Abstract
Scrub typhus is caused by the obligate intracellular rickettsia Orientia tsutsugamushi (previously called Rickettsia tsutsugamushi). The bacterium is maternally inherited in trombicuid mites and transmitted to humans by feeding larvae. We report here the 2,127,051-bp genome of the Boryong strain, which represents the most highly repeated bacterial genome sequenced to date. The repeat density of the scrub typhus pathogen is 200-fold higher than that of its close relative Rickettsia prowazekii, the agent of epidemic typhus. A total of 359 tra genes for components of conjugative type IV secretion systems were identified at 79 sites in the genome. Associated with these are >200 genes for signaling and host–cell interaction proteins, such as histidine kinases, ankyrin-repeat proteins, and tetratrico peptide-repeat proteins. Additionally, the O. tsutsugamushi genome contains >400 transposases, 60 phage integrases, and 70 reverse transcriptases. Deletions and rearrangements have yielded unique gene combinations as well as frequent pseudogenization in the tra clusters. A comparative analysis of the tra clusters within the genome and across strains indicates sequence homogenization by gene conversion, whereas complexity, diversity, and pseudogenization are acquired by duplications, deletions, and transposon integrations into the amplified segments. The results suggest intragenomic duplications or multiple integrations of a massively proliferating conjugative transfer system. Diversifying selection on host–cell interaction genes along with repeated population bottlenecks may drive rare genome variants to fixation, thereby short-circuiting selection for low complexity in bacterial genomes.
Keywords: bacterial genome, duplication, repeats
The obligate intracellular bacterium Orientia tsutsugamushi is the causative agent of scrub typhus (1). This disease is characterized by fever, rash, eschar, pneumonitis, meningitis, and disseminated intravascular coagulation, which, if left untreated, can lead to severe multiple organ failure (2). Mortality rates from scrub typhus in untreated patients range from 1% to 40%, depending on endemic area and strain of O. tsutsugamushi encountered (3). During World War II, Allied forces suffered more fatalities due to scrub typhus than as a direct consequence of the fighting in South-East Asia (4). Scrub typhus is restricted to a well defined geographic region that extends from Eastern Russia and Northern Japan in the north and Northern Australia in the south to Pakistan and Afghanistan in the west (3). This distribution distinguishes scrub typhus from other enzootic rickettsiosis and results from the ecology of the trombicuid mite vector and their vertebrate hosts (3). O. tsutsugamushi is harbored in the salivary glands of mites and is transmitted to humans and other hosts during larval feeding (5).
Scrub typhus cases can effectively be treated by using broad-range antibiotics such as doxycycline or chloramphenicol. However, treatment and disease control are complicated by frequent reinfection and relapses, which may be explained by the high antigenic diversity of this bacterium (6). Recent increases in the number of scrub typhus cases, coupled with observations of reduced effectiveness of antibiotic treatments (7–10), have highlighted the need for effective vaccines. However, vaccine development has been hampered by a limited host immune response (11) and O. tsutsugamushi's ability to immunosuppress the host (12).
We report here the complete genomic sequence of the O. tsutsugamushi Boryong strain, isolated from a Korean patient. The genome has an extraordinary structure, with 37% identical repeats in the form of mobile genetic elements and accessory genes, putatively involved in host–parasite interactions. These striking features provide insights into the evolutionary aspects of intracellular parasitism as well as the pathogenic properties of the scrub typhus agent.
Results and Discussion
Overview of the O. tsutsugamushi Genome.
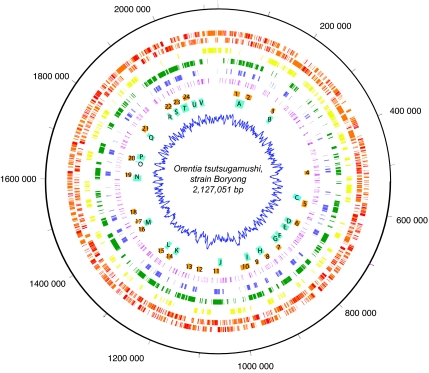
The complete genome of O. tsutsugamushi strain Boryong is a single circular chromosome consisting of 2,127,051 bp with an average G+C content of 30.5% [supporting information (SI) Table 1]. The genome size and estimated number of genes are the largest among the currently sequenced genomes in the order Rickettsiales. We identified 2,179 potential protein-coding sequences, of which an estimated 963 sequences represents fragmented genes (Fig. 1; red first and second circles), as a result of which the overall coding capacity of the genome is estimated to be as low as 49.6%. Despite the larger genome size, there is a core of 512 genes (Fig. 1; yellow third circle) shared with seven other Rickettsia species. The majority of fragmented genes coincided with repeated DNA regions, which are distributed throughout the whole genome (Fig. 1; green fourth circle). The O. tsutsugamushi genome showed little colinearity with the other Rickettsia genomes, and there was no systematic pattern in the GC-skew plot (Fig. 1; innermost circle), indicating that the genome has been extensively shuffled in recent evolutionary time.
Fig. 1.
Circular map of the O. tsutsugamushi genome. The outer circle shows plus-strand genes and the second circle, minus-strand genes; in red, fragmented genes, and in orange, full-length genes. The third circle shows Rickettsial core genes in yellow; the fourth circle, identical repeats >200 bp in green; the fifth circle, tra genes for components of conjugative TFSSs in blue; the sixth circle, transposases in pink; in the seventh circle, numbered color blocks represent the tra gene clusters in Fig. 3; in the eighth circle, colored letters represent tra-associated gene clusters in Fig. 3; and the innermost circle shows the GC skew plot (G−C)/(G+C).
Repeated Genes and Mobile Elements.
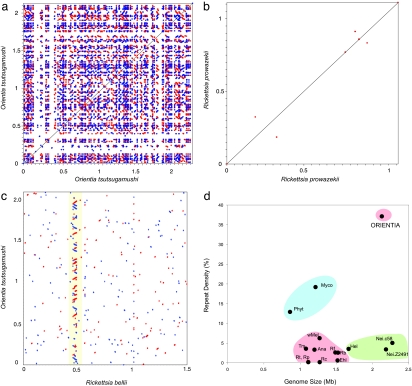
A unique feature is the presence of 4,197 identical repeats >200 bp, which represents 37.1% of the O. tsutsugamushi genome (Fig. 2a). The mean size of the perfect repeats was 947 bp, with a maximum size of 8,909 bp. Approximately 60% are present in three or more copies. The most abundant repeat was 211 bp in size and present in 16 copies. In addition, we identified 147 tandem repeats of cluster sizes 2–27 with 8–470 bp per unit repeat, as well as 14 palindromic repeats, the longest of which was 12,647 bp. As many as 90% of the tandem repeats are located within predicted genes, including reverse transcriptases, transposons, integrases, ankyrin repeat genes, tetratrico peptide-repeat genes and hypothetical genes (SI Table 2). A majority of the high-copy-number sequences represent mobile genetic elements, such as conjugative transfer genes, transposons, integrases, and phage genes (13, 14).
Fig. 2.
Repeat density in obligate intracellular bacteria. The frequency of repeats in the genomes of (a) O. tsutsugamushi and (b) R. prowazekii, with each dot representing a repeat >95% identical and >200 bp in size; blue, direct repeats; red, reverse repeats. (c) Scrambled gene order structures in O. tsutsugamushi and R. bellii. The yellow box indicates the location of the single tra cluster for TFSSs in the R. bellii genome and the homologous amplified tra clusters in the O. tsutsugamushi genomes. (d) Repeat density is plotted against genome size (% identical repeats >200 bp). Bacterial groupings are indicated by circles; red, obligate intracellular members of the Rickettsiales including Orientia, O. tsutsugamushi; Ana, Anaplasma marginale; Ehl, Ehrlichia ruminantium; Rb, R. bellii; Rc, Rickettsia conorii; Rf, R. felis; Rp, R. prowazekii; Rt, Rickettsia typhi; Trs, Wolbachia endosymbiont; TRS, wMel, Wolbachia wMel; green, Hel, Helicobacter pylori; Nei.c58, Neisseria meningitidis C58; Nei.z2491, N. meningitidis z2491; blue, the most highly repeated bacterial genomes until the time of this study are those of Myco, Mycoplasma mycoides; and Phyt, Phytoplasma strain OY M.
In total, we identified 1,146 mobile genetic elements, which correspond to ≈40% of the O. tsutsugamushi genome. Such a high fraction of repeats and mobile elements is all the more impressive in that repeat densities (>200 bp) for members of the Rickettsiales have until now not exceeded 10% (Fig. 2d). For comparison, the repeat density is 200-fold lower in the Rickettsia prowazekii genome (15) (Fig. 2b). The number of long identical repeats (1–7 kb) in O. tsutsugamushi is unprecedented among bacterial genomes (SI Fig. 5). Although tandem repeats represent only a small part of all repeats, tandem repeat copy numbers approach those of Ehrlichia ruminantium (SI Fig. 6), which has an atypical high content of tandem repeats due to selection for antigenic variation. However, the identity and size distribution profiles of the tandem repeats differ (SI Fig. 6), suggesting different strategies for genetic diversification in the two pathogens. With an overall repeat density twice as high as the most repetitive bacterial genomes [19.2% in Mycoplasma mycoides (16) and 12.9% in Phytoplasma (17)] (Fig. 2d), the O. tsutsugamushi genome is outstanding.
Expansion of Genes for Conjugative Type IV Secretion Systems (TFSS).
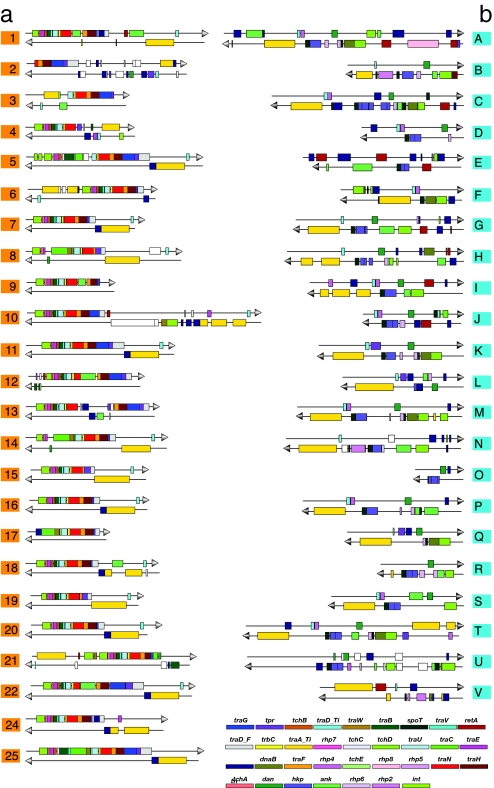
Conjugative TFSS mediate the transfer of DNA among bacteria (18), as well as the transport of bacterial effector proteins into the host cell during the establishment of an infection (19). Conjugation systems are rare in intracellular bacteria, which are estimated to contain only a single plasmid gene per genome on the average (20). The O. tsutsugamushi genome is exceptional in that it has 359 tra genes for conjugative TFSS, which should be compared with four plasmid-encoded tra genes in Rickettsia felis (21) and a single chromosomal copy of a tra operon in Rickettsia bellii (22). A plot of the gene-order structures in O. tsutsugamushi and R. bellii illustrates the conservation and amazing proliferation of the tra operon in O. tsutsugamushi (Fig. 2c) against a background of otherwise scrambled gene order structures. The tra genes are arranged into 24 fragmented repeat clusters with eight or more tra genes per cluster (Fig. 3a), as well as shorter tra clusters at 15 additional sites. Cluster 24 contained the most complete set of genes, with the central part of the repeat (traV-trbC-traN-traF) being preserved in most of the other 23 clusters. As many as 19 of the tra clusters are flanked on one side by integrase genes, another two contain internal integrase genes, and four are flanked on the other side by tRNA genes, consistent with the clusters being the amplified remains of one or more integrated genomic islands.
Fig. 3.
Gene order structure of (a) 24 long repeated tra gene clusters and (b) the flanking tra-associated gene clusters encoding putative effector proteins. The position of the clusters in the genome is indicated in Fig. 1.
Gene rearrangements and losses were seen in all clusters. Upstream of the central traV-trbC-traN-traF motif, 19 clusters showed deletions in the traC gene and 18, in the traB gene. Downstream, 15 exhibited deletions in traG and 4 in traH. The traA/I gene was missing from four clusters, translocated to the upstream region of the tra clusters in two cases, and another two clusters contained large insertions within the traA/I gene. As a result of the fragmentation process, gene and pseudogene sizes in the tra clusters varied extensively (SI Table 3). For example, the size of the traC segment was only 255 nucleotides in tra cluster three as compared with 2,562 nucleotides in tra cluster 24.
The application of phylogenetic methods to the analysis indicated that the amplified tra genes in O. tsutsugamushi were in all cases more similar to each other than either was to its homolog in R. bellii (SI Fig. 7). The tree topologies suggested groups of nearly identical genes, with different genes supporting different clustering schemes. A plot of the Shannon–Weiner diversity index in sliding windows along tra gene alignments revealed extensive sequence similarity across the gene copies, with some of the centrally located tra genes, such as traU, traW, and trbC having no polymorphic sites over most of the alignments (SI Fig. 8). Such a patched similarity pattern across and within the repeated tra genes is consistent with ongoing sequence homogenization by gene conversion and also with multiple insertions in a short time span.
Gene Family Expansion of tra-Associated Genes.
Located within or in the immediate vicinity of the tra clusters in O. tsutsugamushi are >200 genes encoding paralogous proteins putatively involved in signaling and host–cell interaction processes (Fig. 3b). For a list of the proteins in each family, see SI Tables 4–9, and for a representation of their domain structures, see SI Figs. 9–14. These include 27 TPR and 50 ankyrin-repeat proteins (23) and fragments thereof, some of which may mediate DNA and protein interactions with the eukaryotic host cell. Phylogenetic analysis of the TPR-repeat protein family (SI Fig. 15) indicates three groups that share a defined gene order and consists of one full-length “master” gene and several additional nearly identical gene or pseudogene copies. Based on phylogeny (SI Fig. 16), gene order, and domain structure (SI Fig. 10), the 40 ankyrin-repeat proteins formed three broad groups, and the remaining 10 single-copy genes showed a loose association with these groups. All ankyrin-repeat genes were flanked on one or both sides by tra genes, tra-associated genes, transposases, and/or reverse transcriptases.
Additionally, we identified 50 SpoT global regulators. O. tsutsugamushi is the only sequenced species in the Rickettsiales to have a full-length spotT/relA gene that contains both the synthase and hydrolase catalytic residues, and the total number of spoT/relA-like genes is five times that described in R. bellii. The other 49 spoT/relA-like ORFs identified contained either the synthase (n = 1) or hydrolase (n = 48) catalytic residue. Also identified were 56 histidine kinase (HK) and response regulator domains, 10 of which are typical HKs (24) and include pleCD, ntrYX, czcT, and phoB-like genes, most with homologs in Rickettsia and Wolbachia. The remaining 46 HKs were associated with the tra clusters, grouped by phylogenetic analysis with RBE_415 and were organized in the motif spoT-HK-dam. Also located in the vicinity of the tra clusters were gene fragments for 33 DnaB helicases and 34 Dam proteins.
Transposon-Mediated Diversity.
The 414 transposases identified in total were classified into five different families, IS5, IS630, IS982, IS110, and ISNCY, with 27–195 copies per family, with IS982 being the most abundant (SI Table 10 and SI Figs. 17 and 18). Approximately 86% are pseudogenes, with the most-abundant families containing the highest fraction of pseudogenes. Within each family, all copies >600 bp formed a single clade. Located within or in the immediate vicinity of the 24 tra clusters were ≈250 gene fragments for transposases and 51 retA genes for reverse transcriptases, which are probable constituents of retrotransposons. The reverse transcriptases were classified into two different types. The major type has 48 copies in the genome, whereas the minor type contains only three members. Additionally, we identified 61 phage-related ORFs, a majority of which were phage integrases associated with the tra clusters.
To study the mutational mechanisms of decay and expansion, we have compared the sequences of three regions with transposons in the sequenced Boryong strain with the strains Gilliam, Karp, and Kato. One of the selected regions contains a transposase and the short 5′-terminal fragment of the gltA gene in the Boryong genome. Although conserved in size across all four strains, a notable difference is that the gltA gene fragment in the Gilliam strain shows none of the frameshifts or premature stop codons observed in the other three strains (SI Fig. 19). This suggests that the gltA gene has been inactivated by a transposon insertion and is gradually eroding by deletion and nucleotide substitutions, with none of the strains encoding a functional citrate synthase.
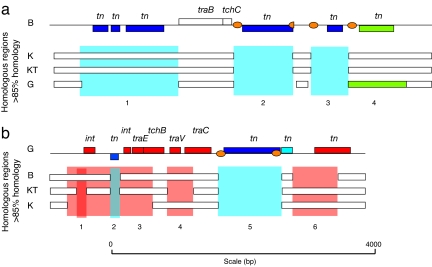
The other two regions cover multiple transposase pseudogenes and show a more extensive size variation among the various strains. The first region (at nucleotide position 172,980-176,345) shows a 48% size difference between strains Gilliam and Boryong, which is due to one vs. four segments with transposon pseudogenes, respectively. Two of the transposon sites (Fig. 4a, regions 2 and 3) are uniquely present in the Boryong strain, perhaps indicative of recent insertions in this strain. All transposons appear to have been rapidly deactivated, with insertion region 3 (Fig. 4a) representing a partially deleted transposon in the Boryong genome. The sole transposon identified in the Gilliam strain is located in segment 4, which is inverted in this strain relative to the corresponding segments in the other strains.
Fig. 4.
A sequence comparison of selected O. tsutsugamushi repeated regions from different clinical isolates. (a) Expansion of region due to transposon insertion. (b) Reduction of the region due to deletion. The top line shows the gene order structure in the Boryong strain (B), and the bars underneath represent homologs regions found in strains Karp (K), Kato (KT), and Gilliam (G). Blue indicates inferred transposon insertions and red color sequence deletions. Green indicates the insertion of a transposon in strain G.
The second region (at nucleotide position 1,316,813–1,322,637) shows a unique transposon insertion in the Gilliam strain that is not present in any of the other strains (Fig. 4b, region 5). Additional differences are due to strain-specific deletions in genes such as int, traE, traV, and traC. Taken together, the in-depth analyses of these regions show the influence of two opposing forces, expansions due to the insertions of transposons and contractions due to short deletions. Both of these processes are disruptive and occur at high frequencies.
Outer Membrane Proteins.
Selection for antigenic variability is a possible evolutionary driving force behind the multiplication of the tra-associated genes. The 56-kDa OmpA strain-specific antigen (1) is uniquely present in O. tsutsugamushi, whereas the 47-kDa OmpA group-specific protein has homologues all of the members of the Rickettsiales. Three additional OmpA-like membrane proteins were identified, two of which are located immediately downstream of the genes for the 47- and 56-kDa proteins, respectively, suggesting they may be coordinately expressed. A multigene family with 18 members also encode OmpA-like motifs and signal peptides, all of which are located within the regions of the integrated conjugative elements and may have been amplified together with the conjugative transfer genes. Additionally, O. tsutsugamushi has multiple proteins in the Sca family; these are known antigenic determinants in Rickettsia that contain autotransporter domains (25) and may play a role in adhesion to host cells (26). Another six genes also encode proteins with autotransporter domains, all of which have the transmembrane domains in the N-terminal region. Two of these are identical and may have been amplified through the action of transposons.
O. tsutsugamushi and the Genome Complexity Hypothesis.
The genome complexity hypothesis suggests that the enormous long-term effective population size of bacteria acts as an efficient barrier to the expansion of mobile elements and repeated sequences (27). This could explain why the content of repeated sequences in prokaryotes is normally much lower than in eukaryotes (27). With a repeat content (>200 bp) approaching 40%, i.e., similar to that of the human genome, the O. tsutsugamushi genome is exceptional among bacterial genomes. We have shown here that the high-repeat density is due to an extreme proliferation of transposons, conjugative elements, and a suite of associated genes coding for ankyrin-repeat proteins, TPR-repeat proteins, HK, and SpoT regulators. Although transposons may amplify trivially by continuous transposition events, we can think of two explanations for the presence of >20 fragmented operons for conjugation systems, namely massive duplications within the genome or repetitive invasions by conjugative integrative plasmids.
In bacterial genomes, gene duplications have so far been considered rare (28), and innovation has mostly been attributed to horizontal gene transfer (29–31). Hence, it is tempting to favor explanations based on multiple integrations of a similar type of plasmid. However, recent genomic studies point toward an important role of gene duplications (32–34). For example, the diversity of HK is driven by gene family expansion, rather than horizontal gene transfers (35). Tandem duplication is a mechanism that could generate many identical sequence copies. Indeed, the duplication-loss model suggests that the background frequency of tandem duplications is high, but that the copy number is normally kept in balance by an equally rapid collapse unless there is selection for more than one copy (36, 37). Speaking against such a scenario is that the tra clusters are not situated in tandem, that there are few tandem gene repeats inside the clusters, and that the sizes of the repeated clusters is >10-fold longer than normally found for tandem repeats, such as for example in E. ruminantium (38, 39). Furthermore, subsequent intragenomic recombination events must be invoked to explain the current distribution of the tra clusters across the genome. Although the presence of genes for recombination processes, such as recA and ruvABC, and sequence homogenization of the tra genes by gene conversion support active recombination processes in the genome, the mere existence of homologous recombination elements does not necessarily imply high recombination frequencies (40) nor does it rule out multiple plasmid invasions as the main source of genome complexity.
Another important issue that deserves discussion is whether the amplifications and rearrangements have contributed to the functional and antigenic diversity of the O. tsutsugamushi population. Previous studies of TFSS have shown that horizontal gene transfers are often associated with functional shifts (41). Likewise, detailed analyses of HK (35) suggest that gene family expansion is linked with domain shuffling and thereby with potentially modified gene functions. However, because many genes in the tra clusters are pseudogenized, their functional status has to await experimental verification; this is an interesting avenue for future research.
Before this study, only sporadic discoveries of mobile elements had been made in obligate intracellular lineages, such as of phages in Wolbachia pipientis (42, 43) and of a plasmid in R. felis (21). Other host-adapted bacteria, such the vertically inherited aphid and psyllid endosymbionts with genomes in the 180- to 600-kb range (44–48) are completely devoid of viral sequences, conjugative transfer systems and repeated sequences above 25 bp. These are also extraordinarily stable genomes with few rearrangements over time periods that span millions of years (45). The loss of repeats and mobile elements has been attributed to the influence of Muller′s ratchet on small bacterial populations that undergo frequent bottlenecks (49–51).
One hypothesis to explain the variability in repeat and mobile DNA content in intracellular bacteria is that the isolation of bacteria adapted to a single host acts as a barrier to horizontal gene transfer, whereas the more permissive lifestyle of host-switching bacteria facilitates the spread of mobile DNA (43). O. tsutsugamushi is a host-switching pathogen, which oscillates between two different types of inheritance patterns, maternal inheritance in its natural host, the mite, and horizontal transmission to small rodents and humans. Thus, conjugation among cells within the population or with other Rickettsia species during coinfections of the same host may facilitate the spread of plasmids and integrative conjugation systems. Different host adaptation strategies, such as the number of hosts and the density and duration of the infection, as well as the mode of transmission, may have a profound effect on bacterial population sizes and may go a long way toward explaining the different levels of genome complexity in bacteria with an obligate intracellular lifestyle.
We hypothesize that the proliferation of the conjugative secretion systems and the associated genes at one stage provided a basis for adapted evolution, possibly driven by positive or diversifying selection on bacterial components that target host proteins and/or trigger the host immune response in incidental hosts. In large free-living bacterial populations, genes that are selected during one growth regime but that are less fit in another environment will be rapidly eliminated. However, the small population sizes of obligate host-associated bacteria may render selection against selfishly reproducing sequences inefficient and slow down the loss of protein family members that no longer confer a selective advantage. Thus, the retention of pseudogenized clusters of tra genes and associated genes in the O. tsutsugamushi genome suggests that genetic drift can be more powerful than selection in shaping bacterial genome complexity, leading to the uncontrolled spread of selfish genetic elements.
Materials and Methods
DNA Preparation.
O. tsutsugamushi Boryong strain was propagated in L929 cells (DMEM, 5% FCS) and bacteria purified on a Percoll gradient (52). Bacteria were washed twice in TS buffer (33 mM Tris·HCl, pH 7.4/250 mM sucrose) and treated with TNE buffer (10 mM Tris·HCl, pH 7.4/150 mM NaCl/10 mM EDTA) supplemented with lysozyme (2 mg/ml) for 3 h at 37°C. Lysis was performed overnight with the addition of 1% SDS, RNase I (25 μg/ml), and proteinase K (2 mg/ml). After three rounds of phenol-chloroform extractions and ethanol precipitation, genomic DNA was resuspended in TE buffer.
Genome Sequencing Strategy.
Five genome shotgun libraries were generated by random shearing of genomic DNA. One fosmid library with mean insert size ≈40 kb (CopyControl Fosmid library production kit, Epicentre, Madison, WI) and four smaller insert libraries of 1.2–1.7, 1.8–2.6 (pBluescript, Stratagene, La Jolla, CA), 3, and 4 kb (pTrueblue, Genomics One, Quebec, ON, Canada). The following strategy was used to assemble a scaffold of 118 contigs (>1 kb) covering ≈2 Mb. First, all repetitive DNA sequences >300 bp that were represented more than three times were masked from the assembled contigs. Gaps between contigs (without repetitive sequences) were filled by using sequence data obtained from 51 sequenced fosmid clones or by primer walking. The final single contig had 14.5-fold coverage of high-quality DNA sequences (total 32 Mb) of the O. tsutsugamushi genome (≈2.2 Mb). Sequences were analyzed and assembled into contigs by using PhredPhrap and Consed software (53).
Because of the complexity of the genome, the integrity of the assembly was validated in three steps: (i) 100 fosmid clones (mean insert size ≈40 kb) that corresponds to a 2-fold coverage were analyzed by restriction mapping by HindIII or NspI and compared with an in silico digestion pattern of the assembled genome. (ii) Genomic regions containing repetitive sequences were randomly selected for amplification by PCR and sequencing. (iii) Genomic DNA was analyzed by pulsed-field gel electrophoresis after digestion with restriction enzymes (ApaI, SmaI, and NotI) and compared with an in silico digestion pattern of the consensus sequence. The whole genomic sequencing process was performed according to the Bermuda standard (error rate of 0.42 per 10 kb and the depth with at least two subclones for all nucleotide positions).
Informatics.
Putative protein-coding regions were identified with Glimmer (54) and Critica software (55). The tRNA genes were detected by tRNA-scan (56). Critica, Psi-Phi (57), BLAST (58), and substitution frequency estimates were used to identify putative pseudogene regions. Repeated genes and regions were detected and clustered by using the REPuter (59) and MUMmer (60) programs. Shared orthologs in seven Rickettsia species were identified by first clustering homologs into Tribe-MCL families and sorting these into true orthologs with the aid of conserved gene order structures, by using a software developed in-house (H.-H.F. and S.G.E.A, unpublished data). Homologs in O. tsutsugamushi were added to these clusters with the aid of Tribe-MCL. KEGG (61), COG (62), InterProScan (63), Pfam (64), and NCBI databases were used to predict putative functions of genes and domains. All predictions where confirmed, corrected, and verified manually.
Acknowledgments
We thank Rezin Dilshad for assistance with phylogenetic analyses. This research was supported by a grant from the Ministry of Health and Welfare, Republic of Korea (Grant 01-PJ10-PG6–01GM01-004, to I.-S.K.); the Swedish Research Council; the Göran Gustafsson Foundation; the Swedish Foundation for Strategic Research; and the Knut and Alice Wallenberg Foundation (to S.G.E.A.).
Abbreviations
- TFSS
type IV secretion system
- HK
histidine kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Molecular Biology database (accession no. AM494475).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611553104/DC1.
References
- 1.Seong SY, Choi MS, Kim IS. Microbes Infect. 2001;3:11–21. doi: 10.1016/s1286-4579(00)01352-6. [DOI] [PubMed] [Google Scholar]
- 2.Walker DH. Biology of Rickettsial Disease. Boca Raton, FL: CRC; 1998. [Google Scholar]
- 3.Kawamura A, Tanaka H, Tamura A. Tsutsugamushi Disease. Tokyo, Japan: University of Tokyo Press; 1995. [Google Scholar]
- 4.Philip CB. J Parasitol. 1948;34:169–191. [PubMed] [Google Scholar]
- 5.Furuya Y, Yoshida Y, Katayama T, Kawamori F, Yamamoto S, Ohashi N, Tamura A, Kawamura A., Jr J Clin Microbiol. 1991;29:2628–2630. doi: 10.1128/jcm.29.11.2628-2630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgeois AL, Olson JG, Fang RC, Huang J, Wang CL, Chow L, Bechthold D, Dennis DT, Coolbaugh JC, Weiss E. Am J Trop Hyg. 1982;31:532–540. doi: 10.4269/ajtmh.1982.31.532. [DOI] [PubMed] [Google Scholar]
- 7.Mathai E, Rolain JM, Verghese GM, Abraham OC, Mathai D, Mathai M, Raoult D. Ann N Y Acad Sci. 2003;990:359–364. doi: 10.1111/j.1749-6632.2003.tb07391.x. [DOI] [PubMed] [Google Scholar]
- 8.Watt G, Chouriyagune C, Ruangweerayud R, Watcharapichat P, Phulsuksombati D, Jongsakul K, Teja-Isavadharm P, Bhodhidatta D, Corcoran KD, Dasch GA, et al. Lancet. 1996;348:86–89. doi: 10.1016/s0140-6736(96)02501-9. [DOI] [PubMed] [Google Scholar]
- 9.Park JI, Han SH, Cho SC, Jo YH, Hong SM, Lee HH, Yun HR, Yang SY, Yoon JH, Yun YS, et al. Teahan Kan Hakhoe Chi. 2003;9:198–204. [PubMed] [Google Scholar]
- 10.Matsui T, Kramer MH, Mendlein JM, Osaka K, Ohyama T, Takahashi H, Ono T, Okabe N. Jpn J Infect Dis. 2002;55:197–203. [PubMed] [Google Scholar]
- 11.Eisenberg GH, Jr, Osterman JV. Infect Immun. 1979;26:131–136. doi: 10.1128/iai.26.1.131-136.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay S, Jiang J, Chan TC, Manetz TS, Chao CC, Ching WM, Richards AL. Infect Immun. 2005;73:5039–5047. doi: 10.1128/IAI.73.8.5039-5047.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrus V, Pavlovic G, Decaris B, Guedon G. Mol Microbiol. 2002;46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 14.Osborn AM, Boltner D. Plasmid. 2002;48:202–212. doi: 10.1016/s0147-619x(02)00117-8. [DOI] [PubMed] [Google Scholar]
- 15.Andersson SGE, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 16.Westberg J, Persson A, Holmberg A, Goesmann A, Lundeberg J, Johansson KE, Pettersson B, Uhlen M. Genome Res. 2004;14:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai X, Zhang J, Ewing A, Miller SA, Jancso Radek A, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, et al. J Bacteriol. 2006;188:3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen I, Christie PJ, Dubnau D. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cascales E, Christie PJ. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bordenstein SR, Reznikoff WS. Nat Rev Microbiol. 2005;3:688–699. doi: 10.1038/nrmicro1233. [DOI] [PubMed] [Google Scholar]
- 21.Ogata H, Renesto P, Audic S, Robert C, Blanc G, Fournier PE, Parinello H, Claverie JM, Raoult D. PloS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D. PloS Genet. 2006;2:e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolanin PM, Thomason PA, Stock JB. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, McNeill TZ, Jiang H, Muzny D, Jacob LS, et al. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux V, Raoult D. Int J Syst Evol Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- 27.Lynch M, Conery JS. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 28.Lerat E, Daubin V, Ochman H, Moran NA. Plos Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence JG, Hendrickson H. Mol Microbiol. 2003;50:739–749. doi: 10.1046/j.1365-2958.2003.03778.x. [DOI] [PubMed] [Google Scholar]
- 30.Doolittle WF. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 31.Lerat E, Daubin V, Moran NA. Plos Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gevers D, Vandepoele K, Simillon C, Van de Peer Y. Trends Microbiol. 2004;12:148–154. doi: 10.1016/j.tim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. Proc Natl Acad Sci USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLeod M, Warren RL, Hsiao WWL, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, et al. Proc Natl Acad Sci USA. 2006;103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alm E, Huang K, Arkin A PloS Comput. Biol. 2006;11:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson DI, Slechta ES, Roth JR. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- 37.Kugelberg E, Kofold E, Reams AB, Anderson DI, Roth JR. Proc Natl Acad Sci USA. 2006;103:17319–17324. doi: 10.1073/pnas.0608309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frutos R, Viari A, Ferraz C, Morgat A, Eychenie S, Kandassamy Y, Chantal I, Bensaid A, Coissac E, Vachiery N, et al. J Bacteriol. 2006;188:2533–2542. doi: 10.1128/JB.188.7.2533-2542.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins NE, Liebenberg J, de Villiers EP, Brayton KA, Louw E, Pretorius A, Faber FE, van Heerden H, Josemans A, van Kleef M, et al. Proc Natl Acad Sci USA. 2005;102:838–843. doi: 10.1073/pnas.0406633102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha EP, Cornet E, Michel B. PloS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank AC, Alsmark CM, Thollesson M, Andersson SGE. Mol Biol Evol. 2005;22:1325–1336. doi: 10.1093/molbev/msi124. [DOI] [PubMed] [Google Scholar]
- 42.Bordenstein SR, Wernegreen JJ. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- 43.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, et al. PLoS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 45.Tamas I, Klasson L, Canback B, Naslund AK, Eriksson AS, Wernegreen JJ, Sandstrom JP, Moran NA, Andersson SG. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 46.van Ham RC, Kamerbeek J, Palacios C, Rausell C, Abascal F, Bastolla U, Fernandez JM, Jimenez L, Postigo M, Silva FJ, et al. Proc Natl Acad Sci USA. 2003;100:581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Brocal V, Gil R, Ramos S, Lamelas A, Postigo M, Michelena JM, Silva FJ, Moya A, Latorre A. Science. 2006;314:312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 48.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 49.Andersson SG, Kurland C. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- 50.Dale C, Moran NA. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Andersson SGE. Science. 2006;314:259–260. doi: 10.1126/science.1133739. [DOI] [PubMed] [Google Scholar]
- 52.Tamura A, Urakami H, Tsuruhara T. Microbiol Immunol. 1982;26:321. doi: 10.1111/j.1348-0421.1982.tb00181.x. [DOI] [PubMed] [Google Scholar]
- 53.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 54.Salzberg SL, Delcher AL, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Badger JH, Olsen GJ. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 56.Lowe TM, Eddy SR. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lerat E, Ochman H. Genome Res. 2004;14:2273–2278. doi: 10.1101/gr.2925604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Nucleic Acids Res. 1990;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtz S, Schleiermacher C. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 60.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 62.Tatusov RL, Koonin EV, Lipman DJ. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 63.Zdobnov EM, Apweiler R. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 64.Sonnhammer ELL, Eddy SR, Birney E, Bateman A, Durbin R. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]