Abstract
Maintenance of ATP levels is a critical feature of all cells. Mitochondria are responsible for most ATP synthesis in eukaryotes. We show here that mammalian cells respond to a partial chemical uncoupling of mitochondrial oxidative phosphorylation with a decrease in ATP levels, which recovers over several hours to control levels. This recovery occurs through an increased expression of the transcriptional coactivator peroxisome proliferator-activated receptor-coactivator 1α (PGC-1α) and mitochondrial genes. Cells and animals lacking PGC-1α lose this compensatory mechanism and cannot defend their ATP levels or increase mitochondrial gene expression in response to reduced oxidative phosphorylation. The induction of PGC-1α and its mitochondrial target genes is triggered by a burst of intracellular calcium, which causes an increase in cAMP-response-element-binding protein and transducer of regulated cAMP-response-element-binding proteins actions on the PGC-1α promoter. These data illustrate a fundamental transcriptional cycle that provides homeostatic control of cellular ATP. In light of this compensatory system that limits the toxicity of mild uncoupling, the use of chemical uncoupling of mitochondria as a means of treating obesity should be re-evaluated.
Keywords: mitochondria, uncoupling
Mitochondria are responsible for producing most of the ATP needed for energy-requiring reactions in eukaryotic cells (1). In the process of oxidative phosphorylation (OXPHOS), a proton gradient across the inner mitochondrial membrane is coupled to the synthesis of ATP by the F1F0ATPase at complex V. Mitochondrial metabolism is dynamic and can be modulated in response to external stimuli. For example, the β-adrenergic pathway is activated upon cold exposure in mice, which causes brown adipose tissue to shift toward more uncoupled respiration (2–4). In uncoupled respiration, the protons pumped into the intermembrane space leak back into the mitochondrial matrix, bypassing ATP production and generating heat.
Mitochondria themselves can elicit intracellular signaling pathways that alter nuclear gene expression, mitochondria number, and function (5, 6). This signaling process is referred to as retrograde signaling. Retrograde signaling has been well studied in the budding yeast, Saccharomyces cerevisiae. Yeast lacking mtDNA show increased expression of many genes important for the function of mitochondria, such as citrate synthase, and several proteins have been identified in yeast that regulate retrograde signaling (7). Little is known about the regulatory proteins involved in the mechanisms of retrograde signaling in higher eukaryotes. Similarly, how and to what extent higher eukaryotic cells can defend their ATP levels during chronic challenges has not been extensively explored.
Peroxisome proliferator-activated receptor-coactivator 1α (PGC-1α) was first identified as a cold-inducible transcriptional coactivator of peroxisome proliferator-activated receptor γ that can activate a program of adaptive thermogenesis (8). PGC-1α has since been shown to be a dominant regulator of mitochondrial function, biogenesis, and respiration in many tissues (9–13). Ectopic expression of PGC-1α in white adipocytes increases cellular respiration and genes important for mitochondrial function, such as uncoupling protein 1, cytochrome c (Cyt c), and Cyt c oxidase subunit II (COX II). Furthermore, increased expression of PGC-1α increases mitochondrial volume density and cristae density (14). PGC-1α also can activate a program of fiber-type switching in skeletal muscle, including increased mitochondrial content and the expression of myofibrillar proteins characteristic of type 1 and type 2a muscle fibers (15). PGC-1β, the closest homolog of PGC-1α, has also been shown to regulate mitochondrial biogenesis and respiration in cells and transgenic animals (13, 16, 17).
2,4-Dinitrophenol (DNP), a chemical mitochondrial uncoupler, was widely used in the 1930s as a treatment for obesity (18–20). Mild doses were amazingly effective, often causing rapid loss of adipose mass and body weight. While unregulated use led to reports of toxicity and even death, it was not as lethal as might have been expected for a compound that disrupted OXPHOS. This tolerance indicates that cells can sustain mild chemical uncoupling, perhaps by using an energy compensatory mechanism. In the present study, we have investigated whether uncoupling of the mitochondrial membrane potential could activate a compensatory pathway controlling ATP homeostasis. Indeed, chemical uncoupling increases the expression of PGC-1α and PGC-1β, as well as the expression of several target mitochondrial genes. Studies using PGC-1α null cells and animals indicate that PGC-1α is required for the induction of mitochondrial gene expression, recovery of ATP levels, and cell survival. Together, these data illustrate a fundamental system of energy homeostasis whereby cells and tissues use PGC-1α to recover energy balance and also demonstrate that a powerful compensatory system exists that limits the toxicity of mild uncoupling.
Results
Chemical Uncoupling Causes a Transient Drop in ATP Levels.
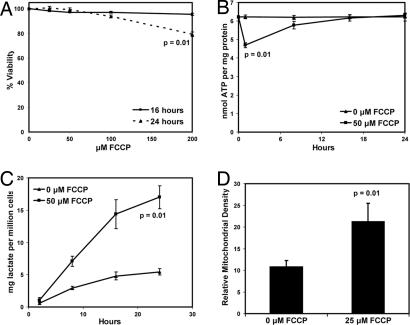
To examine whether and how mammalian cells might adapt to the chemical uncoupling of mitochondrial OXPHOS, we measured ATP levels and cell death in fibroblasts treated with the chemical mitochondrial uncoupler carbonyl cyanide p-(trifluromethoxy)phenyl hydrazone (FCCP) (21). Perhaps surprisingly, fibroblasts treated with up to 200 μM FCCP for 16 h remained 98% viable (Fig. 1A), although viability decreased to ≈80% in the cells treated with 200 μM FCCP for 24 h. Treatment of fibroblasts with FCCP caused a 25% in ATP levels at 30 min of treatment (Fig. 1B), but this drop was transient, as ATP levels later recovered to those of the untreated cells. We also examined the amount of lactate produced during this experiment. Over 24 h, cells treated with FCCP secreted four times more lactate than the untreated cells (Fig. 1C). This finding suggests that glycolytic metabolism helps cells withstand a decrease in OXPHOS caused by chemical uncoupling.
Fig. 1.
Chemical uncoupling induces multiple metabolic changes. (A) Cells were treated with 0, 25, 50, 100, or 200 μM FCCP for 16 or 24 h, and viability was measured by using the LDH assay (P = 0.01 compared with untreated cells at 24 h). (B) ATP levels were measured after fibroblasts were treated with 0 or 50 μM FCCP for 0, 2, 8, 16, or 24 h (P = 0.01 compared with the untreated cells at 0.5 h). (C) Lactate was measured after fibroblasts were treated with 0 or 50 μM FCCP for 2, 8, 16, or 24 h (P = 0.01 compared with the untreated cells). (D) Mitochondrial volume density in cells treated with 0 and 25 μM FCCP for 72 h (P = 0.01).
We asked whether this mitochondrial uncoupling had any affect on mitochondrial density by using electron microscopy to quantify the volume of mitochondria in cells treated with FCCP. Fibroblasts were exposed to 0 or 25 μM FCCP for 72 h, and mitochondrial volume density was measured as described (14, 22). Fibroblasts treated with FCCP displayed a 1.8-fold higher mitochondrial volume than control cells and higher levels of Cyt c protein [Fig. 1D and supporting information (SI) Fig. 7], suggesting that chronic uncoupling induces a compensatory program that not only helps to recover ATP levels, but alters mitochondrial density well.
Partial Chemical Uncoupling of Mitochondria Induces PGC-1α, PGC-1β, and Mitochondrial OXPHOS Genes.
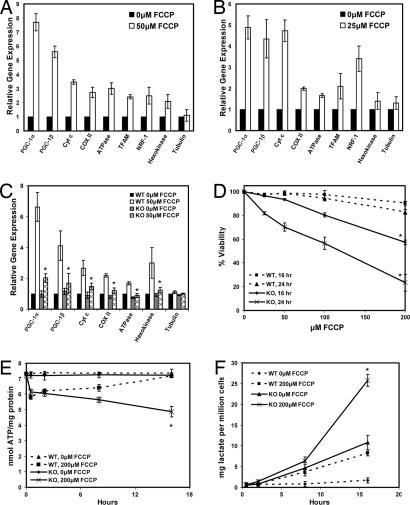
To determine whether changes in the expression of genes related to mitochondrial function accompany the metabolic alterations seen in cells treated with FCCP, we measured the mRNAs encoding the PGC-1 coactivators, powerful regulators of mitochondrial gene expression, and expression of genes of the electron transport system. Using real-time PCR, we observed an induction of mRNAs encoding for Cyt c, COX II, F1β-ATPase, and nuclear respiratory factor 1 (NRF-1). In addition, PGC-1α and PGC-1β mRNA increased 7.5- and 5.5-fold, respectively, upon treatment with FCCP for 16 h (Fig. 2A). In extended treatment with FCCP for 72 h, mRNA for PGC-1α and PGC-1β was elevated 4-fold, and the mRNAs encoding Cyt c, COX II, F1β-ATPase, and NRF-1 were also significantly elevated relative to vehicle-treated cells (Fig. 2B). To determine whether a different mitochondrial uncoupler could also increase these genes, fibroblasts were treated with DNP. Sixteen hours of treatment with DNP increased PGC-1α and PGC-1β mRNA ≈7-fold; mRNAs for Cyt c, COX II, F1β-ATPase, and hexokinase were similarly induced (SI Fig. 8). These data indicate that chemical uncoupling of the mitochondrial electron transport chain induces genes of mitochondrial OXPHOS and mRNA encoding the PGC-1 coactivators.
Fig. 2.
Chemical uncoupling induces OXPHOS genes and the PGC-1 coactivators. (A) Fibroblasts were treated with 0 or 50 μM FCCP for 16 h. RNA was measured by using real-time PCR (all points are significant except for tubulin, P < 0.01). (B) The experiment in A was repeated, but cells were treated for 72 h with 0 or 25 μM FCCP (all points are significant except for tubulin and hexokinase, P < 0.01). (C) WT and PGC-1α−/− preadipocytes (KO) were treated with 0 or 50 μM FCCP for 16 h, and mRNAs were measured. The PGC-1α measurement represents exon 2, which is still present in the KO cells (P < 0.05 compared with the WT FCCP-treated cells). (D) WT and PGC-1α KO preadipocytes were treated with 0, 25, 50, 100, and 200 μM FCCP for 16 or 24 h. Viability was measured by using the LDH cytotoxicity assay (P < 0.01 compared with the WT FCCP-treated cells). (E) ATP levels in the WT and PGC-1α KO cells were measured after treatment with 50 μM FCCP (P = 0.02 compared with the WT FCCP-untreated cells at 16 h). (F) Lactate levels in the media of WT and KO cells treated with 200 μM FCCP for the were measured (P = 0.005 compared with the WT FCCP-treated cells at 16 h).
PGC-1α Is Required for the Induction of Mitochondrial Gene Expression, Recovery of ATP Levels, and Cell Survival.
We investigated whether the induction of PGC-1α is required for the uncoupling-mediated induction of the compensatory metabolic program. Immortalized preadipocytes isolated from the brown fat of WT and PGC-1α−/− mice (23) were treated with FCCP for 16 h. Similar to what was seen in fibroblasts, this treatment induced a significant increase in the expression of mRNAs encoding PGC-1α, PGC-1β, and several genes of the electron transport system (Fig. 2C). Strikingly, cells lacking PGC-1α had an almost complete ablation of the induction of PGC-1β and OXPHOS genes, which indicates that PGC-1α is almost absolutely required for the induction of the mitochondrial electron transport genes induced by partial mitochondrial uncoupling.
To determine whether PGC-1α contributes to cell survival during reduced mitochondrial function, a cytotoxicity assay was used to examine cell viability after treatment with FCCP in both the WT and PGC-1α null cells. Whereas the WT cells showed very little death with up to 200 μM FCCP at either time point, the PGC-1α null cells showed a very substantial loss of viability (Fig. 2D). Only 80% and 60% of the mutant cells survived 100 and 200 μM FCCP, respectively, after 16 h. After 24 h, only 30% of the mutant cells survived 200 μM FCCP compared with 85% of control cells. Thus, PGC-1α contributes substantially to the ability of these cells to survive partial uncoupling of the electron transport chain.
We next studied whether PGC-1α plays a role in the recovery of the ATP levels seen after treatment with FCCP (Fig. 1B). Both WT and PGC-1α−/− cells displayed an initial 20% decrease in ATP levels (Fig. 2E). After 16 h, the WT cells completely recovered their ATP levels to those of the untreated cells, whereas the PGC-1α−/− cells could not restore their ATP levels. By 16 h, the ATP levels in the mutant cells were ≈68% of the ATP levels in the untreated cells. In the same experiments, we measured lactate levels produced. After 16 h, the FCCP-treated WT cells secreted approximately three times more lactate than the untreated WT cells (Fig. 2F). The untreated PGC-1α−/− cells secreted significantly more lactate than both the treated and untreated WT cells, which was also true of the FCCP-treated PGC-1α−/− cells. These experiments together indicate that the recovery of ATP levels and cell survival seen after chronic treatment with FCCP requires PGC-1α.
Intracellular Calcium Plays a Critical Role in the PGC-1α Pathway.
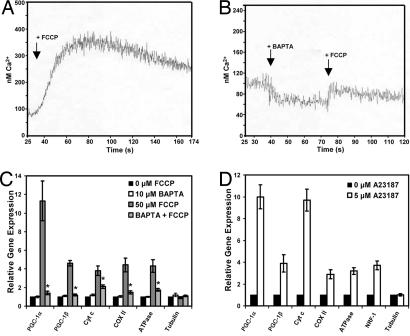
Mitochondria store significant quantities of calcium, and mitochondrial uncoupling is known to cause an increase in intracellular calcium ([Ca2+]i) levels (6, 24–28). These data are of particular interest because the induction of PGC-1α in other contexts has been shown to be regulated by components of the calcium signaling pathways (17, 29). 1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA), a membrane soluble calcium chelator (30), has been shown to block the rise in [Ca2+]i caused by the uncoupling agent FCCP (24, 31–33). Using the calcium-sensitive dye Fura-2 (34), we observed a 3-fold increase in [Ca2+]i when fibroblasts were treated with FCCP (Fig. 3A). Pretreatment of cells with BAPTA largely suppressed this rise in [Ca2+]i (Fig. 3B). To determine whether reducing the FCCP-mediated [Ca2+]i rise with BAPTA could block the expression of PGC-1α and the other OXPHOS genes, fibroblasts were pretreated with BAPTA before treatment with FCCP. Indeed, the rise in PGC-1α mRNA caused by FCCP was completely blocked (Fig. 3C); the increases in the mRNAs for Cyt c, COX II, and F1β-ATPase caused by FCCP were also ablated.
Fig. 3.
The induction of PGC-1α, PGC-1β, and OXPHOS genes requires increased [Ca2+]i levels. (A) Fibroblasts were loaded with Fura-2, and [Ca2+]i levels were measured (34, 54) as 50 μM FCCP was added. (B) [Ca2+]i levels were measured as in A first with the addition 10 μM BAPTA and the subsequent addition of FCCP. (C) Fibroblasts were pretreated for 1 h with 0 or 10 μM BAPTA followed by 16 h of treatment with 0 or 50 μM FCCP, and mRNAs were measured (P < 0.01 compared with the FCCP-treated cells). (D) Fibroblasts were treated with 0 or 5 μM A23187 for 16 h (all points are significant except for tubulin, P < 0.01), and mRNAs were measured.
To test whether compounds that increased [Ca2+]i levels had the same effect on PGC-1α and mitochondrial gene expression as mitochondrial uncouplers, we treated fibroblasts for 16 h with the calcium ionophore A23187. As seen after treatment with FCCP, the mRNAs for PGC-1α, PGC-1β, and the mRNAs for Cyt c, COX II, F1β-ATPase, and NRF-1 all were increased (Fig. 3D). These data strongly suggest that altered calcium flux is a key component in the mechanism of this mitochondrial retrograde pathway regulated by PGC-1α.
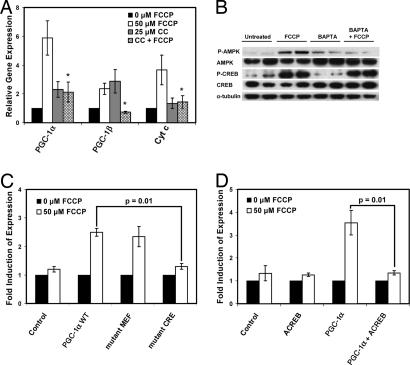
AMP-activated protein kinase (AMPK) is activated during low energy conditions, particularly when the ratio of AMP/ATP increases (35). AMPK activation is also known to induce PGC-1α gene expression in muscle (36, 37). As expected, AMPK is activated upon treatment with FCCP (Fig. 4B) (33, 38). To investigate the role of AMPK in the uncoupling-induced homeostatic pathway, we inhibited AMPK and looked at the subsequent induction of PGC-1α and OXPHOS gene expression. Fibroblasts were pretreated with the AMPK inhibitor compound C for 1 h, followed by treatment with FCCP. Whereas FCCP alone induced PGC-1α, PGC-1β, and Cyt c mRNA levels, the addition of compound C almost completely blocked the further induction of those genes (Fig. 4A). However, pretreatment of cells with BAPTA blocks the activation of AMPK (Fig. 4B), suggesting that calcium signaling is upstream of the activation of AMPK by FCCP.
Fig. 4.
AMPK and CREB are involved in the mechanism inducing PGC-1α. (A) Fibroblasts were pretreated for 1 h with 0 or 25 μM compound C (CC) followed by 16 h of treatment with 0 or 50 μM FCCP. mRNAs were measured (P < 0.02 compared with FCCP-treated cells). (B) Fibroblasts were treated as in Fig. 3C. Western blots were used to measure protein levels. (C) PGC-1α promoter constructs with a luciferase reporter containing mutations in the MEF or CRE site were transfected into fibroblasts (PGC-1α WT is the unmutated PGC-1α 2-kb promoter), then treated with FCCP. Luciferase units were normalized to β-galactosidase and to the cells treated with 0 μM FCCP (P = 0.01). (D) ACREB was cotransfected with the PGC-1α 2-kb promoter into fibroblasts. The cells were treated with FCCP, and luciferase units were measured and normalized as in C (P = 0.01).
Involvement of cAMP-Response-Element-Binding Protein (CREB) in the Induction of the PGC-1α Promoter.
To elucidate the mechanism of this energy cycle further, we investigated the cis- and trans-acting factors that play a role in the induction of the PGC-1α gene upon chemical uncoupling. Activity of a promoter segment containing 2 kb of the 5′ flanking region of the PGC-1α promoter driving a luciferase gene (29) was induced 3-fold upon treatment with FCCP (Fig. 4C). Transfections of the PGC-1α promoter harboring mutations in the MEF2 and CREB binding sites were analyzed. Promoter constructs with a mutation in the MEF2 binding site showed no difference in activity compared with the WT PGC-1α promoter (Fig. 4C). However, mutation of the CREB binding site (39) resulted in an almost complete ablation of the increase in the PGC-1α promoter activity in response to FCCP.
To investigate the role of the transcription factor CREB in the FCCP-mediated induction of the PGC-1α promoter, we examined the induction of the WT 2-kb PGC-1α promoter in the presence of a dominant-negative version of the CREB protein (ACREB) (40). ACREB, a fusion of an acidic amphipathic domain and the leucine zipper dimerization domain of CREB, heterodimerizes with endogenous CREB and prevents it from binding to DNA. ACREB completely blocked the induction of the PGC-1α promoter observed upon treatment with 50 μM FCCP (Fig. 4D). Typically CREB protein requires activation by phosphorylation, often at Ser-133. Fig. 4B shows that treatment with FCCP increases the regulatory Ser-133 phosphorylation of CREB. However, BAPTA, which blocks the induction of PGC-1α mRNA and OXPHOS gene expression, does not block the phosphorylation of CREB. These data together suggest a required role for CREB in the control of the PGC-1α promoter, but also suggest other regulated steps in this pathway must exist.
Involvement of Transducer of Regulated CREB Proteins (TORCs) in the Regulation of PGC-1α.
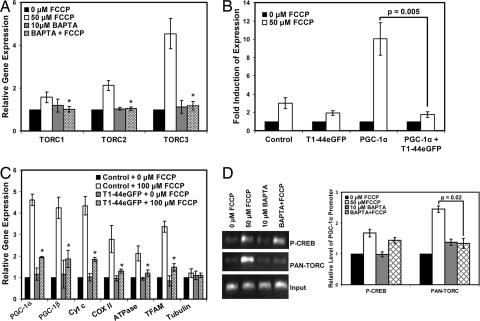
We sought to identify other factors in the calcium signaling pathway that were sensitive to BAPTA. Recently, the TORC coactivators have emerged as important regulators of CREB-dependent transcription, irrespective of the phosphorylation state of CREB (41–48). Given this and their involvement in calcium signaling pathways (43, 44), the TORCs seemed to be plausible regulators in the pathway by which uncoupling induces PGC-1α. To determine whether TORCs are involved in the PGC-1α energy cycle, fibroblasts were treated with FCCP. TORC1 mRNA increased 1.6-fold, TORC2 mRNA increased 2.1-fold, and TORC3 mRNA increased 4.2-fold (Fig. 5A); pretreatment with BAPTA completely blocked the induction of all three TORC mRNAs. To determine whether TORCs play a role in the FCCP-mediated induction of the PGC-1α promoter, we measured the induction of the WT 2-kb PGC-1α promoter in the presence of a dominant-negative version of TORC (43). The dominant-negative TORC protein (T1–44eGFP) is a fusion protein of the conserved CREB binding domain of TORC1 and eGFP. T1–44eGFP has been shown to block the activation of CREB by TORC1, TORC2, and TORC3 (43). In the absence of T1–44eGFP, the PGC-1α promoter was induced 9.5-fold upon treatment with FCCP (Fig. 5B). However, T1–44eGFP completely blocked the FCCP-mediated induction of the PGC-1α promoter, suggesting strongly that TORCs are required for the induction of the PGC-1α promoter under these conditions.
Fig. 5.
TORC plays a critical role in the uncoupling-mediated induction of PGC-1α and mitochondrial genes. (A) Fibroblasts were treated as described in Fig. 3C (P < 0.01 compared with the FCCP-treated samples), and mRNAs for TORC1, TORC2, and TORC3 were measured. (B) Fibroblasts were transfected with the PGC-1α 2-kb promoter, cotransfected with T1–44eGFP, and then treated with FCCP. Luciferase units were normalized to the cells treated with 0 μM FCCP (P = 0.005). (C) Preadipocytes constitutively expressing T1–44eGFP or the eGFP control were treated with either 0 or 50 μM FCCP for 16 h, and mRNAs were measured (P < 0.01 compared with FCCP-treated control cells). (D) ChIP was used to measure phospho-CREB protein and TORC protein bound to the PGC-1α promoter upon treatment with FCCP. Fibroblasts were treated as described in Fig. 3C. Cells were cross-linked, and protein–DNA complexes were harvested by using phospho-CREB or PAN-TORC antibodies. The amount of PGC-1α promoter was then quantified by using PCR and real-time PCR with PGC-1α-specific primers (P = 0.02).
In a similar experiment, preadipocytes constitutively expressing T1–44eGFP were treated with FCCP for 16 h, and expression of several mitochondrial genes was subsequently measured. Control cells showed a 4.6-fold increase in PGC-1α mRNA and increases in PGC-1β, Cyt c, COX II, F1β-ATPase, and mitochondrial transcription factor A (TFAM) mRNAs (Fig. 5C). However, cells expressing T1–44eGFP had a blunted induction of mRNAs for all of these genes, again suggesting that TORCs are required for the PGC-1α energy cycle induced by uncoupling.
ChIP was used to measure the association of TORCs with the endogenous PGC-1α promoter in the presence of FCCP and BAPTA. Fibroblasts were pretreated with BAPTA, followed by treatment with FCCP. DNA was immunoprecipitated by using an anti-phospho-CREB or anti-PAN TORC antibody, and the presence of the PGC-1α promoter was quantified by PCR and real-time PCR with PGC-1α promoter-specific primers. FCCP treatment increased the amount of phospho-CREB and TORC associated with the PGC-1α promoter by ≈1.8- and 2.5-fold, respectively (Fig. 5D). Pretreatment with BAPTA reduced the amount of phospho-CREB to ≈1.4-fold; this same treatment, however, completely blocked the association of TORCs with the PGC-1α promoter. These results indicate that the uncoupling-mediated association of TORCs with the PGC-1α promoter is BAPTA-sensitive and can at least partly explain the mechanism by which the uncoupling of OXPHOS induces PGC-1α.
Uncoupling Induces the PGC-1α Energy Cycle in Vivo.
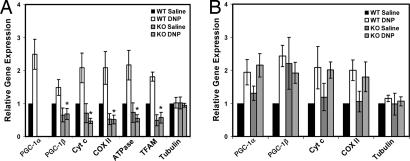
To determine whether chemical uncoupling of mitochondria in vivo also increases the expression levels of PGC-1α and the OXPHOS genes, WT and skeletal muscle-specific PGC-1α knockout mice were treated with 50 mg/kg DNP for 5 h. In the muscle of WT mice, DNP increased the expression of PGC-1α and PGC-1β mRNA 2.5- and 1.4-fold, respectively, in addition to Cyt c, COX II, F1β-ATPase, and TFAM mRNA (Fig. 6A). The mice lacking skeletal muscle PGC-1α had a lower basal level expression of those genes and completely failed to induce PGC-1β, Cyt c, COX II, F1β-ATPase, and TFAM in response to DNP. At the concentrations used, there was a trend toward 10% lower ATP levels in the DNP-treated muscles in mice lacking skeletal muscle PGC-1α, but it fell just below the level of significance (data not shown). Importantly, the genes for PGC-1α, PGC-1β, and the OXPHOS genes were induced in the livers of both WT and muscle-specific knockout mice treated with DNP (Fig. 6B). Furthermore, the mRNA expression levels of PGC-1α, PGC-1β, Cyt c, COX II, and TFAM all were significantly elevated in white adipose tissue as well (data not shown). In brown adipose tissue, only PGC-1α and TFAM were elevated (data not shown). This result indicates that partial mitochondrial uncoupling in vivo induces the genes of OXPHOS, and this response requires the PGC-1α coactivator.
Fig. 6.
The uncoupling-mediated induction of mRNA for PGC-1α, PGC-1β, and mitochondrial genes occurs in vivo. (A and B) WT and muscle-specific PGC-1α−/− female mice were injected with either saline or 50 mg/kg DNP. Skeletal muscle (A) and liver (B) were harvested after 5 h (P < 0.01 compared with the WT DNP-treated mice, n = 8).
Discussion
We show here that cells have a finely tuned homeostatic mechanism to compensate for chronic alteration of the electrochemical gradient across the inner membrane. Perturbation of this gradient by chemical uncouplers results, as expected, in a drop in ATP levels. However, cells recover over several hours to attain their original ATP concentrations. This compensatory mechanism is driven mainly by the induction of PGC-1α mRNA, which then stimulates a rise in PGC-1β and an increase in both mitochondrial gene expression and mitochondrial volume per se. That this mechanism is crucial to the basic survival of cells is illustrated by the cell death and loss of ATP levels that results when PGC-1α−/− cells are challenged with chemical uncoupling. These data also illustrate that increased glycolysis cannot provide sufficient production of ATP because PGC-1α−/− cells actually have a greater rate of glycolysis than control cells in response to uncoupling, but fail to defend their ATP levels.
It is important to note that while some individual components of this pathway have been described in other contexts, such as the release of calcium upon mitochondrial uncoupling (6, 24–28) and the role of TORC proteins on the PGC-1α promoter (47), the merging of these various individual components into a highly integrated regulatory system has not been previously described to our knowledge. Moreover, the striking ability of this system to precisely maintain cellular ATP levels strongly suggests the physiological relevance of this pathway.
The PGC-1α mitochondrial compensatory mechanism appears to be triggered by an early rise in [Ca2+]i levels, which may be further amplified by the release of calcium from the endoplasmic reticulum. Blocking this initial rise in [Ca2+]i with BAPTA stops the induction of PGC-1α completely and also blocks the increases in OXPHOS gene expression. Although AMPK is almost certainly an important player in this mitochondrial system, calcium appears to be the dominant pathway, because the activation of AMPK observed here also depends on the early rise in [Ca2+]i levels. That AMPK can be activated by increases in [Ca2+]i has also been observed by others (33, 49–51). But clearly, both signal transduction pathways, calcium and AMPK, are required for full induction of PGC-1α.
Activated CREB has proven to be an important regulator of the PGC-1α promoter in liver and muscle (17, 29, 39). Although the mitochondrial uncoupling performed in these studies leads to a great increase in phosphorylation of CREB on Ser-133, it is also clear from the BAPTA studies that this phosphorylation is not sufficient to trigger a rise in PGC-1α levels. TORCs, identified recently as important modulators of both CREB function and PGC-1α gene expression (41–48), are induced by chemical uncoupling and absolutely require increased [Ca2+]i levels. Hence, a substantial part of the induction of PGC-1α that triggers this homeostatic mechanism depends on both CREB and the TORC proteins. Activated AMPK is also required for the induction of PGC-1α and mitochondrial gene expression (52). Paradoxically, AMPK has been shown to inhibit TORC2 via preventing nuclear import (45, 46). The role of AMPK on the other TORC proteins and PGC-1α function itself remains to be determined.
Finally, these results strongly suggest a re-evaluation of the potential for using chemical uncoupling as a therapeutic approach to human obesity. Seventy years ago, DNP was widely used to treat obesity (18–20). Unfortunately, its over-the-counter accessibility led to uncontrolled use and reports of toxicity and death, so chemical uncoupling of mitochondria was effectively eliminated as a plausible means of treating obesity. However, mild chronic doses had been remarkably effective and, surprisingly, were much less toxic than might have been expected for a compound that uncoupled OXPHOS. Some of the relatively low toxicity of DNP may be explained by increases in cellular glycolysis and increased electron transport because of altered ADP/ATP ratios, but it is highly likely that the chronic mitochondrial homeostatic mechanisms described here come into play. Fig. 6 shows that these mechanisms exist in muscle and liver of mice, and it is highly likely that they exist in tissues of humans. Mild, but chronic, treatment with mitochondrial uncouplers should cause increased energy expenditure and oxygen consumption while causing little or no change in ATP levels. This old, but very simple, idea for treating human obesity should be revisited with an eye to the molecular compensatory mechanism revealed here.
Materials and Methods
Reagents.
FCCP, DNP, and BAPTA were obtained from Sigma (St. Louis, MO). Compound C and A23187 were from Calbiochem (San Diego, CA). Antibodies were from Cell Signaling (Danvers, MA) except for anti-Pan TORC (Calbiochem). The 2-kb PGC-1α promoter and promoter mutants have been described (29). ACREB was provided by Charles Vinson (40) (National Institutes of Health, Bethesda, MD). The dominant-negative TORC (T1–44eGFP) and control (eGFP) were provided by Novartis (Basel, Switzerland) (43). Immortalized preadipocytes from PGC-1α WT and null mice (23, 53) and preadipocytes expressing T1–44eGFP were provided by Marc Uldry (Dana–Farber Cancer Institute).
Cell Culture, Transfection, and ChIP.
10T 1/2 fibroblasts were grown in DMEM (10% FBS). Immortalized preadipocytes from PGC-1α WT and −/− mice were cultured in DMEM (20% FBS). For the reporter gene assays, cells were transfected overnight with SuperFect (Qiagen, Valencia, CA) and treated with FCCP for 24 h. Luciferase activity was normalized to β-galactosidase (Promega, Madison, WI), and then compared with the empty pGL3basic vector. PGL3basic vector, pSV vector, and eGFP served as the controls for the PGC-1α 2-kb promoter, ACREB, and T1–44eGFP, respectively. ChIP was performed with a ChIP-IT kit (Active Motif, Carlsbad, CA).
Analysis of Gene Expression.
RNA was isolated by using TRIzol (Invitrogen, Carlsbad, CA) and measured by using iSCRIPT and SYBRGreen (Bio-Rad, Hercules, CA). mRNA levels were normalized to actin mRNA, and then relative mRNA levels were determined by using the ΔΔCt.
Electron Microscopy, Lactate, ATP, and Viability Measurements.
ATP levels were measured by using the ATP Determination Kit (Invitrogen). Lactate in the media was quantified with Lactate Reagent (Trinity Biotech, Bray, Ireland). Viability was measured by using the LDH cytotoxicity kit (Roche, Indianapolis, IN). Electron microscopy was performed, and mitochondrial volume density was measured as described (14, 22).
Calcium Measurements.
Fura-2 ratios were used to calculate [Ca2+] as described (34, 54). For the measurements, EGTA was added to chelate the extracellular calcium. FCCP was then added followed by digitonin and EGTA/Tris for calibration. When BAPTA was used, it was added before the addition of FCCP.
Animal Experiments.
All animal experiments were performed according to a protocol approved by the Institutional Animal Care and Use Committee. Mice (see SI Text) were injected with 50 mg/kg DNP (90% saline, 10% DMSO solution) or 0 mg/kg DNP (90% saline, 10% DMSO solution). Mice were killed after 5 h.
Statistical Analysis.
Results are expressed as ±SD. Two-tailed Student's t tests were used to determine P values.
Acknowledgments
We thank Sherry Chin for assistance generating the PGC-1α muscle-specific −/− mice. This work was supported by National Institutes of Health Grants R01DK060837, NIDDK-DK54477, and DK61562 (to B.M.S). L.M.R. was supported by National Institutes of Health Training Grant/National Research Service Award 2-T32-GM07226-27.
Abbreviations
- PGC-1
peroxisome proliferator-activated receptor-coactivator 1
- OXPHOS
oxidative phosphorylation
- CREB
cAMP-response-element-binding protein
- ACREB
dominant-negative CREB
- TORC
transducer of regulated CREB protein
- Cyt c
cytochrome c
- COX II
Cyt c oxidase subunit II
- DNP
2,4-dinitrophenol
- FCCP
carbonyl cyanide p-(trifluromethoxy)phenyl hydrazone
- NRF
nuclear respiratory factor
- [Ca2+]i
intracellular calcium
- BAPTA
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate
- AMPK
AMP-activated protein kinase
- TFAM
mitochondrial transcription factor A.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702683104/DC1.
References
- 1.Scheffler IE. Mitochondria. New York: Wiley-Liss; 1999. [Google Scholar]
- 2.Nicholls DG, Locke RM. Physiol Rev. 1984;64:1–64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Lowell BB, Spiegelman BM. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 4.Klingenberg M, Huang SG. Biochim Biophys Acta. 1999;1415:271–296. doi: 10.1016/s0005-2736(98)00232-6. [DOI] [PubMed] [Google Scholar]
- 5.Butow RA, Avadhani NG. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 6.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Butow RA. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 8.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 9.Kelly DP, Scarpulla RC. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 10.Vega RB, Huss JM, Kelly DP. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Handschin C, Spiegelman BM. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 14.St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 18.Parascandola J. Mol Cell Biochem. 1974;5:69–77. doi: 10.1007/BF01874175. [DOI] [PubMed] [Google Scholar]
- 19.Harper JA, Dickinson K, Brand MD. Obes Rev. 2001;2:255–265. doi: 10.1046/j.1467-789x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 20.Cutting W, Mehrtens HG, Tainter ML. J Am Med Assoc. 1933;101:193–195. [Google Scholar]
- 21.Heytler PG, Prichard WW. Biochem Biophys Res Commun. 1962;7:272–275. doi: 10.1016/0006-291x(62)90189-4. [DOI] [PubMed] [Google Scholar]
- 22.Weibel E. Stereological Methods: Practical Methods for Biological Morphometry. London: Academic; 1979. [Google Scholar]
- 23.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, Bond JD, Ingram VM. Proc Natl Acad Sci USA. 1997;94:9705–9710. doi: 10.1073/pnas.94.18.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernardi P, Paradisi V, Pozzan T, Azzone GF. Biochemistry. 1984;23:1645–1651. doi: 10.1021/bi00303a010. [DOI] [PubMed] [Google Scholar]
- 26.Pozzan T, Bragadin M, Azzone GF. Biochemistry. 1977;16:5618–5625. doi: 10.1021/bi00644a036. [DOI] [PubMed] [Google Scholar]
- 27.Sandoval ME. Brain Res. 1980;181:357–367. doi: 10.1016/0006-8993(80)90618-6. [DOI] [PubMed] [Google Scholar]
- 28.Kessler RJ, Tyson CA, Green DE. Proc Natl Acad Sci USA. 1976;73:3141–3145. doi: 10.1073/pnas.73.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsien RY. Biochemistry. 1980;19:2396–2404. doi: 10.1021/bi00552a018. [DOI] [PubMed] [Google Scholar]
- 31.Yuan XJ, Sugiyama T, Goldman WF, Rubin LJ, Blaustein MP. Am J Physiol. 1996;270:C321–C331. doi: 10.1152/ajpcell.1996.270.1.C321. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JD, Chang JP. Cell Calcium. 2005;37:573–581. doi: 10.1016/j.ceca.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Thors B, Halldorsson H, Thorgeirsson G. FEBS Lett. 2004;573:175–180. doi: 10.1016/j.febslet.2004.07.078. [DOI] [PubMed] [Google Scholar]
- 34.Grynkiewicz G, Poenie M, Tsien RY. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 35.Hardie DG, Carling D. Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 36.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. Proc Natl Acad Sci USA. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwa M, Nakano H, Kumagai S. J Appl Physiol. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi T, Hirshman MF, Fujii N, Habinowski SA, Witters LA, Goodyear LJ. Diabetes. 2000;49:527–531. doi: 10.2337/diabetes.49.4.527. [DOI] [PubMed] [Google Scholar]
- 39.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 40.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, et al. Proc Natl Acad Sci USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M. Curr Biol. 2004;14:2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, III, Takemori H, et al. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 45.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 46.Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Proc Natl Acad Sci USA. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canettieri G, Koo SH, Berdeaux R, Heredia J, Hedrick S, Zhang X, Montminy M. Cell Metab. 2005;2:331–338. doi: 10.1016/j.cmet.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 50.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Kahn BB, Alquier T, Carling D, Hardie DG. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 54.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]