Abstract
Signaling by Smoothened (Smo) plays fundamental roles during animal development and is deregulated in a variety of human cancers. Smo is a transmembrane protein with a heptahelical topology characteristic of G protein-coupled receptors. Despite such similarity, the mechanisms regulating Smo signaling are not fully understood. We show that Gprk2, a Drosophila member of the G protein-coupled receptor kinases, plays a key role in the Smo signal transduction pathway. Lowering Gprk2 levels in the wing disc reduces the expression of Smo targets and causes a phenotype reminiscent of loss of Smo function. We found that Gprk2 function is required for transducing the Smo signal and that when Gprk2 levels are lowered, Smo still accumulates at the cell membrane, but its activation is reduced. Interestingly, the expression of Gprk2 in the wing disc is regulated in part by Smo, generating a positive feedback loop that maintains high Smo activity close to the anterior–posterior compartment boundary.
Keywords: Smoothened, wing disc, imaginal discs
Smo is the key transducer of a conserved signaling pathway regulating many developmental processes in vertebrates and invertebrates (1, 2). The transmembrane protein Patched (Ptc) is the receptor for the ligand Hedgehog (Hh) and represses Smo activity in the absence of ligand (2). The binding of Hh to Ptc relieves this repression and allows Smo to signal to a protein complex that includes the transcription factor Ci/Gli (1). Smo controls the activation of Ci in the presence of the Hh ligand in part by preventing Ci proteolytic processing into a transcriptional repressor. In the Drosophila wing disc, the epithelium giving rise to the wing and thorax of the fly, Smo signaling controls the expression of several genes in anterior cells close to the anterior–posterior (A/P) compartment boundary and promotes the growth and patterning of the wing (3–5).
The cytoplasmic tail of Drosophila Smo is a target for phosphorylation by protein kinase A and casein kinase I, and it has been shown that Smo phosphorylation by these kinases is essential for its activity and membrane accumulation (6–9). However, most of these phosphorylated residues are not conserved in its vertebrate counterparts (10). Recently, the G protein-coupled receptor kinase 2 (Grk2) has been shown to phosphorylate mammalian Smo (11). G protein-coupled receptor kinases (GRKs) selectively phosphorylate the ligand-activated form of G protein-coupled receptors (12). This phosphorylation promotes uncoupling from G proteins and also the recruitment of β-arrestins, which target the receptor for clathrin-mediated endocytosis. In addition, GRKs and β-arrestins also participate in signal propagation by recruiting additional proteins to the receptor complex (12–14). There are two Drosophila GRKs, GPRK1 and GPRK2. GPRK1 modulates the amplitude of the visual response acting as a Rhodopsin kinase, whereas GPRK2 regulates the level of cAMP during Drosophila oogenesis (15, 16). Phosphorylation of mammalian Smo by GRK2 promotes its endocytosis in clathrin-coated pits in a process dependent on β-arrestin2 (11). However, whether this form of Smo internalization is part of a desensitization mechanism, as is the case for different G protein-coupled receptors (12), or if it participates in Hh signaling is still not known. To address the participation of GRKs during Smo signaling in Drosophila, we have analyzed the function of Gprk2 during imaginal wing disc development. We found that Gprk2 activity is required for Smo activation. Thus, the reduction of Gprk2 expression by interference RNA, or its elimination by a genetic mutation, causes the accumulation of Smo in wing disc anterior cells exposed to Hh. The accumulation of Smo is, however, correlated with reduced activity, because Smo high-level targets are not correctly activated and flies expressing Gprk2-RNAi display Hh loss-of-function phenotypes. Interestingly, the reduction in Gprk2 expression is able to antagonize the activity of Smo mutant forms that mimic its phosphorylation by protein kinase A and casein kinase 1, suggesting that additional phosphorylation by Gprk2 is a necessary step to obtain the correct activation of Smo to promote the expression of its targets requiring high levels of signaling.
Results and Discussion
Regulation of Gprk2 Expression by Hh Signaling in the Wing Disc.
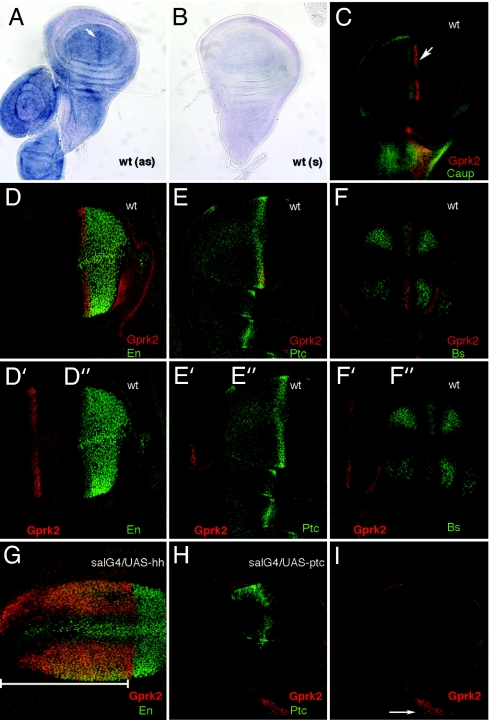
The expression of Gprk2 mRNA in the wing disc is generalized but appears increased in a stripe of cells located close to the A/P compartment boundary (white arrow in Fig. 1A). To better characterize this pattern, we used the P-lacZ insertion Gprk206936, which is localized in the 5′ untranslated region of the gene (17). Interestingly, β-gal expression is restricted to the A/P compartment boundary of the wing disc during the third larval instar (white arrow in Fig. 1C). The cells expressing β-gal were further identified by using a combination of region-specific markers such as Engrailed (En), Patched (Ptc), Blistered (Bs), and Caupolican (Caup) (Fig. 1). This analysis places the stripe of maximal expression of Gprk2 to anterior cells abutting the A/P boundary. These cells express Ptc and En in the anterior compartment and are localized in the region exposed to high-level Hh signaling. In fact, Hh signaling regulates the expression of Gprk2 in these anterior cells, because β-gal expression in Gprk206936 discs is expanded to the entire anterior compartment when hh is ectopically expressed (Fig. 1G), and it is repressed when the activity of the pathway is reduced by ectopic expression of Ptc (Fig. 1 H and I). The regulation of Gprk2 accumulation in anterior cells by Hh suggests that Hh signaling and Gprk2 might be functionally related.
Fig. 1.
Enhanced GPRK2 accumulation in regions of high-level Hh signaling in the wing disc. (A and B) In situ hybridization of third instar wing imaginal discs with Gprk2 antisense (as) (A) and sense (s) (B) probes. The expression of Gprk2 is detected at higher levels in cells abutting the A/P compartment boundary (white arrow in A). (C) Expression of β-gal (red) and Caupolican (Caup, green) in a third instar wing disc heterozygous for the P-lacZ insertion PZ-Gprk2[06936]. The white arrow indicates the stripe of β-gal accumulation. (D) Expression of β-gal/Gprk2 (Gprk2, red) and Engrailed (En, green) in wild-type wing disc (wt), showing that β-gal expression occurs in anterior cells expressing En (nuclei in orange). Individual channels are shown in D′ (Gprk2) and D″ (En). (E) Expression of β-gal/Gprk2 (Gprk2, red) and Patched (Ptc, green). Individual channels are shown in E′ (Gprk2) and E″ (Ptc). (F) Expression of β-gal/Gprk2 (Gprk2, red) and Blistered (Bs, green). Individual channels are shown in F′ (Gprk2) and F″ (Bs). (G) Expression of β-gal/Gprk2 (Gprk2, red) and Engrailed (En, green) in sal-Gal4/+; PZ-Gprk2[06936]/UAS-hh wing discs. The extent of β-gal and En expression is included in the bottom white line and appears as orange nuclei. (H) Expression of β-gal/Gprk2 (Gprk2, red) and Patched (Ptc, green) in sal-Gal4/UAS-ptc; PZ-Gprk2[06936]/+ wing discs, showing that β-gal expression is eliminated in the wing blade upon ectopic Ptc expression. (I) Individual red channel of H, showing that the expression of β-gal/Gprk2 in the thorax (where sal-Gal4 in not expressed) is not affected (white arrow). wt, wild type.
Gprk2 Is Required for Hh Signaling in the Wing Disc.
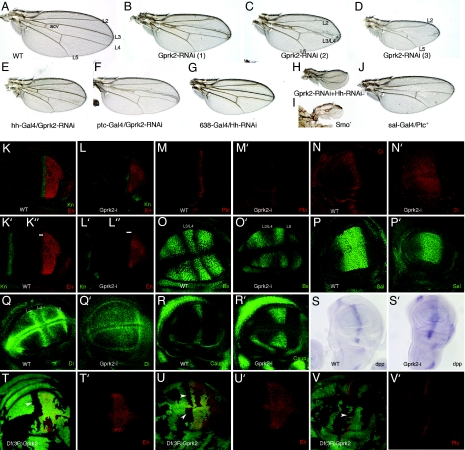
All available Gprk2 alleles are P element insertions in the 5′ region of the gene (17). These alleles are homozygous viable, and the mutant wings do not display any visible phenotype (data not shown). We generated stronger loss-of-function conditions of the gene by (i) expressing Gprk2 interference RNA (Gprk2i) under the control of yeast upstream activator sequences (UAS; UAS-Gprk2i) and (ii) constructing a synthetic deletion of the gene [see Materials and Methods and supporting information (SI) Fig. 6]. In wing discs of the combination Gal4–638/UAS-Gprk2i, we found a reduction of Gprk2 mRNA levels of 66 ± 1.5% (SI Fig. 6A). The corresponding adult wings show a range of striking phenotypes similar to loss of Hh function, displaying a reduction of the L3/L4 intervein, fusion of the L3 and L4 veins, and in a lower percentage of wings, the loss of the L3 and L4 veins (Fig. 2 A–D and Table 1). These veins and the L3/L4 intervein correspond to the territory specified by Hh signaling (18). In fact, reduction of Gprk2 levels results in wings very similar to those with a moderate loss of Hh signaling, generated either by ectopic expression of Ptc (Fig. 2J) or by expression or hh-interference RNA (Fig. 2G). This phenotype is very different from that observed upon increased activity of the pathway (see Fig. 5A). The Gal4–638 line is expressed in the entire wing (SI Fig. 6B), and to distinguish between the effects of lowering Gprk2 levels in cells producing or responding to Hh, we used three other Gal4 lines expressed in either the anterior (Gal4-Ci and Gal4-ptc) or the posterior (Gal4-hh) compartments. We found that only the expression of Gprk2i in anterior cells recapitulates the reduction of the L3/L4 intervein observed in Gal4–638/UAS-Gprk2i wings (Fig. 2 E and F and data not shown). Thus, the combinations Gal4-Ci/UAS-Gprk2i (data not shown) and Gal4-ptc/UAS-Gprk2i (Fig. 2F) show a reduction or elimination of the L3/L4 intervein, whereas the wings of the Gal4-hh/UAS-Gprk2i combination display a normal pattern of veins (Fig. 2E). The phenotypes observed upon a reduction of Gprk2 unambiguously indicate that Gprk2 function is necessary for the transduction of the Hh signal. Furthermore, when the expression of Gprk2 is reduced in flies expressing lower levels of the ligand Hh (Gal4–638/+; UAS-hhi / UAS-Gprk2i; Fig. 2H), the resulting wings have stronger hh loss-of-function phenotypes, and a previously unrecognized phenotypic class indistinguishable to those of wings formed by smo mutant cells (Gal4–638/+; FRT42 smo2/FRT42 M (2)l2; UAS-FLP/+; Fig. 2I) is now observed (Fig. 2H and Table 1).
Fig. 2.
Gprk2 is required for Hh signaling. (A) Wild-type wing showing the position of the longitudinal veins L2 to L5. acv, anterior cross vein. (B–D) Adult wings resulting from ectopic expression of interference Gprk2 RNA in Gal4–638/UAS-Gprk2i flies. Mild phenotypes consist of a moderate reduction of wing size and the shortening of the distance between the L3 and L4 veins [B; Gprk2-RNAi (1)]. Medium phenotypes are characterized by partial fusion of the L3 and L4 veins [C; Gprk2-RNAi (2)]. Strong phenotypes consist in greater reductions of wing size and the disappearance of the L3 and L4 veins [D; Gprk2-RNAi (3)]. (E) Adult wing from Gal4-hh/UAS-Gprk2i flies, expressing interference Gprk2 RNA in the posterior compartment. (F) Adult wing from Gal4-ptc/UAS-Gprk2i flies, expressing interference Gprk2 RNA in anterior cells close to the A/P compartment boundary (compare to B and C). (G) Hh loss-of-function phenotype in 638-Gal4/UAS-hh-RNAi flies consists of wing size reduction, shortening of the distance between the veins L3 and L4, and loss of the anterior cross vein (acv in A). (H) Severe hh loss-of-function phenotype in 638-Gal4/UAS-hh-RNAi; UAS-Gprk2i/+; compare to the expression of only hh-RNAi (G) or only Gprk2-RNAi (B–D). (I) Severe hh loss-of function phenotype in Gal4–638/+; FRT42 smo2/FRT42 M (2)l2; UAS-FLP/+. These wings, formed by homozygous smo2 cells, are extremely reduced in size and have lost most pattern elements. (J) Hh loss-of-function phenotype caused by ectopic expression of ptc (sal-Gal4/UAS-ptc) is very similar to loss of Gprk2 function (compare to D). (K–K″) Expression of Knot (Kn; K and K′, green) and Engrailed (En; K and K″, red) in a third instar wild-type wing imaginal disc. (L–L″) Loss of Gprk2 reduces Knot expression (Kn; L and L′, green) and eliminates anterior expression of Engrailed (En; L, L″, red). The white lines in K″ and L″ delimit the anterior domain of En expression. (M and M′) Expression of Ptc in wild-type discs (wt; M) and in Gal4–638/UAS-Gprk2i (Gprk2-i; M′). Loss of Gprk2 reduces the expression of Ptc. (N and N′) Expression of Ci in wild-type discs (wt; N) and in Gal4–638/UAS-Gprk2i wing discs (Gprk2-i; N′). (O and O′) Expression of Bs in wild-type discs (wt; O) and in Gal4–638/UAS-Gprk2i wing discs (Gprk2-i; O′). Loss of Gprk2 eliminates the L3/L4 intervein domain of Bs expression. (P and P′) Expression of the Dpp target gene Sal in wild-type (wt; P) and Gal4–638/UAS-Gprk2i discs (Gprk2-i; P′). Overall, the level and anterior-posterior extent of the Sal domain is not affected. (Q and Q′) Expression of Dl in wild-type discs (wt; Q) and in Gal4–638/UAS-Gprk2i (Gprk2-i; Q′). Loss of Gprk2 reduces Dl expression in the L3 and L4 veins. (R and R′) Wing discs showing the expression of Caupolican (Caup) in wild-type (wt; R) and Gal4–638/UAS-Gprk2i wing discs (Gprk2-i; R′). Loss of Gprk2 expands Caup. (S and S′) In situ hybridization with a dpp RNA probe in wild-type (wt; S) and Gal4–638/UAS-Gprk2i (Gprk2-i; S′) wing discs. Upon a reduction in Gprk2 levels, dpp expression occurs at lower levels and in an expanded domain. (T–V) Clones of Df(3R)Gprk2 cells induced in hs-FLP1.22; FRT82 Df(3R)Gprk2/ FRP82 M (3)w Ubi-GFP discs at 48–72 h after egg laying. Homozygous Gprk2− cells are labeled by the absence of GFP and appear as black spots. In T and T′ and U and U′, the expression of En (En; red) is shown, and in V and V′, the expression of Ptc is shown in red. In anterior Gprk2 mutant cells located close to the A/P compartment boundary, the expression of En (T–U′) and Ptc (V and V′) is not detected. T′, U′, and V′ show the red channels of T, U, and V, respectively.
Table 1.
Percentage of phenotypic classes observed in genetic combinations involving the overexpression of Gprk2-RNAi
| Genetic combination | Phenotypic class |
No. of wings | En expression |
Ptc expression |
Smo expression |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0, % | 1, % | 2, % | 3, % | 4, % | Wild type | Reduced | Eliminated | Wild type | Reduced | Eliminated | Wild type | Increased | ||
| 638/+; Gprk2i/+ | 29 | 58 | 10 | 3 | 0 | 1,145 | 0 | 4 | 10 | 0 | 3 | 11 | 0 | 20 |
| 638; Gprk2i/+ | 51 | 49 | 0 | 0 | 0 | 37 | 0 | 4 | 6 | 0 | 7 | |||
| 638/+; Gprk2i/+; Gprk2i/+ | 2 | 29 | 54 | 15 | 0 | 299 | ||||||||
| 638/+; Gprk2i/+; Df(3R)Gprk2/+ | 15 | 60 | 16 | 11 | 0 | 123 | ||||||||
| 638/+; Gprk2i/+; Hhi/+ | 0 | 34 | 32 | 28 | 7 | 224 | ||||||||
Phenotypic classes are represented as 0 (weaker) to 4 (stronger). Data are shown as percentage of wings. En and Ptc columns show the number of wing discs with wild-type, reduced, or eliminated expression of Engrailed (En) and Patched (Ptc) in anterior cells. The Smo column represents the number of wing discs with wild-type or increased expression of Smo in anterior cells. All crosses were done at 29°C with the driver 638-Gal4.
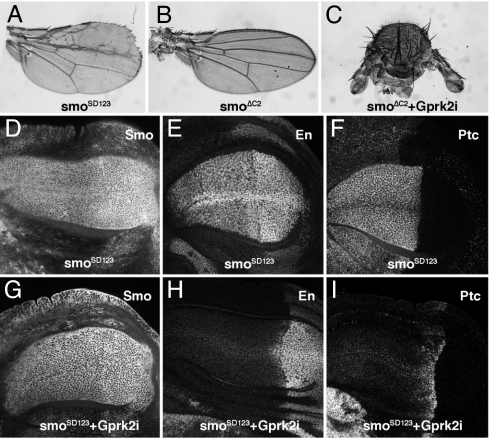
Fig. 5.
Interactions between Gprk2 and the intracellular domain of Smo. (A) Phenotype caused by the expression of the Smo phosphomimic form SmoSD123 in the wing blade at 25°C. (B and C) Phenotype caused by ectopic expression of a Smo intracellular deletion (638-Gal4/UAS-SmoΔC2; B) and severe loss-of-Hh signaling phenotype resulting from the coexpression of Gprk2 RNAi in this genetic background (638-Gal4/UAS-SmoΔC2+UAS-Gprk2i; C). (D–F) Expression of Smo (D), En (E), and Ptc (F) in wing discs of 638-Gal4/+; UAS-SmoSD123 /+ genotype. Note the expansion of Smo, En, and Ptc domains of expression to the entire anterior compartment. (G–I) Expression of Smo (G), En (H), and Ptc (I) in wing discs of 638-Gal4/+; UAS-SmoSD123 /UAS-Gprk2i genotype. The anterior expression of En (H) and Ptc (I) is now suppressed upon a reduction in Gprk2 levels. Despite of the presence of the SmoSD123 protein in the cell membrane of anterior cells (G), the pathway does not activate the expression of the high-level targets En and Ptc.
To directly monitor the activity of the Hh pathway, we studied the expression of several Hh targets in Gal4–638/UAS-Gprk2i discs. The expression of En (Fig. 2 K, L, and L″ and Table 1) and Ptc (Fig. 2 M and M′ and Table 1) in anterior cells is always impaired when Gprk2 levels are reduced. These two genes correspond to Hh targets activated by a high level of signaling (18). The expression of Knot (Kn) is also reduced in Gal4–638/UAS-Gprk2i discs (Fig. 2 K′–L′), and the stripe of maximal accumulation of Ci is also modified in Gal4–638/UAS-Gprk2i discs compared with wild-type ones (Fig. 2 N and N′). We also analyzed in Gal4–638/UAS-Gprk2i discs the expression of other genes regulated directly or indirectly by Hh signaling. The expression of the Notch ligand Delta (Dl) is very weak or absent in the primordia of the veins L3 and L4 (Fig. 2Q′), where it accumulates at high levels in normal discs (Fig. 2Q). Similarly, the expression of Bs in the L3/L4 intervein is reduced or absent in Gal4–638/UAS-Gprk2i discs (Fig. 2 O and O′). The expression of the low-level Hh signaling targets caup and decapentaplegic (dpp) is also modified in Gal4–638/UAS-Gprk2i discs. Caup expression in the presumptive L3 vein is generally expanded toward the A/P compartment boundary in Gal4–638/UAS-Gprk2i discs (Fig. 2 R and R′), most likely because En, a repressor of Caup in anterior cells (19), is not expressed upon a reduction of Gprk2 levels. We found that the domain of Caup expression in the L3 vein is reduced or lost compared with wild-type discs in only a small fraction of discs (7%) (data not shown). The expression of dpp is detected in Gal4–638/UAS-Gprk2i discs at lower levels but in a domain broader than the characteristic of normal discs (Fig. 2 S and S′). Taken together, our data suggest that Gprk2 plays a positive role in the Hh signaling pathway. The lowering of Gprk2 levels reduces very efficiently high-level Hh signaling and much less efficiently low-level Hh signaling. Thus, a complete elimination of Hh signaling is only observed when Gprk2 levels are reduced in wing discs with lower hh (Gal4–638/+; UAS-hhi / UAS-Gprk2i; Fig. 2H). Finally, the expression of spalt, a target of the Dpp/BMP4 pathway (20), is almost normal upon Gprk2 reduction (Fig. 2 P and P′), indicating specificity of Gprk2 function toward Hh signaling.
To confirm the specificity of the Gprk2 RNAi, we also analyzed the expression of two Hh-targets, En and Ptc, in wing disc cells homozygous for a deficiency that removes all of the Gprk2 coding region [Df(3R)Gprk2; see SI Fig. 6 and Materials and Methods]. In both cases we found that anterior Gprk2− clones eliminate, in a cell-autonomous manner, the anterior expression of En (70% of 22 clones; Fig. 2 T–U′) and Ptc (Fig. 2 V and V′). Gprk2− clones located in the posterior compartment did not affect the expression of En, confirming that Gprk2 activity is required in cells receiving Hh.
Gprk2 Participates in the Reception of the Hh Signal.
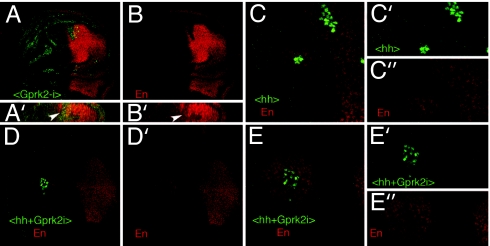
To further analyze where Gprk2 function is required in the Hh signaling pathway, we studied the expression of En in clones of cells ectopically expressing hh or both hh and Gprk2i (Fig. 3). We found that clones expressing Gprk2i located in the domain of En expression in the anterior compartment cell-autonomously suppress the expression of En (Fig. 3 A–B′). The expression of En is induced by Hh signaling in hh-expressing clones, both within the clone and in the surrounding cells (Fig. 3 C–C″ and ref. 5). However, in the hh+Gprk2i-expressing clones, the expression of En is only induced in wild-type anterior cells that do not express Gprk2i (Fig. 3 D–E″). These observations confirm that Gprk2 activity is required for transducing the Hh signal in Hh-receiving cells and not for Hh secretion.
Fig. 3.
GPRK2 function is required downstream of Hh. (A and B) Expression of Engrailed (En, red) in wing discs carrying clones of Gprki-expressing cells induced in hs-FLP; Ubx/abx Gal4-lacZ/UAS-Gprk2i (B) corresponds to the red channel, and the clones are labeled in green. The expression of En is lost in anterior cells. A′ and B′ show Z sections of the clones shown in A and B, respectively. (C) Expression of Engrailed (En, red) in wing discs carrying clones of Hh-expressing cells induced in hs-FLP; Ubx/abx Gal4-lacZ/UAS-hh larvae (<hh>, green). En is activated in anterior cells both within hh-expressing cells (green) and in the surrounding wild-type cells. Individual channels are shown for En (red, C″) and Hh-expressing cells (green, C″). (D–E) Expression of Engrailed (En, red) in wing discs carrying clones of cells expressing Hh and Gprk2i (<hh+Gprk2i>, green). En is activated in anterior cells surrounding the clones but not within the clone itself. Individual channels are shown below for En (D′ and E″) and (Hh+Gprk2i)-expressing cells (E′). E–E″ are higher magnifications of the clone shown in C.
Smo Protein Expression in Gprk2 Mutant Discs.
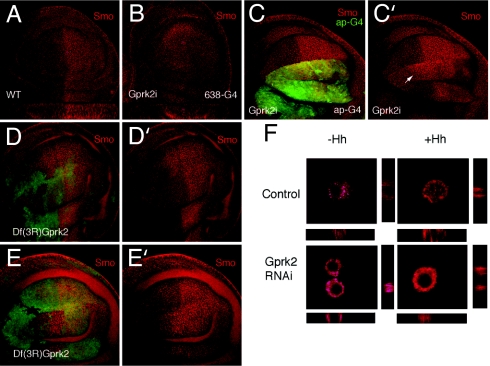
Experiments in mammalian cells in culture have shown that β-arrestin2 and GRK2 mediate internalization of active Smo (11). Consequently, we studied the expression and subcellular localization of Smo in wing discs where Gprk2 activity is reduced. In wild-type discs, smo RNA is expressed in all cells, but Smo protein accumulates associated to cell membranes only in the posterior compartment and in some anterior cells exposed to Hh (7, 9, 21) (Fig. 4A). Intriguingly, the reduction in Gprk2 levels in the entire wing blade eliminates the distinction in Smo accumulation between anterior and posterior cells, and Smo is detected at similar levels in both compartments (Fig. 4 A and B). When the levels of Gprk2 are reduced only in the dorsal compartment (Gal4-ap/UAS-Gprk2i; Fig. 4C) or in clones of Gprk2− homozygous cells (Fig. 4 D–E), the changes in Smo expression in anterior cells are more evident. Thus, we observed that Smo accumulates at high levels associated to cell membranes in a broader anterior domain of cells within the range of Hh (Fig. 4 C–E). The extension of Smo accumulation in anterior cells might be due to an extension of the Hh diffusion range because Ptc is not expressed in Gprk2 mutant cells (see ref. 22). This is, to our knowledge, a previously unrecognized instance in which Smo accumulation and signaling can be uncoupled, because it was thought that, at least in Drosophila, Smo membrane accumulation leads to signaling. The same effects are observed when S2 cells were used (Fig. 4F). Thus, Smo is expressed in S2 cells in intracellular vesicles at low levels (Fig. 4F Upper Left). Upon Hh treatment, Smo translocates close to the plasma membrane in these cells (Fig. 4F Upper Right). In cells that have been treated for 4 days with Gprk2 dsRNA (causing a reduction of Gprk2 mRNA levels of 77 ± 1.5%; data not shown) the levels of Smo are higher independently of Hh (Fig. 4F Lower).
Fig. 4.
Smo accumulation depends on Gprk2 activity. (A and B) Expression of Smo in wild-type (WT; A) and 638-Gal4/UAS-Gprk2i (GPRK2i; B) wing discs visualized by using mouse anti-Smo (red). Below each disc is a Z section through the middle of the wing blade. The apical side of the epithelium is up. (C and C′) Expression of GFP (green) and Smo (red) in ap-Gal4/UAS-Gprk2i UAS-GFP. Smo is accumulated in a broader domain in anterior-dorsal cells (white arrow). (D–E) Examples of Df(3R)gprk2 clones (labeled by the absence of green) showing increased expression of Smo (red) in anterior cells located close to the A/P compartment boundary. (F) Control S2 cells and S2 cells treated with dsRNAgprk2 for 4 days were plated on poly(lysine)-coated slides and incubated for 6 h with S2 or S2HhN-conditioned medium prior to fixation and immunoassayed for Smo.
To further analyze the relationship between Smo and Gprk2 functions, we expressed, in the same wing, Gprk2i with different N-terminal (smoΔN; extracellular) and C-terminal (smoΔC2 and smoΔC4; intracellular) deletions of Smo (9). The expression of Smo proteins bearing either N-terminal or C-terminal deletions fails to rescue Smo mutants (9), but their overexpression does not interfere significantly with Smo signaling (638-Gal4/UAS-smoΔC2; Fig. 5B and data not shown). We found a strong synergic genetic interaction when Smo C-terminal deletions (ΔC2; AT724 and ΔC4; AS939) (9) were coexpressed with Gprk2i (638-Gal4/UAS-smoΔC2+UAS-Gprk2i; Fig. 5C and data not shown). Thus, wings expressing C-terminal deletions of Smo with reduced Gprk2 levels display a strong hh loss-of-function phenotype that is comparable to the elimination of smo (see Fig. 2I). Gprk2i combined with UAS-smoΔN resulted in additive phenotypes (data not shown). We suggest that the reduction of Gprk2 uncovers a dominant-negative effect of SmoΔC proteins, reducing the efficiency of Smo signaling. The basis for this dominant negative effect could be the inclusion of a form of Smo, SmoΔC, unable to be phosphorylated by Gprk2, in the Smo complexes that have been postulated to mediate Smo activity (23). Therefore, we propose that Gprk2 function, acting through the C-terminal tail of Smo, is involved in an activation step promoting Smo interaction with the Costal2/Fused/Su(fu) complex to prevent Ci processing into a repressor form and to accumulate Ci in an activating form. Based on the effects of mammalian GRK2 and β2-arrestin on Smo (11, 24), it is possible that Gprk2-mediated activation of Smo involves the recycling of Smo from the cell membrane to an intracellular signaling compartment.
The interaction between SmoΔC and Gprk2 indicates a critical role of the Smo intracellular C-terminal domain for its relationship with Gprk2 function. Interestingly, the Smo intracellular C-terminal domain is where all of the consensus phosphorylation sites by casein kinase 1 and protein kinase A are located, as well as other serine and threonine residues in the vicinity of acidic residues that are similar to mammalian GRK2 phosphorylation consensus (25, 26). We expressed in the wing disc a form of Smo that mimics its phosphorylation by these kinases (SmoSD123; ref. 8) and analyzed whether this Smo-activated form is sensitive to Gprk2 levels. The expression of SmoSD123 in the wing disc causes overgrowth of the anterior compartment and defects in the L3 and L2 veins (Fig. 5A). In the corresponding wing discs, the accumulation of Smo and the expression of its targets En and Ptc are expanded to occupy the entire anterior compartment (Fig. 5 D–F). When Gprk2 levels are reduced in discs expressing SmoSD123, Smo accumulation is still observed in all anterior cells (Fig. 5, compare G with D). In contrast, the expression of both En and Ptc is now restricted to their normal domains adjacent to the A/P compartment boundary (Fig. 5, compare H and I with E and F). The overgrowth phenotype characteristic of Gal4–638/+; UAS- SmoSD123 discs (Fig. 5 D–F) is not rescued by the reduction of Gprk2 expression (Fig. 5 G–I), suggesting that the low-level Hh target dpp is still expressed through the anterior compartment. These data suggest that to generate the high levels of Smo activity required to activate the expression of its targets En and Ptc, the SmoSD123 protein has to be phosphorylated by Gprk2.
Conclusions
Drosophila Gprk2 is critically required to generate high levels of Hh signaling in the wing disc. The genetic interactions between Gprk2 and Smo proteins bearing C-terminal deletions or Smo phosphomimic variants suggest that Smo is a target of Gprk2. The modifications in Smo protein accumulation detected in wing discs and S2 cells with reduced Gprk2 expression suggests that a likely step affected by Gprk2 is the activation of Smo by a phosphorylation step that could prime Smo for internalization to a signaling compartment. GRK2 has recently been shown to play a positive role in Shh transduction in mammalian cells (24). Taken together, these findings and our data indicate that Smo phosphorylation by GRK homologues constitute a conserved component of the Smo signal transduction cascade.
Materials and Methods
Genetic Strains.
We used the Gprk2 alleles Gprk2EY09213, Gprk206936, and Gprk2PL00297, the Smo2 null mutation, the Gal4 lines Gal4–638, Gal4-ptc, Gal4-hh, Gal4-Ci, and Gal4-sal, and the UAS lines UAS-hh, UAS-ptc, UAS-SmoSD123, UAS-FLP, UAS-GFP, UAS-smoΔC2, UAS-smoΔC4, and UAS-smoΔN (8, 9). UAS-hhi flies were provided by the National Institute of Genetics (Mishima, Japan) stock center. The expression of Gal4–638 is restricted to the wing pouch since the second larval instar (see SI Fig. 6). Lines not described in the text can be found in FlyBase (17). Unless otherwise stated, crosses were done at 29°C.
Generation of UAS-Gprk2i.
The EST LD42147 was used as template for amplification of a 500-bp Gprk2 C-terminal fragment. The amplified fragment was cloned into pSTBlue-1 (Novagen, Madison, WI), from which the fragments BamHI+SacI and SphI+NotI were purified. The BamHI-SacI fragment was cloned in pHIBS and digested with SphI and XhoI. The fragments SphI-NotI and SphI-XhoI were cloned in pBluescript II SK+ (Stratagene, La Jolla, CA). The insert was liberated with KpnI and NotI and cloned in pUAST (27). Several UAS-Gprk2i were established after germ-line transformation following standard procedures.
Generation of a Gprk2 Deficiency.
We used the Exelixis flanking insertions f00526 and d09952 (28) separated by 54 kb including Gprk2 and the 5′ untranslated end of CG11337. Flipase (FLP)-induced recombination target recombination was induced by a daily 1-h heat shock at 37°C to the progeny of hsFLP1.22/+; f00526/d09952 females and w; TM2/TM6b males. Eight putative w; f00526-d09952/TM2 offspring males of white phenotype were individually crossed to w; TM2/TM6b females and after 3 days were used to extract genomic DNA to confirm by PCR the existence of FLP recombination target recombination. The position of the Exelixis flanking insertions f00526 and d09952 and the extent of the Gprk2 deficiency are described in SI Fig. 6.
Generation of FLP–FLP Recombination Target Clones.
We induced clones of cells expressing hh or Gprk2i and hh by a 10-min heat shock in larvae of hsFLP; abx/Ubx<f+>lacZ-Gal4/UAS-hh and hsFLP; abx/Ubx<f+>lacZ-Gal4/UAS-hh UAS-Gprk2i, respectively. The elimination of the f+ cassette by FLP-mediated recombination allows the expression of a dicistronic lacZ-Gal4 gene (29). Clones were identified by the expression of β-gal. Wings homozygous for smo2 were generated in Gal4–638/+; FRT42 smo2/FRT42 M (2)l2; UAS-FLP/+. Homozygous Df(3R)Gprk2 M+ clones were induced in larvae of genotype hsFLP1.22; FRT82 Df(3R)Gprk2 / FRT82 M (3)w Ubi-GFP. Homozygous Df(3R)Gprk2 cells were recognized in the wing disc by the absence of the GFP marker.
Cell Culture.
S2 cells were cultured in the Schneider's Drosophila serum-free medium (Invitrogen, Carlsbad, CA) with 10% FCS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transfection was carried out by using the Cellfectin Reagent Kit (Invitrogen) following the manufacturer's instructions. HhN inducible vector was provided by Stephen M. Cohen (European Molecular Biology Laboratory, Heidelberg, Germany). Stable cell lines overexpressing HhN were generated by puromycin treatment, and Hh S2-conditioned medium was obtained by incubation with 0.7 mM CuSO4 for 24 h as described (21). To reduce Gprk2 levels using an RNAi approach in S2 cells, we incubated them with 30 μg/ml of Gprk2 dsRNA (see below) in serum-free medium for 1 h followed by addition of 10% FCS and incubated for an additional 3–4 days to allow protein turnover before treatment with control or Hh-conditioned medium for an additional 6 h. The cells were plated and assayed for Gprk2 RNA quantification and processed for immunofluorescence.
Double-Stranded Gprk2 RNA Preparation.
We used the Gprk2 EST as template for amplification of Gprk2 DNA template flanked by T7 RNA polymerase binding sites. dsRNA was generated by in vitro transcription by using the Ambion MEGAsript T7 kit (Ambion, Austin, TX).
RNA Isolation and Quantitative Real-Time RT-PCR.
Total RNA was prepared from a pool of 30 wing discs (both wild type and Gal4–638/UAS-GPRk2i) or S2 cells by using the TRIzol reagent RNA protocol following Life Technologies (Grand Island, NY) instructions. Total RNA (0.7 μg) was used for a first round of reverse transcription by using the Gene Amp RNA PCR kit (Applied Biosystems, Foster City, CA). Quantitative PCR analysis was performed in a ABIPRISM 7.000 (Applied Biosystems) by using the TaqMan probe from Applied Hs99999901_s1 for 18S rRNA and the TaqMan UP probe no. 61 from Universal Probe Library (Roche, Indianapolis, IN) for Gprk2 and 20 ng of the corresponding cDNA. Quantification of mRNA reduction was performed with the ΔΔCt method.
Immunocytochemistry.
We used rabbit anti-Kn (18), anti-Sal (30), antiactivated Cas3 (Cell Signaling, Beverly, MA), mouse monoclonal anti-Bs (31), rat anti-Caup (32), and anti-Ci (33). From the University of Iowa Developmental Studies Hybridoma Bank (Iowa City, IA), we used the mouse monoclonals anti-En, anti-Ptc, and anti-Smo. Secondary antibodies were from Jackson ImmunoResearch (West Grove, PA) (used at 1:200 dilution). Imaginal wing discs were dissected, fixed, and stained as described in ref. 34. Confocal images were captured by using a confocal microscope (Bio-Rad, Hercules, CA). In situ hybridization with dpp and Gprk2 RNA probes was carried out as described in ref. 34. We used the EST LD42147 as a template to synthesize the Gprk2 probe.
Acknowledgments
We thank A. López-Varea and R. Hernández for their skillful technical help and P. Ingham (Centre for Developmental and Biomedical Genetics, University of Sheffield, Sheffield, U.K.), S. Cohen, and the National Institute of Genetics for providing tools necessary for this work. This work was supported by Dirección General de Investigación Científica y Técnica Grants BCM2003-1191 and GEN2001-4846-C05-01 (to J.F.d.C.) and SAF2005-03053 (to F.M.) and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.”
Abbreviations
- UAS
upstream activator sequence
- GRK
G protein-coupled receptor kinase
- A/P
anterior–posterior
- FLP
Flipase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702374104/DC1.
References
- 1.Lum L, Beachy PA. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 2.Hooper JE, Scott MP. Nat Rev. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 3.Tabata T, Kornberg TB. Cell. 1994;76:89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 4.Zecca M, Basler K, Struhl G. Development (Cambridge, UK) 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- 5.Strigini M, Cohen SM. Development (Cambridge, UK) 1997;124:4697–4705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Proc Natl Acad Sci USA. 2004;101:17900–17907. doi: 10.1073/pnas.0408093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu AJ, Zheng L, Suyama K, Scott MP. Genes Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia J, Tong C, Wang B, Luo L, Jiang J. Nature. 2004;432:1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 9.Nakano Y, Nystedt S, Shivdasani AA, Strutt H, Thomas C, Ingham PW. Mech Dev. 2004;121:507–518. doi: 10.1016/j.mod.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Huangfu D, Anderson KV. Development (Cambridge, UK) 2005;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 12.Lefkowitz RJ, Shenoy SK. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 13.Kohout TA, Lefkowitz RJ. Mol Pharmacol. 2003;63:9–18. doi: 10.1124/mol.63.1.9. [DOI] [PubMed] [Google Scholar]
- 14.Penela P, Ribas C, Mayor F., Jr Cell Signal. 2003;15:973–981. doi: 10.1016/s0898-6568(03)00099-8. [DOI] [PubMed] [Google Scholar]
- 15.Schneider LE, Spradling AC. Development (Cambridge, UK) 1997;124:2591–2602. doi: 10.1242/dev.124.13.2591. [DOI] [PubMed] [Google Scholar]
- 16.Lannutti BJ, Schneider LE. Dev Biol. 2001;233:174–185. doi: 10.1006/dbio.2001.0219. [DOI] [PubMed] [Google Scholar]
- 17.The FlyBase Consortium. Nucleic Acids Res. 2005;33:390–395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crozatier M, Glise B, Vincent A. Development (Cambridge, UK) 2002;129:4261–4269. doi: 10.1242/dev.129.18.4261. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Skarmeta JL, Modolell J. Genes Dev. 1996;10:2935–2945. doi: 10.1101/gad.10.22.2935. [DOI] [PubMed] [Google Scholar]
- 20.Barrio R, de Celis JF. Proc Natl Acad Sci USA. 2004;101:6021–6026. doi: 10.1073/pnas.0401590101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denef N, Neubuser D, Perez L, Cohen SM. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Struhl G. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 23.Hooper JE. Development (Cambridge, UK) 2003;130:3951–3963. doi: 10.1242/dev.00594. [DOI] [PubMed] [Google Scholar]
- 24.Reya RJ, Lefkowitz RJ, Caron MG. Mol Cell Biol. 2006;26:7550–7560. doi: 10.1128/MCB.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida N, Haga K, Haga T. Eur J Biochem. 2003;270:1154–1163. doi: 10.1046/j.1432-1033.2003.03465.x. [DOI] [PubMed] [Google Scholar]
- 26.Deupree JD, Borgeson CD, Bylund DB. BMC Pharmacol. 2002;2:9. doi: 10.1186/1471-2210-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 28.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, et al. Nat Genet. 2004;3:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 29.de Celis JF, Bray S. Development (Cambridge, UK) 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- 30.de Celis JF, Barrio R, Kafatos FC. Development (Cambridge, UK) 1999;126:2653–2662. doi: 10.1242/dev.126.12.2653. [DOI] [PubMed] [Google Scholar]
- 31.Nussbaumer U, Halder G, Groppe J, Affolter M, Montagne J. Mech Dev. 2000;96:27–36. doi: 10.1016/s0925-4773(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 32.Gómez-Skarmeta JL, Díez del Corral R, de la Calle E, Ferrer-Marcó D, Modolell J. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 33.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 34.de Celis JF. Development (Cambridge, UK) 1997;124:1007–1018. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]