Abstract
MicroRNAs (miRNAs) are tiny noncoding RNAs whose function as modulators of gene expression is crucial for the proper control of cell growth and differentiation. Although the profile of miRNA expression has been defined for many different cellular systems, the elucidation of the regulatory networks in which they are involved is only just emerging. In this work, we identify a crucial role for three neuronal miRNAs (9, 125a, and 125b) in controlling human neuroblastoma cell proliferation. We show that these molecules act in an additive manner by repressing a common target, the truncated isoform of the neurotrophin receptor tropomyosin-related kinase C, and we demonstrate that the down-regulation of this isoform is critical for regulating neuroblastoma cell growth. Consistently with their function, these miRNAs were found to be down-modulated in primary neuroblastoma tumors.
Keywords: miR-9, miR-125a, miR125b, tyrosine kinase receptor
Over the past few years microRNAs (miRNAs) have emerged as a class of regulatory trans-acting factors that function as crucial determinants of cell fate specification. In animals, their role as negative regulators is exerted mainly at the translational level and is mediated by miRNA binding to cis-regulatory elements present in the 3′ UTR of mRNAs. The mode of action of miRNAs has a vast regulatory potential, because a single mRNA can be controlled by multiple miRNAs, and each miRNA may have hundreds of different targets (1).
Many mammalian miRNAs are expressed in a tissue- or cell type-specific manner, implying crucial roles in differentiation. Interestingly, the brain was identified as the organ expressing the highest variety of such molecules, suggesting an important role in nervous system development (2, 3).
In non-mammalian models, the functions of certain miRNAs during development of the nervous system have been identified. In Caenorhabditis elegans miR-273 and lys-6 participate in a complex regulatory network that ensures the stability and irreversibility of the terminal differentiated state of taste receptor neurons (4); in zebrafish a single family of miRNA, miR-430, is able to rescue normal brain formation in a Dicer-deficient organism (5).
In mammals, although several datasets are available on miRNA expression profiling, circuits in which they function are just beginning to be elucidated. For example, it has been reported that miR-132 regulates neuronal morphogenesis by decreasing levels of the GTPase-activating protein, p250GAP (6), and a role has been proposed for multiple brain-related miRNAs in regulating the transcription factor REST that mediates neuronal identity (7). In humans, which of the 66 brain-expressed miRNAs are associated with neuronal differentiation has been established by analyzing their expression in the totipotent embryonic NT2/D1 carcinoma cell line induced to differentiate upon retinoic acid (RA) treatment (8).
Neuroblastoma (NB) is a highly malignant pediatric tumor arising from an aberrant development of neural crest embryonic cells (9): it retains several features of neural crest progenitors as, for instance, the ability to respond to retinoids. The retinoid derivative trans-RA exerts potent antitumoral effects by inhibiting cell proliferation and inducing differentiation and apoptosis (10, 11). RA-induced growth arrest of N-Myc-amplified SK-N-BE NB cells is well documented, and 13-cis-RA is being used clinically to treat NB (12).
In this study, we identified specific subsets of miRNAs that are up-regulated during the response of the SK-N-BE cells to RA treatment. For three of them, miR-9, miR-125a, and miR-125b, we described an interesting correlation with cell proliferation: they were able to decrease cell growth when ectopically expressed in SK-N-BE cells, and, in agreement with this finding, they were down-regulated in freshly dissected human primary NBs. The truncated isoform of the signal-transducing neurotrophin receptor tropomyosin-related kinase C (trkC) was identified as one of the targets of such miRNAs, and RNAi against this factor proved an important role for this protein in the control of NB cell proliferation.
Results and Discussion
Expression Profiles of miRNAs in SK-N-BE Cells.
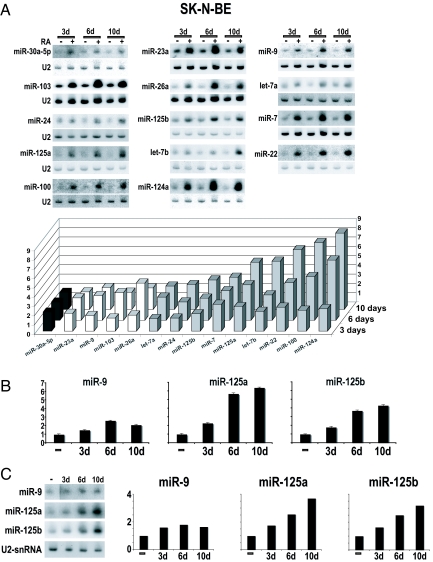
RA is known to induce growth arrest of NB cells, resulting in terminal neuronal differentiation. The expression pattern of 70 miRNAs, described as being expressed in neuronal cells (ref. 8 and C. Presutti, personal communication), was analyzed at specific time points during the RA-induced growth arrest of human SK-N-BE cells. Results of Northern blot analysis are summarized in Table 1: 23 miRNAs were undetectable, 33 species did not change upon RA treatment, and 14 miRNAs were up-regulated. Fig. 1A shows the Northern blot analysis of the up-regulated miRNAs: they include two brain-specific (miR-9 and miR-124a), two brain-enriched (miR-125a and miR-125b), and 10 brain nonenriched (let-7a, let-7b, miR-7, miR-22, miR-23a, miR-24, miR-26a, miR-30a-5p, miR-100, and miR-103) miRNAs. Most of them were induced 3 days after RA treatment and progressively accumulated at higher levels after terminal differentiation (10 days). Some of them, instead, displayed a different timing of expression with a peak at 3 or 6 days followed by a decrease in accumulation at later times (see the histogram in Fig. 1A). These profiles indicate that the miRNAs up-regulated during the response to RA treatment can be grouped into subfamilies based on their expression at specific time points of neuronal differentiation.
Table 1.
Three subfamilies of the 70 miRNAs in which expression was tested by Northern blot analysis in RA-treated SK-N-BE cells
| Not detectable | Expressed | Up-regulated |
|---|---|---|
| miR-1 | miR-9* | let-7a |
| miR-29a | miR-15a | let-7b |
| miR-30a-3p | miR-15b | miR-7 |
| miR-30c | miR-16 | miR-9 |
| miR-30c-3p | miR-17-5p | miR-22 |
| miR-95 | miR-19b | miR-23a |
| miR-96 | miR-20 | miR-24 |
| miR-99a | miR-26 | miR-26a |
| miR-126 | miR-26b | miR-30a-5p |
| miR-128a | miR-27a | miR-100 |
| miR-134 | miR-28 | miR-103/107 |
| miR-135a | miR-29b | miR-124a |
| miR-142-3p | miR-29c | miR-125a |
| miR-142-5p | miR-30b | miR-125b |
| miR-140a | miR-30d | |
| miR-196 | miR-30e-5p | |
| miR-197 | miR-92 | |
| miR-211 | miR-93 | |
| miR-221 | miR-98 | |
| miR-223 | miR-99b | |
| miR-296 | miR-101 | |
| miR-342 | miR-130a | |
| miR-424 | miR-130b | |
| miR-132 | ||
| miR-137 | ||
| miR-139 | ||
| miR-145 | ||
| miR-148b | ||
| miR-151 | ||
| miR-153 | ||
| miR-181a | ||
| miR-195 | ||
| miR-218 |
Fig. 1.
Expression profiles of miRNAs up-regulated in RA-treated SK-N-BE cells. (A) (Upper) Northern blots of the 14 miRNAs up-regulated in SK-N-BE cells upon RA treatment for 3 (lane 3d), 6 (lane 6d), or 10 (lane 10d) days. The probes are indicated on the left side of each panel; data were normalized to U2 snRNA hybridization signals. (Lower) The histogram depicts the distribution of the subsets of the 14 up-regulated miRNAs, clustered on the basis of their maximum induction times. Fold changes in miRNA expression are shown as the mean of the ratio of the RNA levels in RA-treated vs. untreated cells from three independent experiments. (B) RT-PCR quantification of miR-9, miR-125a, and miR-125b expression in SK-N-BE cells upon RA treatment for 3 (3d), 6 (6d), or 10 (10d) days. (C) Northern blots and histograms illustrating miRNA induction in KCNR NB cells, treated with RA for the same time points described in A and B. In B and C fold changes in miRNA expression are reported as the ratio of the RNA levels in RA-treated vs. untreated cells.
Ectopic Expression and Knockdown of miR-9, miR-125a, and miR-125b.
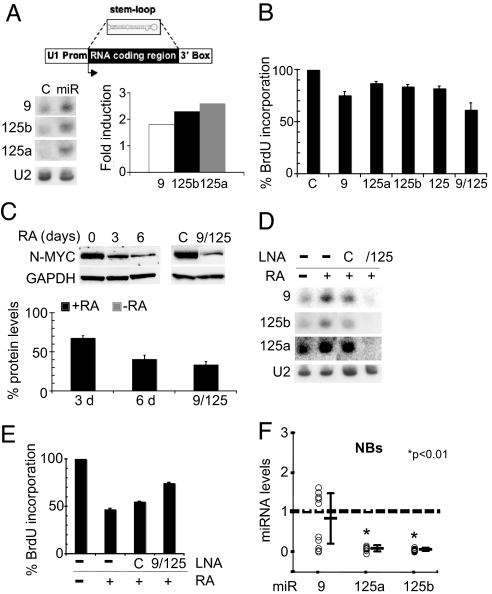
Among RA-induced miRNAs, we focused on miR-9, miR-125a, and miR-125b because their up-regulation, validated in SK-N-BE cells by RT-PCR (Fig. 1B), was observed also in other neuronal cell lines, the NB KCNR (Fig. 1C) and the medulloblastoma D283 (P.L., L.D.M., U.G., E.F., A.G., I.B., and E.C., unpublished work). To produce constitutive expression of the miRNAs in the absence of RA, the coding regions of the three miRNAs were cloned between the polII promoter and termination regions of the U1 snRNA gene (see schematic representation in Fig. 2A) (13). Fig. 2A shows that, upon transfection of the resulting constructs into SK-N-BE cells, the expression levels of miR-9, miR-125b, and miR-125a increased 1.7-, 2.2-, and 2.6-fold, respectively, compared with control cells. These expression values are comparable to the endogenous levels observed at 3 days after RA induction (see histogram of Fig. 1A).
Fig. 2.
Overexpression and knockdown of miR-9, miR-125a, and miR-125b. (A) Northern blot of the ectopically expressed miRNAs (lanes miR); in lane C, RNA from cells transfected with the control plasmid was analyzed. In the upper part, a schematic representation of the construct expressing the miRNAs is reported. The histogram on the right shows the increased levels of miRNA expression: Fold induction was reported as the ratio of the RNA levels in cells transfected with the miRNAs relative to control cells. (B) The effect of miRNA overexpression on cell proliferation was evaluated by BrdU assay; values, reported as the percentage of BrdU incorporation in cells transfected with the miRNAs relative to control cells, are the means ± SD of three separate experiments. (C) Western blot analysis of N-Myc protein levels after RA treatment for 0 (lane 0), 3 (lane 3), and 6 (lane 6) days or after miRNA ectopic expression (lane 9/125) in SK-N-BE cells. In lane C, protein extracts from cells transfected with the control plasmid were analyzed. GAPDH protein levels were used as a loading control. The histogram below reports the N-Myc protein levels as the ratio between N-Myc values in RA-treated for 3 (3 d) or 6 (6 d) days vs. untreated cells or between cells overexpressing the miRNAs vs. control cells (9/125). (D) Northern blot of RNA from cells untreated (lane −) or treated with RA (lanes +) and transfected with a control LNA (lane C) or with anti-miR LNAs (lane 9/125). (E) The effect of miRNA knockdown on cell proliferation was assessed by evaluating the percentage of BrdU incorporation in RA-treated cells transfected with specific LNAs (9/125) or control LNA (C) relative to untreated cells. (F) Quantitative PCR of miR-9, miR-125a, and miR-125b levels in 10 primary human NBs. Expression levels are reported as fold changes relative to control tissue (dashed line). ∗, P < 0.01 vs. control RNA. In A and D, the probes are indicated on the left side of each panel; data were normalized to U2 snRNA hybridization signals.
The effect of ectopic expression of such miRNAs on cell proliferation, in the absence of RA, was evaluated through the incorporation of the thymidine analogue 5-bromo-2′-deoxyuridine (BrdU). Fig. 2B shows that, compared with control cells, 25, 13, and 17% reductions in BrdU incorporation were detected after transfection of miR-9 (9), miR-125a (125a), and miR-125b (125b), respectively. When miR-125a and miR-125b were cotransfected (125), a reduction of 18% was observed, a value very close to that obtained by the corresponding single transfections. This result, together with the fact that the two miRNAs share the same seed (they differ only at positions 13, 14, 19, and 24), induced us to use a mixture of both constructs (reported as 125) in the following experiments. Notably, when the three miRNAs were cotransfected (9/125), a decrease of ≈40% was observed. Altogether, these results indicate that the ectopic expression of miR-9, miR-125a, and miR-125b produces a strong decrease of cell proliferation in vitro and suggest that these miRNAs function in an additive manner.
SK-N-BE cells display N-Myc amplification; because this gene is relevant for neuronal tumorigenesis, promoting cell growth (14), and its down-regulation correlates with reduced cell proliferation (15), we used N-Myc as a marker for assessing the effect of miRNA ectopic expression on cell proliferation. Fig. 2C shows that RA-treated cells exhibited lower levels of N-Myc relative to untreated cells. Notably, the N-Myc levels followed a 60% reduction when the three miRNAs were ectopically expressed in the absence of RA; this reduction is supposed to be an indirect effect because the N-Myc mRNA does not contain target sites for any of the three miRNAs overexpressed.
The effect of miRNAs on cell growth was further analyzed by their knockdown through locked nucleic acid (LNA) oligonucleotides (16, 17). LNAs complementary to miR-9, miR-125a, and miR-125b were cotransfected into SK-N-BE cells treated with RA. Depletion of miRNAs was assessed by Northern blot (Fig. 2D); compared with RA-treated cells transfected with a control LNA (lane C), miR-9 levels decreased ≈75%, whereas miR-125a and -125b almost disappeared. The effects of such depletions, assessed by BrdU assay, are shown in Fig. 2E: BrdU incorporation was 20% higher for cells transfected with LNAs, arguing that the depletion of the three miRNAs counteracted the effect of RA on cell growth. These data confirm that the expression levels of the three miRNAs influence cell growth and that they have an antiproliferative function.
The ability of miR-9, miR-125a, and miR-125b to inhibit both the proliferation of SK-N-BE cells and the expression of N-Myc, a master oncogene in NB (18–20), suggests that a specific deregulation of these miRNAs might occur in this malignancy. Therefore, the levels of miR-9, miR-125a, and miR-125b were monitored in a series of freshly dissected human primary NB samples. Although miR-125a and -125b were consistently down-regulated in all of the cases tested, miR-9 expression was significantly down-regulated in 50% of the tumors (Fig. 2F). These findings demonstrate a remarkable correlation between decreased expression of these miRNAs and neuronal tumorigenesis. It will be interesting to analyze the molecular basis for the miR-9 variability and whether this reflects any heterogeneity in the genetic background of the different NB tumors.
Identification of miRNA Target.
To analyze the molecular mechanisms in which the miRNAs are involved we looked for their target gene(s). An online search of miR-9, miR-125a, and miR-125b targets by miRanda (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl) and TargetScan databases (http://genes.mit.edu/targetscan.test/ucsc.html) provided a large number of putative mRNA targets. Among them, we focused on trkC for the following reasons: (i) it is a potential common target for the three miRNAs, displaying a binding site for miR-9 and another one for both miR-125a and -125b, that, as already stated, share the same seed; (ii) it was reported to be involved in neuronal differentiation (21, 22), and (iii) its expression levels are associated with a favorable outcome in NB (23, 24). trkC is the membrane-bound tyrosine kinase receptor with high-affinity binding for the neurotrophin-3; trkC and neurotrophin-3 contribute to the regulation of proliferation, survival, and differentiation of several cell population in the mammalian nervous system (25, 26).
As is the case for the other members of the trk receptor family (trkA and trkB), the trkC gene undergoes alternative splicing to produce isoforms that differ in functional capacity. These include a full-length, enzymatically active isoform (fl-trkC) and a truncated isoform (t-trkC), lacking the kinase domain. Interestingly, only the mRNA for the t-trkC isoform possesses the target sequences for miR-9, miR-125a, and miR-125b (Fig. 3A).
Fig. 3.
Analysis of trkC expression in SK-N-BE cells. (A) Schematic representation of the trkC gene. The boxes represent the exons, continuous lines the introns; products of alternative splicing are depicted below, together with the target sites for the miRNAs. (B) Western blot analysis of the fl-trkC and t-trkC protein isoform levels from SK-N-BE cells treated with RA, for the times indicated above. GAPDH protein levels were used as a loading control. The histogram illustrates trkC protein levels, reported as arbitrary units (AU), in untreated (0) or RA-treated [3 (3 d), 6 (6 d), and 10 (10 d) days] SK-N-BE cells. The table summarizes the different truncated/full-length protein ratios in cells untreated (column C) or treated with RA (column RA).
So far, the role for the kinase-deficient isoforms remains elusive; they could act as inhibitors by sequestering the full-length receptor or the neurotrophic factor. In this regard, it has already been shown that the t-trkB receptors, which are activated by the neurotrophin brain-derived neurotrophic factor and neurotrophin-4/5, indeed have dominant inhibitory effects on brain-derived neurotrophic factor signaling, by forming nonfunctional heterodimers with the full-length receptors (27). In this scenario, different truncated/full-length protein ratios would affect the neurotrophin signaling.
We analyzed trkC expression in RA-treated SK-N-BE cells; whereas the levels of the full-length protein (fl-trkC) increased during RA treatment, those of the truncated isoform (t-trkC) decreased, with the lowest value at 3 days (Fig. 3B). Interestingly, these values are inverted in proliferating SK-N-BE cells (see table in Fig. 3B). The fact that only the truncated form can be targeted by miR-9, miR-125a, and miR-125b together with the decreased levels of this isoform observed in cells treated with RA strongly suggested that its expression might be modulated by miRNAs.
trkC Is the Target of miR-9 and miR-125a and -125b.
To validate trkC as the target of miR-9 and miR-125a and -125b, we set up a luciferase reporter assay. A portion of the 3′ UTR of the truncated trkC isoform, including the two miRNA target sites, was cloned downstream of the r-luc ORF (see schematic representation in Fig. 4A). As a control, a construct derivative mutated in the miRNA binding sites, thus preventing miRNA-3′ UTR interactions, was also produced. The histogram in Fig. 4A shows reduction in luciferase activities of 30% and 17%, after transfection of 3′ WT trkC along with the single plasmids expressing miR-9 (9) or miR-125a and -125b (125). Interestingly, luciferase activity decreased by 50% when the three miRNAs were coexpressed (9/125, white bar). These results prove the additive effect of the three miRNAs and are in agreement with the cell proliferation data (Fig. 2B). In the presence of mutated target sites (9/125, black bar) no reduction of luciferase activity was observed, demonstrating the specificity of miRNA interaction.
Fig. 4.
Validation of trkC as a miRNA target (A) Schematic representation of the construct generated for the luciferase assay; binding sites for miR-9 and miR-125b on t-trkC 3′ UTR are indicated. The histogram reports the levels of luciferase activity in cells overexpressing the miRNAs indicated below and transfected with the wild-type 3′ UTR (3′ wt) or with its mutant derivative lacking the miRNA binding sites (3′ mut). (B) Western blot analysis of fl-trkC and t-trkC in cells overexpressing miR-9, miR-125a, and miR-125b (lane 9/125) or in control cells (lane C). The histogram depicts the fl-trkC and t-trkC levels in cells overexpressing the three miRNAs relative to control cells. (C) Western blot analysis of t-trkC protein after knockdown of miRNAs. Cells were untreated (lane −) or treated with RA (lanes +) and transfected with a control LNA (lane C) or with anti-miR LNAs (lane 9/125). The histogram reports the relative changes in t-trkC protein levels evaluated as the ratio of the protein in cells transfected with the anti-miRNA LNAs (9/125) or with the control LNA (C) relative to the untreated cells (−). (D) Western blot analysis of fl-trkC and t-trkC after specific silencing of the truncated isoform (lane αt-trkC). (E) The effect of t-trkC silencing on cell proliferation was evaluated by BrdU assay; values, reported as the percentage of BrdU incorporation in cells expressing the siRNAs relative to control cells, are the means ± SD of three separate experiments. (F) Western blot analysis of t-trkC overexpression (lane t-trkC) compared with control cells (lane C). (G) The effect of t-trkC overexpression on cell proliferation was evaluated by BrdU assay on RA-treated cells; values, reported as the percentage of BrdU incorporation in cells overexpressing the protein relative to control cells, are the means ± SD of three separate experiments. In B–D and F, GAPDH protein levels were evaluated as a loading control.
Further evidence of trkC repression through the miRNA pathway came from overexpression and knockdown experiments. Whereas the ectopic expression of miRNAs caused a decrease by 70% of the t-trkC isoform (Fig. 4B), LNA-directed miRNA depletion in RA-treated SK-N-BE cells (Fig. 4C) showed a partial recovery of the truncated isoform (≈3-fold).
Alteration of the t-trkC Protein Expression Affects Cell Proliferation.
To verify whether the amount of the t-trkC isoform correlates with cell proliferation, we altered its cellular levels and analyzed the effect on cell growth. The down-regulation of the protein was performed by RNAi. SK-N-BE cells were transfected with a plasmid expressing siRNAs against exon 14b, which specifically marks the truncated isoform. In these conditions, a 60% reduction of the t-trkC levels was observed (Fig. 4D). The effect of the knockdown experiment on cell proliferation was then assessed by the BrdU assay; compared with control cells, a 30% reduction in BrdU incorporation was detected (Fig. 4E), indicating that the down-regulation of t-trkC functionally mimicked the miRNA overexpression.
At variance with the previous experiment, t-trkC overexpression (Fig. 4F) produced, in RA-treated cells, a 20% increase of cell proliferation with respect to control cells (Fig. 4G). Altogether these results strongly suggest a role for this protein in the control of neuronal cell proliferation.
In conclusion, a new regulatory circuitry involving miR-9, miR-125a, miR-125b, and trkC was shown to play an important role in controlling cell proliferation of the SK-N-BE NB cell line. We demonstrated that these miRNAs modulate the expression of the truncated neurotrophin receptor trkC whose down-regulation correlates with cell growth repression (Fig. 5). This is in agreement with the observed underexpression of such miRNAs in human primary NB tumors and with their up-regulation during in vitro neuronal differentiation.
Fig. 5.
A model depicting the interplay between miRNAs and the t-trkC isoform in RA-induced neuronal differentiation. In undifferentiated SK-N-BE cells, the t-trkC protein level is 2-fold higher than that of the full-length isoform (Left). Upon RA treatment the overexpression of miRNAs causes a reduction of the truncated form, which correlates with a decrease in cell proliferation (Right).
Finally, our data not only support the involvement of these miRNAs in the control of cell growth and, therefore, their potential role as oncosuppressors but also suggest their possible use as diagnostic markers for tumorigenesis.
Materials and Methods
Cell Cultures and Treatments.
SK-N-BE cells were cultured in RPMI medium 1640 (Gibco, Carlsbad, CA), supplemented with 10% fetal bovine serum, 1× l-glutamine, and penicillin/streptomycin, and induced to differentiation by 10 μM all-trans-RA (Sigma, St. Louis, MO).
Human Tissue Samples.
Surgical specimens of primary NBs were collected from 10 patients with institutional review board approval. RNAs of normal human dorsal ganglia were from Clontech (Mountain View, CA).
RNA Extraction and Northern Blot Analysis.
Total RNA, extracted from untreated and RA-treated SK-N-BE cells, was fractionated on a 10% polyacrylamide-urea gel and transferred to a nylon membrane. DNA oligonucleotides complementary to the sequences of mature miRNAs and to U2 snRNA (U2R: 5′-GGGTGCACCGTTCCTGGAGGTAC-3′) were 32P-labeled and used as probes.
Densitometric Analysis.
The expression levels of miRNAs in treated vs. untreated cells were quantified by the InstantImager software package (Packard, Palo Alto, CA) as follows. miRNAs with hybridization ratios ≥2 were reported as up-regulated. The value of the background (measured adjacent to each band) was subtracted from the signal of the mature miRNA. The values obtained were normalized against the U2 snRNA in the same lane.
Cloning and Overexpression of miR-9, miR-125a, and miR-125b.
The genomic fragments containing the premiR-9-3 (from −21 to + 93 relative to the 5′ end of miR-9), the premiR-125a (from −14 to + 72), and the premiR-125b-2 (from −16 to + 89) were PCR amplified and cloned into a vector carrying the constitutive-expression cassette of the snRNA U1 gene (13), to generate the constructs overexpressing miRNAs. A plasmid producing an unrelated 21-nt-long RNA, bearing no homology to any known miRNA or mRNA sequence in human, was used as a control in the transfection experiments. The plasmids were transfected in SK-N-BE cells by Lipofectamine and Plus Reagent (Invitrogen, Carlsbad, CA).
miRNA Knockdown.
FITC-labeled LNA oligonucleotides (Exiqon, Vedbaek, Denmark), with a ratio miR-9 to miR-125a/b of 1:1, were transfected into SK-N-BE cells at a final concentration of 40 nM by Hyperfect reagent (Qiagen, Hilden, Germany). After 6 h, RA was added to the culture medium to induce cell differentiation, and 72 h later, cell proliferation was evaluated and miRNA or protein levels were analyzed.
Cell Proliferation Assay.
Cells were cultured for 72 h and incubated with 10 μM 5-BrdU (Labeling and Detection Kit I, Roche, Basel, Switzerland) for 3 h before they were fixed in 4% paraformaldehyde. For immunocytochemistry, cells were permeabilized with 0.2% Triton X-100, treated with 2 M HCl, and blocked in PBS-buffered 3% BSA. Texas red dye-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) was used as a secondary antibody. Nuclei were counterstained with Hoechst reagent. Two hundred nuclei were counted in triplicate, and the number of BrdU-positive nuclei was recorded.
Luciferase Activity Assays.
A 1011-nt-long region of the 3′ UTR of the human t-trkC isoform was PCR-amplified and cloned downstream of the stop codon in pRL-TK vector. From this construct a mutant derivative, lacking the miRNA target sites, was generated by inverse PCR. Such constructs were cotransfected in SK-N-BE cells together with the control plasmid or with plasmids overexpressing miR-9, miR-125a, and miR-125b and with firefly luciferase expression vector pGL3. Cells were harvested 24 h posttransfection and assayed with the Dual-Luciferase Assay (Promega, Madison, WI). All of the assays were performed in triplicate in three independent experiments.
Protein Extraction and Western Blot Assay.
Protein extracts were prepared from SK-N-BE cells lysed in radioimmunoprecipitation assay buffer, fractionated onto NuPAGE 4–12% polyacrylamide gels (Invitrogen), blotted onto nitrocellulose membranes, and reacted overnight with rabbit anti-trkC or anti-N-Myc antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). The GAPDH signal was used as a loading control.
RNAi Assay.
A 98-nt-long DNA sequence encoding for siRNAs designed to target the 3′ UTR of the t-trkC isoform [nucleotides 3210–3230 of sequence NM_001007156 (GenBank)] was cloned in the psiUx plasmid (28). Seventy-two hours after transfection, cells were treated for BrdU incorporation and immunofluorescence, or protein extracts were prepared.
Ectopic Expression of t-trkC.
Total RNA, extracted from SK-N-BE cells, was used to synthesize first-strand cDNA by the SuperScript System (Invitrogen) using dT oligonucleotides. t-trkC ORF was amplified by nested PCRs on the cDNA template (oligonucleotides: 5′ UTR t-trkC, 5′-gattttgcatctgatcgctcg-3′; 3′ UTR t-trkC, 5′-aaggagcacagtgatgattgg-3′; ATG t-trkC, 5′-atggatgtctctctttgcccagc-3′; and STOP t-trkC, 5′-ttaaaagccatgacgtcctttgc-3′), cloned in the pCDNA3.1 vector, and transfected in RA-treated NB cells as described above.
RNA Isolation and miR Quantification by RT-PCR Analysis.
RNA isolation from tissue samples was performed as described (29). Quantitative analysis of miR-9, miR-125a, and miR-125b was carried out on RNA samples using the specific stem-loop primers for reverse transcription followed by real-time TaqMan reagents (Applied Biosystems, Foster City, CA). All values were normalized to endogenous control U6. miRNA quantification was expressed in arbitrary units.
Acknowledgments
We thank A. Tacconelli and A. R. Farina for kindly providing RNA from KCNR cells and M. Arceci and M. Marchioni for technical help. This work was partially supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC) and AIRC-Roman Oncogenomic Center and from the Sixth Research Framework Program of the European Union, Project RIGHT Grant LSHB-CT-2004 005276, SIROCCO Grant 037900, Ministero dell'Università e della Ricerca Scientifica e Tecnologica Grants FIRB-p.n. RBNE015MPB and RBNE01KXC9 (to I.B.), and Programmi di Ricerca di Interesse Nazionale and “Centro di Eccellenza di Biologia e Medicina Molecolare.” P.L. was supported by a fellowship from AIRC/Fondazione Italiana per la Ricerca sul Cancro.
Abbreviations
- miRNAs
microRNAs
- RA
retinoic acid
- NB
neuroblastoma
- trkC
tropomyosin-related kinase C
- LNA
locked nucleid acid
- t-trkC
truncated trkC
- fl-trkC
full-length trkC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. Proc Natl Acad Sci USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 6.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Xie X. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abemayor E, Sidell N. Environ Health Perspect. 1989;80:3–15. doi: 10.1289/ehp.89803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolande RP. Surv Synth Pathol Res. 1985;4:296–311. doi: 10.1159/000156982. [DOI] [PubMed] [Google Scholar]
- 11.Thiele CJ. Cancer Metastasis Rev. 1991;10:311–319. doi: 10.1007/BF00554793. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds CP, Lemons RS. Hematol Oncol Clin North Am. 2001;15:867–910. doi: 10.1016/s0889-8588(05)70256-2. [DOI] [PubMed] [Google Scholar]
- 13.De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, Cossu G, Bozzoni I. Proc Natl Acad Sci USA. 2002;99:9456–9461. doi: 10.1073/pnas.142302299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Oncogene. 1996;13:803–812. [PubMed] [Google Scholar]
- 15.Thiele CJ, Reynolds CP, Israel MA. Nature. 1985;313:404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 16.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 17.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Schwab M. Cancer Lett. 2004;204:179–187. doi: 10.1016/S0304-3835(03)00454-3. [DOI] [PubMed] [Google Scholar]
- 19.Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 20.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. EMBO J. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessarollo L, Tsoulfas P, Donovan MJ, Palko ME, Blair-Flynn J, Hempstead BL, Parada LF. Proc Natl Acad Sci USA. 1997;94:14776–14781. doi: 10.1073/pnas.94.26.14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henion PD, Garner AS, Large TH, Weston JA. Dev Biol. 1995;172:602–613. doi: 10.1006/dbio.1995.8054. [DOI] [PubMed] [Google Scholar]
- 23.Ryden M, Sehgal R, Dominici C, Schilling FH, Ibanez CF, Kogner P. Br J Cancer. 1996;74:773–779. doi: 10.1038/bjc.1996.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashiro DJ, Nakagawara A, Ikegaki N, Liu XG, Brodeur GM. Oncogene. 1996;12:37–41. [PubMed] [Google Scholar]
- 25.Ernfors P, Rosario CM, Merlio JP, Grant G, Aldskogius H, Persson H. Brain Res Mol Brain Res. 1993;17:217–226. doi: 10.1016/0169-328x(93)90005-a. [DOI] [PubMed] [Google Scholar]
- 26.Klein R, Silos-Santiago I, Smeyne RJ, Lira SA, Brambilla R, Bryant S, Zhang L, Snider WD, Barbacid M. Nature. 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- 27.Eide FF, Vining ER, Eide BL, Zang K, Wang XY, Reichardt LF. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denti MA, Rosa A, Sthandier O, De Angelis FG, Bozzoni I. Mol Ther. 2004;10:191–199. doi: 10.1016/j.ymthe.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Ferretti E, Di Marcotullio L, Gessi M, Mattei T, Greco A, Po A, De Smaele E, Giangaspero F, Riccardi R, Di Rocco C, Pazzaglia S, Maroder M, Alimandi M, Screpanti I, Gulino A. Oncogene. 2006;25:7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]