Abstract
Heterotrimeric G proteins function as molecular relays that mediate signal transduction from heptahelical receptors in the cell membrane to intracellular effector proteins. Crystallographic studies have demonstrated that guanine nucleotide exchange on the Gα subunit causes specific conformational changes in three key “switch” regions of the protein, which regulate binding to Gβγ subunits, receptors, and effector proteins. In the present study, nitroxide side chains were introduced at sites within the switch I region of Gαi to explore the structure and dynamics of this region throughout the G protein cycle. EPR spectra obtained for each of the Gα(GDP), Gα(GDP)βγ heterotrimer and Gα(GTPγS) conformations are consistent with the local environment observed in the corresponding crystal structures. Binding of the heterotrimer to activated rhodopsin to form the nucleotide-free (empty) complex, for which there is no crystal structure, causes prominent changes relative to the heterotrimer in the structure of switch I and contiguous sequences. The data identify a putative pathway of allosteric changes triggered by receptor binding and, together with previously published data, suggest elements of a mechanism for receptor-catalyzed nucleotide exchange.
Keywords: G protein receptor complex, site-directed spin-labeling, switch I, visual signal transduction
Heterotrimeric G protein α subunits function in the cell as molecular switch proteins, cycling between an inactive GDP-bound heterotrimeric conformation and an active GTP-bound conformation. Heptahelical receptors in the cell membrane activate G proteins by catalyzing GTP for GDP exchange on Gα, leading to the activation of downstream effector proteins by Gα(GTP) and Gβγ. The signal is terminated upon the hydrolysis of GTP to GDP by Gα and its reassociation with Gβγ (1).
Nucleotide-dependent conformational changes in Gα have been identified by x-ray crystallography. A comparison of the GDP-bound (2) and GTPγS-bound (3) structures of the transducin α subunit (Gαt) identified three segments of the protein that undergo rearrangement upon activation, named switches I–III. Similarly, the corresponding structures of Gαi demonstrate a GTPγS-dependent conformational change in switch I, whereas switches II and III, which are disordered in the GDP-bound structure (4), become ordered upon GTPγS binding (Fig. 1A) (5). Switches I and II interact directly with the γ-phosphate of GTP and form part of the Gβγ-interacting surface in the heterotrimer (Fig. 1B) (6, 7). Switch II is an important effector-binding site on Gα (8–11), and a recent site-directed spin-labeling (SDSL) study showed that nucleotide exchange results in an increase in switch II dynamics, an event that may play a role in effector recognition/binding (12). Switch I plays a more critical role in binding RGS (regulators of G protein signaling) proteins (11, 13). Clearly, these switch regions form the basis of the conformational switching mechanism in Gα by sensing the identity of the bound nucleotide and regulating G protein interactions with other molecules.
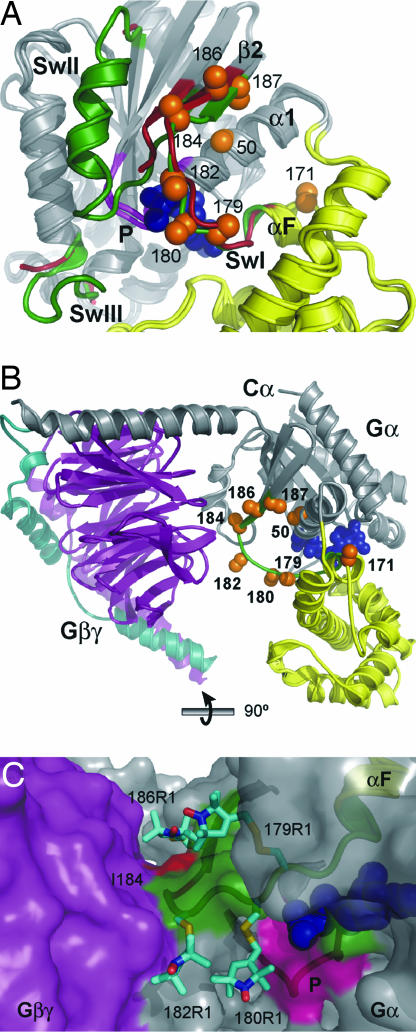
Fig. 1.
Models of Gαi and Gαiβγ crystal structures showing the spin-labeled sites. (A) Overlay of Gαi(GDP) [Protein Data Bank (PDB) ID code 1BOF] and Gαi(GTPγS) (PDB ID code 1GIA) structures identifying the switch sequences (red in GDP and green in GTPγS) and sites where R1 was introduced with orange spheres (Cα and Cβ; GTPγS-bound structure only). Spheres at Cβ show the projection of the native side chain within the structure. The GTPγS ligand is shown as a space-filling model in blue. Switches II (SwII) and III (SwIII) are not resolved (disordered) in the Gαi(GDP) structure and are not shown. Homologous sequences are resolved in the Gαt(GDP) crystal structure and show conformational changes upon nucleotide exchange. The helical domain and P-loop (P) are indicated in yellow and pink, respectively. (B) Ribbon model of Gαi(GDP)βγ (PDB ID code 1GP2) showing positions within Gαi (gray) where R1 was introduced. The switch I segment is green, and Gβ and Gγ are purple and cyan, respectively. (C) Surface representation of the heterotrimer showing stick models of R1 at sites 179, 180, 182, and 186 in Gαi that lie in the cleft between the Gα and Gβ subunits. The contributions of switch I and P-loop residues to the surface are shown in green and pink, respectively, and the Gβ surface is purple. The nearest intramolecular contacts for 180R1 are residues in the P-loop. The I184 side chain makes direct contact with the Gβ subunit and is highlighted in red.
Gα subunits of heterotrimeric G proteins consist of two domains: a GTPase domain, which resembles monomeric G proteins, such as Ras, and a helical domain that buries the guanine nucleotide-binding pocket in the core of the protein. Switch I is a loop that forms one of the two linkers between these domains by connecting the αF-helix of the helical domain to the β2-strand of the GTPase domain (Fig. 1A). Upon exchange of GDP for GTPγS, switch I is drawn toward the nucleotide-binding pocket, establishing stabilizing interactions with Mg2+ and the γ-phosphate (2, 4).
Switch I is located at sites of crystal contact in both the GDP- and GTPγS-bound structures for all Gα structures determined thus far, suggesting the possibility that the conformation of this region may be different in solution. Moreover, crystal structures necessarily hide the important role that protein dynamics can play in regulating protein function. Because switch I plays a crucial role in the structural changes leading to G protein activation, it is important to investigate the native structure and dynamics of this region in solution. Toward this end, SDSL was used to monitor changes in the switch I sequence of Gαi through each transition leading to G protein activation using activated rhodopsin (R*) as the receptor. For each of eight spin-labeled mutants containing the nitroxide side chain R1 (12), EPR spectra were recorded for four states: Gαi(GDP), Gαi(GDP)βγ, R*·Gαi (0)βγ (the “empty complex”), and Gαi(GTPγS). The spectra are characteristic of the local protein structure (14, 15) and encode information on backbone dynamics (16). Thus, the spectra provide information on the solution structure and functional dynamics of the switch I region at each step in G protein activation, enabling comparisons with the crystal structures where available. Most importantly, the data provide insight into conformational changes involving switch I that accompany the formation of the receptor-bound empty complex.
Results
Characterization of Spin-Labeled Mutants.
Sites selected for the introduction of R1 in the switch I sequence (177–187) were residues 179, 180, and 182 in the switch I loop and residues 184, 186, and 187 in the β2-strand. In addition, R1 was introduced at residue 171 in a contiguous sequence of the helical domain on the αF-helix and at residue 50 on the adjacent α1-helix. Fig. 1A shows the position of these residues in the Gαi subunit. Both Cα and Cβ carbons are shown as spheres to illustrate the direction that the native side chain projects in the structure. Except for buried residues 50 and 187, these sites are on the protein surface. Importantly, none of the side chains mutated in the switch I loop contribute directly to nucleotide binding.
All of the spin-labeled mutants formed stable R*–G protein complexes, and all mutants demonstrate R*-catalyzed increases in GTPγS binding, except site 184R1 [supporting information (SI) Fig. 4]. Because residue 184 contributes directly to the Gβ-binding surface (Fig. 1C), this result is likely due to the reduced affinity of this spin-labeled mutant for Gβγ (see below) and the low protein concentrations used in the nucleotide exchange assays. Clearly, this mutant does bind Gβγ and R* in a GTPγS-dependent manner, as demonstrated both by the rhodopsin binding assay (SI Fig. 4) and EPR spectral changes (see below), where higher protein concentrations are used. Interestingly, three of the spin-labeled mutants (residues 171, 182, and 184) had substantially faster rates of basal nucleotide exchange compared with the cysteine-depleted Gαi base mutant (SI Fig. 4). After biochemical characterization, a series of EPR spectra were recorded for these mutants at each step in the G protein cycle from Gαi(GDP) to Gαi(GTPγS) (Fig. 2).
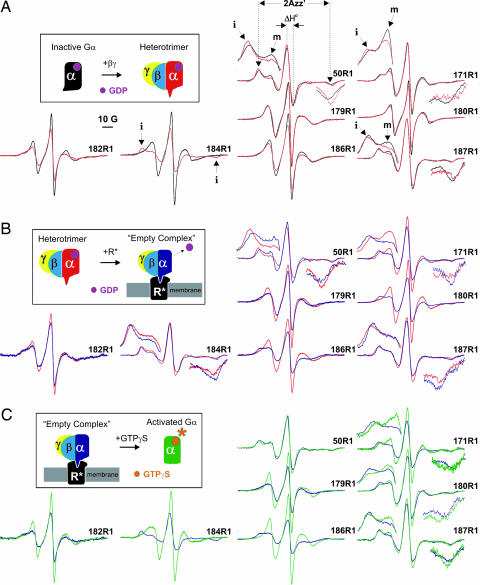
Fig. 2.
EPR spectra for each spin-labeled Gαi mutant along the activation pathway. The diagrams show each Gαi conformation in a different color; the spectra are colored to match the corresponding Gαi conformation. The spectra in each pair are normalized to the same number of nitroxide spins so that comparing relative intensities reveals changes in line width. In some cases, low and high field regions of the spectra have been expanded to clearly illustrate changes in the outer hyperfine extrema. The x axis (magnetic field) was expanded by a factor of two, whereas y-axis (intensity) expansion was arbitrary. (A) EPR spectral changes observed upon heterotrimer formation. (B) Receptor-induced changes in the heterotrimer. (C) Formation of the activated Gαi(GTPγS) subunit.
In the present study, we qualitatively interpret the EPR spectra in terms of a generalized “mobility” of R1 on the nanosecond time scale as reflected by the peak-to-peak width of the central resonance lines (ΔH°) in the first derivative spectra and the splitting of hyperfine extrema where resolved (2Azz′; Fig. 2A) (17, 18). Increases in either of these quantities are correlated with decreases in mobility and vice versa. For each pair of spectra compared, the spectra are normalized to the same number of nitroxide spins, and increases and decreases in ΔH° are recognized by decreases and increases in spectral intensity.
Gαi(GDP).
The spectra for Gαi(GDP) mutants 179R1, 180R1, 182R1, 184R1, and 186R1 of switch I all have dominant components corresponding to fast (τ ≈ 2 ns) motion of R1 (Fig. 2A, black traces), and, with the exception of 179R1, the motion is essentially isotropic. The crystal structure predicts that the R1 side chain at each of these sites projects into solution (Fig. 1A), consistent with the highly mobile nitroxide observed by EPR. If the loop structure containing residues 180 and 182 and the edge strand of the β-sheet containing residues 184 and 186 were rigid, the spectra would be expected to reflect an ordered anisotropic motion (19–21) similar to that for R1 on rigid helices (22). Thus, the essentially isotropic motion of R1 at these sites suggests a flexible backbone in solution (16). Although the switch I sequence is at a lattice contact in the crystal structure of Gαi(GDP) (4), the thermal B factors for Cα are relatively high through the sequence. Indeed, residue 184 has one of the highest B factors in the structure. Thus, the EPR spectra are generally compatible with expectations based on the crystal structure and emphasize the flexibility of the backbone throughout the 179–186 sequence in solution.
The remaining sites, 50R1, 171R1, and 187R1, exhibit complex, multicomponent spectra reflecting both immobilized (i) and mobile (m) states of R1 (Fig. 2A, arrows) (14, 15). Again, this finding is consistent with the location of these sites in the structure; both 50R1 and 187R1 project into the same interior space in the protein fold (Fig. 1B) and are largely immobilized by tertiary interactions (14, 15). Residue 171R1 is the most solvent-exposed of these three residues and has the largest fraction of a mobile component but still forms immobilizing interactions with the α1-helix across the interdomain cleft (Fig. 1A).
Heterotrimer Formation.
Switch I contributes to the Gβ-binding surface on Gα (Fig. 1B), and changes are observed in the EPR spectra of R1 at some sites in the switch I sequence upon heterotrimer formation. In particular, residues 182R1, 184R1, and 186R1 report a decrease in side chain mobility as revealed by an increase in ΔH° and a concomitant decrease in intensity of the normalized spectra (Fig. 2A, red trace). These features are most evident for residue 184R1, which reports the addition of a new spectral component corresponding to an immobilized state of R1 (Fig. 2A, arrow). The spectral changes at 182R1 and 186R1 are more subtle, where the spectra remain dominated by components corresponding to mobile states of the nitroxide. Whereas 184R1 makes direct contact with the Gβ subunit, 182R1 and 186R1 project into the cleft between Gα and Gβ without forming contacts with Gβ (Fig. 1C). The small decrease in mobility of these latter two residues may be accounted for by a damping of backbone motion due to Gβ contacts at adjacent residues (182–184). Not surprisingly, the presence of R1 at the 184 contact site reduces the affinity of Gαi for Gβγ. A titration of 184R1 with increasing amounts of Gβγ revealed that a 10-fold excess of Gβγ was required to produce the full spectral change shown in Fig. 2A, whereas stoichiometric quantities sufficed for the others.
Residues 179R1 and 180R1 do not report significant changes in mobility upon addition of Gβγ (Fig. 2A), consistent with the crystal structure where these residues project into the intersubunit cleft far from the contact surface (Fig. 1C). The immobilized components in the spectra of 50R1, 171R1, and 187R1 show a small increase in 2Azz′ corresponding to a reduction in mobility upon heterotrimer formation (Fig. 2A). Interpreting this change is problematic for these immobilized states of R1 because the decrease in the rotational correlation time of the Gαi subunit (τR ≈ 16 ns) upon formation of the heterotrimer (τR ≈ 66 ns) likely contributes to the observed spectral changes. On the other hand, for mobile states of R1, where the correlation time of the internal motion of R1 (τi ≈ 2 ns) is substantially shorter than the rotational diffusion time of the Gαi subunit, protein rotational diffusion does not substantially influence the spectral line shape. This point is illustrated by the absence of spectral changes upon heterotrimer formation at 179R1 and 180R1, sites where R1 has high mobility. Thus, the decreases in intensity of the mobile components in 50R1, 187R1, and 171R1 suggest decreases in mobility due to changes in protein structure. Inspection of the heterotrimer crystal structure shows that Gαi binding to Gβγ moves the β2-strand with residue 187R1 closer to the α1-helix and residue 50R1 (7), which may account for the decrease in mobility of these sites in the more mobile state. No obvious structural changes near residue 171 due to heterotrimer formation are evident from a comparison of the crystal structures.
Receptor Activation-Dependent Conformational Changes.
Photoactivation of rhodopsin results in G protein binding and GDP release. The R*–G protein complex containing nucleotide-free Gα conformer, for which there is no crystal structure, is stable in the absence of guanine nucleotides. Each of the spin-labeled mutants shows wild-type levels of binding under the conditions of these experiments (SI Fig. 4).
An interesting pattern of spectral changes involving the mobile R1 surface residues emerges upon formation of the R*–G protein complex (Fig. 2B). The mobility of the nitroxide at sites 182R1 and 186R1 decreases upon receptor binding. Near the empty nucleotide-binding pocket, there are striking decreases in the mobility of R1 at residues 180 and 179. As discussed above, the decreases in the mobility of these initially mobile sites cannot be due to changes in τR upon formation of the R*–G protein complex, but must be due to changes in the internal structure or interactions of the Gαi subunit.
Changes in the complex multicomponent spectra of 50R1, 171R1, 184R1, and 187R1 also are evident upon empty complex formation. Spectra of buried residues 50R1 and 187R1 show decreases in 2Azz′ that suggest increased mobility. Because this effect is opposite that expected from an increase in τR, these changes reflect alterations in structure consistent with decreased packing in the cavity shared by these residues. For 184R1, immobilized at the Gαi–Gβ interface, an increase in 2Azz′ is observed that may have contributions from the increase in τR. However, the persistence of an immobilized state of 184R1 indicates that the contact interface of the heterotrimer is retained in the empty complex. Most importantly, residue 171R1 becomes further immobilized upon binding R*. Although the increases in 2Azz′ of the immobilized component may contain some contribution from the increase in τR, the decreased intensity of the mobile component signals a decrease in mobility due to a change in the internal structure. This change in environment at 171R1 demonstrates that the receptor-mediated conformational changes are propagated to the αF-helix distant from the receptor-binding surface.
Activated Gαi(GTPγS).
Addition of GTPγS to the R*-bound, nucleotide-free G protein causes a conformational change leading to the dissociation of the activated Gα(GTPγS) subunit from Gβγ and the receptor. Upon dissociation of Gα(GTPγS) from the complex, the EPR spectra for all sites except for 50R1 reveal an increase in R1 mobility (Fig. 2C). A comparison of the resulting Gαi(GTPγS) spectra (Fig. 2C, green traces) with those of Gαi(GDP) (Fig. 2A, black traces) reveals changes that accompany activation of the Gαi subunit. Because this comparison is made between species of the same molecular weight and, hence, the same τR, EPR spectral changes reflect exclusively changes in protein structure. For ease of comparison, overlays of the spectra for Gαi(GDP) and Gαi(GTPγS) are provided in SI Fig. 5.
The spectra of 171R1, 182R1, and 184R1 are very similar or identical in the Gαi(GDP) and Gαi(GTPγS) forms (Fig. 3 and SI Fig. 5). For 50R1 and 187R1, a slight mobility decrease and increase, respectively, is indicated by changes in 2Azz′. The spectra of 179R1 and 186R1 have similar line shapes in both forms but reflect an increased mobility in Gαi(GTPγS) relative to Gαi(GDP). Residue 180R1 is unique among the switch I residues because R1 is substantially more immobilized in Gαi(GTPγS) compared with Gαi(GDP). We consider a model to account for these differences in Discussion.
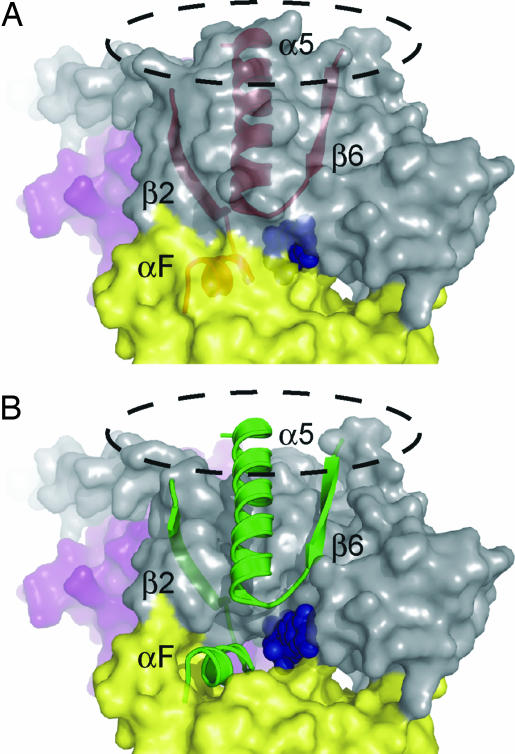
Fig. 3.
Opening the door for GDP release. (A) Transparent surface model of the Gi heterotrimer with the α5/β6 and switch I/αF motifs shown as ribbons. Bound GDP is shown as blue spheres buried in the binding pocket between the GTPase (gray) and helical (yellow) domains. The R*-binding surface is indicated with a dashed line. (B) Removing the side chains from the surface rendering of α5/β6 and αF clearly exposes the nucleotide, suggesting that a movement that rearranges the side chains of these regions could provide a possible exit route for GDP from the interdomain cleft.
Discussion
Crystallographic studies have identified conformational changes in the switch I, II, and III sequences of Gα subunits of heterotrimeric G proteins that accompany G protein activation. However, the mechanism of catalyzed nucleotide exchange is unknown, and elucidation of the mechanism requires information on the conformation of the Gα subunit in the R*–G protein complex. Crystal structures for the complex have not yet been reported, but data regarding the properties of the complex in solution have recently been obtained from NMR (23). The disappearance of many unassigned resonances in the 2D heteronuclear single quantum correlation spectra of Gα in the empty complex relative to the heterotrimer was interpreted as arising from severe broadening of the resonances due to conformational exchange, presumably on the microsecond to millisecond time scale. Although the NMR data may reveal the existence of important conformational exchange processes, they have not identified the specific regions of the protein involved or the molecular details of the conformations involved. In contrast, SDSL can map conformational changes to specific regions of the protein, as illustrated in Results and discussed below.
The present SDSL study is focused on identifying structural changes in the switch I sequence in Gαi accompanying complex formation and complements a similar previous study on switch II (12). For convenience of discussion, the sites examined in this study can be classified in four topographical groups based on the G protein crystal structures (Fig. 1). The first group consists of residues 182R1, 184R1, and 186R1, which are solvent-exposed in Gα(GDP) and are at or near the Gα–Gβ subunit interface in the heterotrimer. The second group is composed of residues 179R1 and 180R1 in the flexible switch I loop away from the Gα–Gβ contact surface and close to the nucleotide-binding pocket. Residues 50R1 and 187R1 of the third group project into the interior of the protein fold of Gαi(GDP) between the α1-helix and β2-strand. Finally, the fourth group contains only residue 171R1, located near the interdomain hinge on the αF-helix. Next, we discuss the structural features of each state of Gαi with respect to these groups.
Switch I Structure and Dynamics in Gαi(GDP), the Heterotrimer, and Gαi(GTPγS).
The high mobility of R1 at sites 182, 184, and 186 in Gαi(GDP) implies a flexible backbone structure, and, as expected, the mobility of these residues decreases upon heterotrimer formation. However, only the spectrum of 184R1 reveals a strongly immobilized component upon Gβγ binding that indicates direct contact with the Gβ subunit, whereas the remaining sites have small decreases in mobility. Modeling the R1 side chain (see Materials and Methods) at 182R1 and 186R1 in the crystal structure reveals that the nitroxides project into the cleft between Gα and Gβ without making contact with Gβ (Fig. 1C), whereas 184R1 must be buried at the interface. As the subunits dissociate after GTPγS binding, the spectra of all three residues return to a state of high mobility similar to that in the Gαi(GDP) state.
In Gαi(GDP), residues 179R1 and 180R1 exhibit a very high degree of side chain mobility that is relatively unaffected by heterotrimer formation, consistent with their location in the crystal structures. Interestingly, the spectra for these residues in the activated, Gαi(GTPγS) conformation are distinct from the spectra for the Gαi(GDP) state and reflect structural differences due to the identity of the bound nucleotide. Indeed, 179R1 is considerably more mobile in Gαi(GTPγS) relative to Gαi(GDP), a result that may be accounted for by the absence of a structural water (HOH 802; PDB ID code 1BOF) in Gαi(GTPγS), which links the backbone of 179 to the Mg2+ ion and the β-phosphate of GDP in Gαi(GDP) (SI Fig. 6). In contrast, 180R1 is considerably more immobilized in the Gαi(GTPγS) state relative to Gαi(GDP). Modeling of 180R1 in the Gαi(GTPγS) structure shows that both the disulfide and nitroxide of R1 can make direct contacts with the sulfur atom of GTPγS, which is absent in Gαi(GDP) (SI Fig. 6), thus accounting for the decreased mobility.
Residues 50R1 and 187R1 of the third group are predominately immobilized due to their location in the interior of the protein fold of Gαi(GDP), but each also has a small population of a mobile state that may arise from a second rotamer of R1 (14). These mobile states decrease mobility upon heterotrimer formation. As we have discussed, the apparent decrease in mobility of the mobile component at both sites, although small, signals internal structure changes. This result may be due to the movement of the β2-strand closer to the α1-helix as observed in the crystal structures. The mobility difference of 50R1 and 187R1 in Gαi(GTPγS) compared with Gαi(GDP) implies structural differences involving the space between the α1-helix and β2-strand that cannot be specified from the limited data.
Finally, residue 171R1 in Gαi(GDP) makes direct contacts with the α1-helix across the interdomain cleft that account for the immobile component of the spectrum. Formation of the heterotrimer results in a general decrease in the mobility of this residue, identifying allosteric changes propagated from the Gβ interaction surface to the distant αF-helix. GTPγS binding reverses the changes observed with heterotrimer formation, indicating that the local structure around 171R1 is similar in the two forms.
Receptor Activation-Dependent Conformational Changes Near Switch I.
The GTPase and helical domains clamp down on the guanine nucleotide, and it has been speculated that to exchange guanine nucleotides there must be a conformational change that opens the cleft (3, 24, 25). Switch I is one of the linkers between the two domains and is connected to the receptor-binding domain at the C terminus and the β2–β3 loop by the β2-strand. Nearly every site examined in the switch I region demonstrated receptor activation-dependent conformational changes. In addition, there are increases in the basal nucleotide exchange for sites 171R1, 182R1, and 184R1, consistent with previous studies by Majumdar et al. (24) showing that Gly→Pro mutations in switch I increase basal GDP release rates in a Gαt/Gαi chimera. Comparing the heterotrimeric versus receptor-bound EPR spectra for sites 182R1, 184R1, and 186R1 near the Gβ interface demonstrates a clear structural rearrangement of the Gαi–Gβ interface. Although the slight decrease in mobility of 184R1 may have contributions from slower protein rotational diffusion, the decreases in mobility of 182R1 and 186R1 unambiguously arise from changes in structure. The location of 182R1 and 186R1 in the structure (Fig. 1C) suggests that new contact interactions may be formed with Gβ, a contention that is supported by the reversal of the spectral changes upon the dissociation of Gαi(GTPγS). These new contact interactions would require a change in the orientation or structure of the subunits in the empty complex compared with the heterotrimer.
In addition to conformational changes at the Gαi–Gβ interface, empty complex formation causes a marked decrease in the mobility of 179R1 and 180R1 near the nucleotide-binding pocket distant from the Gβ contact site. Indeed, residue 180R1 establishes new interactions that give rise to an immobilized state of the side chain. Based on the orientation of the R1 side chain, one likely interaction is with the phosphate-binding loop (P-loop) at the N terminus of the α1-helix (Fig. 1), which would occur, for example, if switch I were to move in a direction to partially occupy the empty nucleotide-binding site. The decrease in the mobility of 179R1 may be due to damping of the backbone motion caused by contact interactions formed at 180R1 with P-loop residues.
Residues 50R1 and 187R1 show distinct increases in mobility upon formation of the empty complex that suggest a decrease in packing in the fold between the α1-helix and β2-strand. Each of the changes discussed above provides direct evidence for structural changes at the Gαi–Gβ interface.
Two models have previously suggested that the activated receptor uses Gβγ to open the nucleotide-binding pocket for GDP release. In the lever-arm model, R* rotates Gβγ away from Gα, pulling switches I and II along with it, thereby causing GDP release (26, 27). The gear-shift model proposes a rotation in the opposite direction that causes close-packing of switches I and II with the core of Gα (28). Although the spectral changes described above do not uniquely describe a particular motion, they generally support a role for intersubunit structural rearrangements in switch I in the formation of the R*-bound, nucleotide-free G protein complex. However, the comparatively small spectral changes observed in switch II upon receptor binding seem to argue against a global rearrangement of the Gα–Gβ interface as in the models described above, but rather suggest allosteric changes as a result of R* binding to Gα.
Site 171R1 is perhaps the most interesting to demonstrate receptor activation-dependent conformational changes. Residue 171 is located in the αF-helix in the hinge between the helical and GTPase domains (Fig. 3). The location of GDP buried deep between these two domains apparently requires an opening in the interdomain cleft to allow GDP release, and one exciting possibility is that 171R1 senses a motion of the αF-helix directly involved in such an opening.
Although the specific structural rearrangement of switch I and contiguous αF-helix cannot be determined from the current data, the changes reported by R1 may constitute one component of a molecular mechanism leading to GDP release from the R*–G protein complex. Specifically, the data presented here suggest that structural changes at the Gα–Gβ interface triggered by receptor binding, either through changes propagated along the α5–β6 loop, down the β2-strand from the β2–β3 loop, or coupled through movements of Gβ, are propagated to the αF-helix. Previously, a rigid body movement of the α5-helix, initiated by direct receptor interaction with the C terminus of Gα, was shown to play a critical role in G protein activation (29). As demonstrated in Fig. 3, the αF-helix and the end of the α5/β6-loop both serve to occlude the nucleotide-binding site. Concerted motion of the αF-helix, perhaps coupled to movement of the entire helical domain, and the α5/β6 motif may thus cooperate in opening a portal for GDP release (Fig. 3). The importance of the αF region in regulating nucleotide exchange is supported by the increased basal nucleotide exchange rate produced by the T171R1 mutation (SI Fig. 4). Although 182R1 and 184R1 had similar effects on the basal exchange rates, their distance from the nucleotide-binding pocket and solvent-facing side chain orientations suggests an indirect effect on GDP binding. Collectively, the results highlight specific allosteric changes at the distant nucleotide-binding pocket triggered by receptor binding.
Summary.
To the extent that they can be compared, SDSL data on the solution structures of Gαi(GDP), Gαi(GDP)βγ, and Gαi(GTPγS) are in excellent agreement with details of the corresponding crystal structures. Most significantly, this study provides structural data for switch I in the empty complex formed with the activated receptor. The results identify a possible allosteric pathway propagated along switch I at the Gα–Gβ interface to the αF-helix, which, like the α5-helix and the α5/β6-loop, forms part of a putative entrance to the nucleotide-binding site. Together with earlier studies that identified movement of the α5-helix coupled directly to receptor binding, the data presented here provide key components of a concerted mechanism of receptor-catalyzed nucleotide exchange.
Materials and Methods
Materials.
GDP and GTPγS were from Sigma–Aldrich (St. Louis, MO). The sulfhydryl spin-label reagent, S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate, was a generous gift from Kalman Hideg (University of Pecs, Pecs, Hungary). All other reagents and chemicals were of the highest available purity.
Preparation of Rod Outer Segment (ROS) Membranes and Gβγ Subunits.
Urea-washed ROS membranes and Gb1g1 were prepared as previously described (28) and stored at −80°C. All ROS and Gβγ samples were buffer-exchanged into 20 mM Mes (pH 6.8)/100 mM NaCl/2 mM MgCl2/10% glycerol before EPR experiments.
Construction, Expression, and Purification of Mutant Proteins.
Briefly, we used a plasmid encoding Gαi that contained six amino acid substitutions at solvent-exposed cysteine residues (C3S-C66A-C214S-C305S-C325A-C351I) and a hexahistidine tag between amino acid residues M119 and T120 (30). This construct served as a template for introducing individual cysteine substitutions by using the QuikChange system (Stratagene, La Jolla, CA). All mutations were confirmed by DNA sequencing (DNA Sequencing Facility, Vanderbilt University). The mutant constructs were then transformed in Escherichia coli BL21-Gold (DE3) (Stratagene), expressed, and purified as previously described (30).
Spin-Labeling, EPR Spectroscopy, and Modeling of the R1 Side Chain.
Spin-labeling was carried out in a buffer containing 20 mM Mes (pH 6.8), 100 mM NaCl, 2 mM MgCl2, 50 μM GDP, and 10% (vol/vol) glycerol. The Gαi mutants were incubated with S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate at a 1:1 molar ratio at room temperature for 5 min. Under these conditions, only the most reactive cysteine residues were modified, and the remaining buried native cysteine residues were unreactive (30). Any excess spin-labeling reagent was removed by extensive washing with buffer using a 30-kDa molecular mass concentrator. For EPR spectroscopy, a series of spectra were recorded for each spin-labeled mutant. First, Gαi mutants (30 μM) were loaded into a sealed quartz flat cell, and spectra were recorded at room temperature on an E580 spectrometer (Bruker BioSpin, Billerica, MA) using a high-sensitivity resonator at X-band microwave frequency. The data were typically averages of 20 to 50 scans. Except where noted otherwise, Gβγ was then added in a 1:1 molar ratio to form heterotrimers. The diluted samples were concentrated to the same concentration as the initial Gαi mutants, and the EPR spectra were recorded both alone in solution and upon addition of urea-washed ROS membranes in the dark (150 μM). The sample was subsequently irradiated for 30 sec by using a tungsten lamp (cutoff filter; λ > 500 nm), and the EPR spectra were recorded immediately after bleaching. Finally, 200 μM GTPγS was added to the samples, and the EPR spectra were recorded.
The R1 side chain was modeled by using rotamers defined by the dihedral angles of the first two bonds of the side chain, starting at the backbone (X1, X2). The preferred rotamers, obtained in crystal structures of R1 and derivatives in T4 Lysozyme, are (−60°, −60°) and (180°, +60°) (refs. 14 and 19 and M. Fleissner, D. Cascio, K. Hideg, and W.L.H., unpublished data). Several structures from the unpublished work that illustrate the rotamers have been deposited (PDB ID codes 1ZWN, 1ZYT, 2A4T, and 2CUU). The particular rotamer and the value of X3 (the disulfide dihedral) were selected to minimize steric overlaps in the protein.
Acknowledgments
This work was supported by grants from the National Institutes of Health (to W.L.H. and H.E.H.), a Public Heath Service Award for the Medical Scientist Training Program (to W.M.O.), the Pharmaceutical Research and Manufacturers of America Foundation (W.M.O.), a Ruth L. Kirschstein National Research Service Award (to N.V.E.), and the Jules Stein Professorship (to W.L.H.).
Abbreviations
- PDB
Protein Data Bank
- P-loop
phosphate-binding loop
- R*
activated rhodopsin
- ROS
rod outer segment
- SDSL
site-directed spin-labeling.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702623104/DC1.
References
- 1.Oldham WM, Hamm EH. Q Rev Biophys. 2006:1–50. doi: 10.1017/S0033583506004306. [DOI] [PubMed] [Google Scholar]
- 2.Lambright DG, Noel JP, Hamm HE, Sigler PB. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 3.Noel JP, Hamm HE, Sigler PB. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 4.Mixon MB, Lee E, Coleman DE, Berghuis AM, Gilman AG, Sprang SR. Science. 1995;270:954–960. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 5.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 6.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 7.Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Singer WD, Sternweis PC, Sprang SR. Nat Struct Mol Biol. 2005;12:191–197. doi: 10.1038/nsmb888. [DOI] [PubMed] [Google Scholar]
- 9.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 10.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 11.Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. Nature. 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 12.Van Eps N, Oldham WM, Hamm HE, Hubbell WL. Proc Natl Acad Sci USA. 2006;103:16194–16199. doi: 10.1073/pnas.0607972103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 14.Langen R, Oh KJ, Cascio D, Hubbell WL. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 15.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 16.Columbus L, Hubbell WL. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 17.Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. J Mol Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Kusnetzow AK, Altenbach C, Hubbell WL. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Protein Sci. 2007 doi: 10.1110/ps.062739107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lietzow MA, Hubbell WL. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 21.Columbus L, Hubbell WL. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 22.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 23.Abdulaev NG, Ngo T, Ramon E, Brabazon DM, Marino JP, Ridge KD. Biochemistry. 2006;45:12986–12997. doi: 10.1021/bi061088h. [DOI] [PubMed] [Google Scholar]
- 24.Majumdar S, Ramachandran S, Cerione RA. J Biol Chem. 2004;279:40137–40145. doi: 10.1074/jbc.M405420200. [DOI] [PubMed] [Google Scholar]
- 25.Hamm HE. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 26.Iiri T, Farfel Z, Bourne HR. Nature. 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 27.Rondard P, Iiri T, Srinivasan S, Meng E, Fujita T, Bourne HR. Proc Natl Acad Sci USA. 2001;98:6150–6155. doi: 10.1073/pnas.101136198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherfils J, Chabre M. Trends Biochem Sci. 2003;28:13–17. doi: 10.1016/s0968-0004(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 29.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 30.Medkova M, Preininger AM, Yu NJ, Hubbell WL, Hamm HE. Biochemistry. 2002;41:9962–9972. doi: 10.1021/bi0255726. [DOI] [PubMed] [Google Scholar]