding.
It is reported that cofilin binds to actin in a pH-dependent manner (37). Both actin and cofilin are acidic overall (pI ≈5.1). Although actin's cofilin-binding surface contains a substantial number of acidic residues (26 in number as opposed to 8 basic residues), the α3 helix of cofilin contains five basic residues (Arg-96, Lys-98, Lys-105, Arg-109, and Arg-110) as opposed to one acidic residue (Asp-106), suggesting the potential for favorable ionic interactions upon binding. We have identified several pairs of residues in the actin/cofilin interface that meet the criteria for hydrogen bonding or salt bridging (Fig. 4B and Table 2) (38). The most important is the possible hydrogen bonding between His-87 in the cleft helix of actin and Ser-89 in the α3 helix of cofilin; this interaction may play a critical role in the charge dependence of cofilin binding. Hydrogen bonding is also likely between Arg-95 from actin's cleft helix and Ser-125, located in the loop between β6 and α4 of cofilin. In addition, Lys-359 from the C terminus of actin may form a hydrogen bonding/salt bridge with Asp-106 of cofilin's α3 helix. These two residues can have other intrachain salt bridges (thus forming an extended salt-bridge network); Lys-359 may interact with Asp-363 within actin, whereas Asp-106 may salt-bridge with Lys-105 within cofilin. The two acidic residues of cofilin Asp-123 and Glu-126 predicted to be involved in the G-actin binding (8) are located opposite to two polar side chains in the cleft helix of actin (Tyr-90, Arg-95). A hydrogen bonding between Asp-123 with Arg-95 seems favorable.
Apart from these proposed ionic interactions contributing to complex formation, hydrophobic interactions that drive out water typically contribute to increasing the value of ion-pair formation (39). The actin-binding surface in cofilin contains a wealth of hydrophobic residues. In the long α3 helix, these include Val-95, Met-99, Val-100, Ala-102, Ala-107, Leu-108, Ala-111, and Leu-112, which may be interacting with the hydrophobic residues in actin. Although the two cleft helices are lined mostly with hydrophilic residues, the N termini (peptide 1–18), the C termini (peptide 360–372), and peptide 96–113 connecting the two helices in the cleft are lined with many hydrophobic residues.
The cofilin interface regions are distinct for actin monomer (G/F-site) and filament (F-site) as established by various studies (Fig. 1A). Consistently, our model has G-actin located at the G/F-site of cofilin. Image reconstructions of cryo-EM data (40) predict cofilin binding between two actin monomers located at the surface of the actin filament. On the other hand, biochemical experiments suggest cofilin to be intercalated between two actin monomers (41). Our model (Fig. 4 B and C) establishes that the case for monomer binding is different from either of these two models.
Conclusion
Although high-resolution structures for both monomeric actin and cofilin are described individually in the literature, the structure of the actin/cofilin complex is unknown. We have used hydroxyl radical-mediated oxidative protein footprinting to identify the G-actin residues that contribute to the cofilin binding surface and to probe the conformational changes induced by cofilin binding. We have demonstrated how these data can be used in conjunction with state-of-the art comparative modeling and docking algorithms to provide an atomic model of the actin/cofilin complex. This method provides a general approach to determining the structure of protein binary complexes. Several conclusions from this model are described. First, the cofilin binding site is conclusively seen to be distinct from that of GS1 and profilin and involves a tight binding to the cleft between subdomains 1 and 2. Second, actin binding segments of cofilin are consistent to the previous data from mutation, footprinting, and NMR studies. Third, the proposed actin/cofilin interface has a number of potential hydrogen bonding and salt links; the proposed His-87–Ser-89 interaction may mediate the charge dependence of cofilin binding. Fourth, cofilin-induced changes in actin monomers are consistent with closure of the nucleotide cleft, and thus the inhibition of nucleotide exchange in the actin/cofilin binary complex in the cell. These results provide a structural basis for interpreting cofilin structural and functional data.
Methods
Sample Preparation.
Rabbit skeletal muscle actin and yeast mutant actin (T203C/C374A) were prepared in the form of Ca-ATP–G-actin as described (see SI Text) (25, 42). Yeast cofilin, GS1, and human platelet profilin were generous gifts from Steven C. Almo (Albert Einstein College of Medicine, Bronx, NY). Bovine pancreatic DNaseI was obtained from Worthington Biochemical Corporation (Lakewood, NJ). Methionine amide (Met-NH2) was purchased from Bachem Bioscience Inc. (King of Prussia, PA); proteolytic enzymes (trypsin, Asp-N, Glu-C) were from Promega (Madison, WI) and Roche (Indianapolis, IN); acrylodan (6-acryloyl-2-(dimethylamino) naphthalene) was from Molecular Probes (Eugene, OR), and Latrunculin A was from Calbiochem (San Diego, CA).
Synchrotron X-Ray Radiolysis and Mass Spectrometry.
Radiolysis experiments of rabbit G-actin and G-actin/cofilin complex at 10 μM concentration were performed at the X-28C beam line of the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY, as described (9, 25). Met-NH2 (10 mM) was added right after exposure to quench secondary oxidation (20), and samples were frozen immediately thereafter. Radiolyzed samples were subjected to enzymatic proteolysis (9, 25), and the resulting peptide mixtures were separated and analyzed by using a coupled, HPLC–electrospray ion source mass spectrometer equipped with quadrupole ion trap (LCQ classic; LCQ DECAXP plus; Finnigan, Breman, Germany) (see SI Text). The detailed analytic procedures have been described (20, 23).
Solvent-Accessible Surface Area Calculation.
The solvent-accessible surface areas of amino acids were calculated by using the computer program GETAREA 1.1 (www.pauli.utmb.edu/cgi-bin/get_a_form.tcl) from the crystal structures with a probe radius of 1.4 Å with additional atomic parameter database and residue-type library entries.
Fluorescence Spectroscopy.
Labeling rabbit muscle G-actin and the mutant yeast G-actin with acrylodan was carried out by incubating 1.1- to 1.2-fold excess of acrylodan over actin for a specific period followed by removal of the excess reagent (see SI Text) (43). Actin/actin binding factor complexes were prepared by mixing 1 μM labeled muscle actin or 5 μM labeled yeast actin with 10 μM actin-binding factors. Fluorescence measurements were carried out in a Flurolog-3 spectrofluorometer (Spex, Edison, NJ), Fluoromax-3 spectrofluorometer (Spex), or a spectrofluorometer from PTI (Lawrenceville, NJ). An excitation wavelength, λex = 360 nm, was used to excite acrylodan in the rabbit actin, whereas λex = 375 nm was used for yeast mutant actin. The time course of the acrylodan labeling of Cys-203 yeast actin mutant was monitored via fluorescence changes at 510 nm, at 20°C. A 10-fold molar excess of yeast actin (5 μM) over acrylodan (0.5 μM) was used, and the concentration of the dimethylformamide (acrylodan's solvent) in the samples did not exceed 1%. Cys-203 modification followed a first-order process, and the rates of labeling were obtained from the slopes of the plots of ln(Fmax − F) versus time (Fig. 3B Inset), where Fmax is the maximum fluorescence at the plateau of the modification and F is the observed fluorescence at a given time.
Modeling of G-Actin/Cofilin Binary Complex.
Actin in the “closed nucleotide cleft” conformation was obtained by homology modeling by using the SWISS-MODEL protein modeling server (http://swissmodel.expasy.org) (32), inputting the rabbit actin sequence as “target” and the 3D structure of bovine actin (2BTF chain A, 2.55-Å resolution) as “template.” Docking of cofilin to G-actin, energy filtering, clustering, and ranking were done by using the ClusPro Web server (http://nrc.bu.edu/cluster) (see SI Text) (34). The output of the docking program, DOT, is 25,000 structures with the best shape complementarity scores. These structures are then energy filtered (electrostatics or desolvation) to retain the top 2,000 structures, which are then clustered (9-Å clustering radius) and ranked according to their cluster sizes. Footprinting constraints were set up within the docking to provide positive weighting to the experimentally derived interfacial residues of G-actin (refer to ref. 34 for details).
Acknowledgments
We thank Steven C. Almo for supplying some of the proteins used in this study. This work was supported by National Institutes of Health Grant NIBIB-P41-01979 (to M.R.C.), Public Health Service Grant GM-077190 (to E.R.), and National Science Foundation Grant MCB 0316269 (to E.R.).
Abbreviations
- ABP
actin binding protein
- GS1
gelsolin segment 1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611283104/DC1.
References
- 1.Amann KJ, Pollard TD. Curr Biol. 2000;10:R728–R730. doi: 10.1016/s0960-9822(00)00751-x. [DOI] [PubMed] [Google Scholar]
- 2.Paavilainen VO, Bertling E, Falck S, Lappalainen P. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD, Blanchoin L, Mullins RD. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 4.Bamburg JR. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 5.DesMarais V, Ghosh M, Eddy R, Condeelis JS. J Cell Sci. 2005;118:19–26. doi: 10.1242/jcs.01631. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 7.Nishida E. Biochemistry. 1985;24:1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- 8.Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan JQ, Vorobiev S, Almo SC, Chance MR. Biochemistry. 2002;41:5765–5775. doi: 10.1021/bi0121104. [DOI] [PubMed] [Google Scholar]
- 10.Pope BJ, Zierler-Gould KM, Kuhne R, Weeds AG, Ball LJ. J Biol Chem. 2004;279:4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 11.Wriggers W, Tang JX, Azuma T, Marks PW, Janmey PA. J Mol Biol. 1998;282:921–932. doi: 10.1006/jmbi.1998.2048. [DOI] [PubMed] [Google Scholar]
- 12.Blanchoin L, Pollard TD. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- 13.Mannherz HG, Ballweber E, Galla M, Villard S, Granier C, Steegborn C, Schmidtmann A, Jaquet K, Pope B, Weeds AG. J Mol Biol. 2007;366:745–755. doi: 10.1016/j.jmb.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 15.Chik JK, Lindberg U, Schutt CE. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 16.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 17.Hambly DM, Gross ML. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Sharp JS, Becker JM, Hettich RL. Anal Chem. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- 19.Sharp JS, Guo JT, Uchiki T, Xu Y, Dealwis C, Hettich RL. Anal Biochem. 2005;340:201–212. doi: 10.1016/j.ab.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Takamoto K, Chance MR. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 21.Takamoto K, Kamal JKA, Chance MR. Structure (London) 2006;15:39–51. doi: 10.1016/j.str.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Xu G, Liu R, Zak O, Aisen P, Chance MR. Mol Cell Proteomics. 2005;4:1959–1967. doi: 10.1074/mcp.M500095-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Guan JQ, Chance MR. Trends Biochem Sci. 2005;30:583–592. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Guan JQ, Almo SC, Chance MR. Acc Chem Res. 2004;37:221–229. doi: 10.1021/ar0302235. [DOI] [PubMed] [Google Scholar]
- 25.Guan JQ, Almo SC, Reisler E, Chance MR. Biochemistry. 2003;42:11992–12000. doi: 10.1021/bi034914k. [DOI] [PubMed] [Google Scholar]
- 26.Guan JQ, Takamoto K, Almo SC, Reisler E, Chance MR. Biochemistry. 2005;44:3166–3175. doi: 10.1021/bi048021j. [DOI] [PubMed] [Google Scholar]
- 27.Kiselar JG, Janmey PA, Almo SC, Chance MR. Proc Natl Acad Sci USA. 2003;100:3942–3947. doi: 10.1073/pnas.0736004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiselar JG, Mahaffy R, Pollard TD, Almo SC, Chance MR. Proc Natl Acad Sci USA. 2007;104:1552–1557. doi: 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies MJ, Dean RT. Radical-Mediated Protein Oxidation: From Chemistry to Medicine. Oxford: Oxford Univ Press; 1997. [Google Scholar]
- 30.Nosworthy NJ, Kekic M, dos Remedios CG. Proteomics. 2001;1:1513–1518. doi: 10.1002/1615-9861(200111)1:12<1513::aid-prot1513>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 31.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 32.Schwede T, Kopp J, Guex N, Peitsch MC. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- 34.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 35.Dominguez R. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Muhlrad A, Pavlov D, Peyser YM, Reisler E. FEBS J. 2006;273:1488–1496. doi: 10.1111/j.1742-4658.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 37.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Cell Motil Cytoskeleton. 2006;63:533–542. doi: 10.1002/cm.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald IK, Thornton JM. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 39.Bogan AA, Thorn KS. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 40.McGough A, Pope B, Chiu W, Weeds A. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renoult C, Ternent D, Maciver SK, Fattoum A, Astier C, Benyamin Y, Roustan C. J Biol Chem. 1999;274:28893–28899. doi: 10.1074/jbc.274.41.28893. [DOI] [PubMed] [Google Scholar]
- 42.Kudryashov DS, Sawaya MR, Adisetiyo H, Norcross T, Hegyi G, Reisler E, Yeates TO. Proc Natl Acad Sci USA. 2005;102:13105–13110. doi: 10.1073/pnas.0506429102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamal JKA, Zhao L, Zewail AH. Proc Natl Acad Sci USA. 2004;101:13411–13416. doi: 10.1073/pnas.0405724101. [DOI] [PMC free article] [PubMed] [Google Scholar]