Abstract
We investigated patterns of genetic diversity of Plasmodium falciparum associated with its two main African vectors: Anopheles gambiae and Anopheles funestus. We dissected 10,296 wild-caught mosquitoes from three tropical sites, two in Cameroon (Simbock and Tibati, separated by 320 km) and one in Kenya (Rota, >2,000 km from the other two sites). We assayed seven microsatellite loci in 746 oocysts from 183 infected mosquito guts. Genetic polymorphism was very high in parasites isolated from both vector species. The expected heterozygosity (HE) was 0.79 in both species; the observed heterozygosities (HO) were 0.32 in A. funestus and 0.42 in A. gambiae, indicating considerable inbreeding within both vector species. Mean selfing (s) between genetically identical gametes was s = 0.33. Differences in the rate of inbreeding were statistically insignificant among sites and between the two vector species. As expected, because of the high rate of inbreeding, linkage disequilibrium was very high; it was significant for all 21 loci pairs in A. gambiae and for 15 of 21 pairs in A. funestus, although only two pairwise comparisons were between loci on the same chromosome. Overall, the genetic population structure of P. falciparum, as evaluated by F statistics, was predominantly clonal rather than panmictic, a population structure that facilitates the spread of antimalarial drug and vaccine resistance and thus may impair the effectiveness of malaria control efforts.
Keywords: malaria, epidemiology, evolutionary genetics, Cameroon, Kenya
Malaria is the most significant and widespread vector-transmitted human disease, accounting yearly for several hundred million clinical cases and >2 million deaths, mostly affecting young children and pregnant women in subSaharan Africa (1). Prevention and cure of the disease are major public-health challenges that are tackled by using several strategies, among them vector control. The transmission of Plasmodium falciparum, the agent of malignant malaria, involves a complex vectorial system consisting of ≈10 Anopheles species, colonizing different ecoclimatic settings, regions, and seasons in strongly variable relative abundances (2–5).
The two most important vectors of malignant malaria in Africa are Anopheles gambiae and Anopheles funestus because of their widespread distribution, highly anthropophilic and endophilic behavior, and long life spans (6, 7). A. gambiae is the most important vector throughout Africa and the most extensively studied Anopheles species (8). The effectiveness of malaria transmission emerges from the complementary ecoclimatic attributes and seasonal patterns of both species. A. funestus breeds in permanent larval sites that enable this species, in regions of seasonal transmission, to extend parasite transmission far into the dry season, after the temporary breeding pools of A. gambiae have dried out (7, 9–11).
Differences in the biology and ecology of these two main vectors might entail a differential impact on the genetic composition of the P. falciparum populations they harbor and transmit. We compared patterns of genetic diversity of P. falciparum isolated from midguts of wild-collected A. gambiae and A. funestus. The parasite population genetic structure in its mosquito vectors is of primary relevance to understanding the disease's epidemiology and its evolution (including predicting and monitoring the spread of drug resistance alleles) and to devising effective means for its control.
Most population genetic investigations of P. falciparum have focused on the haploid blood stages of the parasite in its human host (12–16). In contrast, we investigated the parasite's genetic diversity with oocysts dissected from mosquito guts, which allows the observation of the products of meiosis and, thus, the direct estimation of the population structure of the parasite. A few earlier studies considered oocysts' genetic diversity but targeted a small number of antigen-coding loci subjected to strong host immune selection (17–18), thus giving a possibly biased view of P. falciparum population structure. Using seven presumably neutral microsatellite loci, we have recently demonstrated that P. falciparum population structure is predominantly clonal in A. gambiae from Kenya (19). We now extend this approach to parasite populations from the two major African vector species, A. gambiae s.s. and A. funestus, collected in Central (Cameroon) and East Africa (Kenya) across successive years.
Results
Infection Rate and Parasite Distribution.
We dissected 10,296 wild-caught sympatric A. gambiae s.s. (71%) and A. funestus (29%) in three localities: Simbock and Tibati, located in distinct ecoclimatic areas of Cameroon (Central Africa), and Rota in western Kenya (East Africa). Mosquitoes were collected in 2002 (Simbock and Rota), 2003 (all three sites), and 2004 (Simbock). The midguts of 221 [2.1%, 95% confidence interval (C.I.) = 1.88–2.45] mosquitoes were infected with P. falciparum oocysts. The proportion of infected mosquitoes was higher for A. funestus than for A. gambiae in Simbock (4.6 vs. 1.8%, respectively, Fisher's exact test, P < 10−3) and Tibati (1.3 vs. 0.5%, respectively, P = 0.006), whereas no significant difference between the two species was observed in Rota (4 vs. 4.9% respectively, P = 0.3).
The number of oocysts per infected mosquito (Table 1) did not differ significantly between the two species: There were 6.8 oocysts per midgut (C.I. = 5.6–8.6) for A. gambiae and 5.9 (C.I. = 4.5–7.9) for A. funestus, either overall or within each site (bootstrap two-sample t test, P = 0.4). The frequency distribution of oocysts per mosquito was similar for the two species (stochastic equality test, P = 0.2). The index of discrepancy was 0.99 for both species; the variance-to-mean ratios were 17.7 and 17.6 (Table 1). These values are characteristic of the highly aggregated oocyst distributions generally observed in anopheline mosquitoes, with a high proportion of the parasites concentrated in a small fraction of hosts (19–21).
Table 1.
Distribution of P. falciparum oocysts in A. gambiae and A. funestus in three African sites and different years
| Population |
A. gambiae s.s. |
A. funestus |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mosquito host sample |
P. falciparum oocysts within mosquitoes |
Mosquito host sample |
P. falciparum oocysts within mosquitoes |
|||||||||
| Total | No. infected | Percent infected(95% C.I.) | Total(range) | Mean per mosquito(95% C.I.) | Var/mean ratio | Total | No. infected | Percent infected(95% C.I.) | Total (range) | Mean per mosquito(95% C.I.) | Var/mean ratio | |
| Simbock 2002 | 867 | 17 | 2.0 (1.2, 3.1) | 85 (1–30) | 5.0 (2.9, 10.3) | 14.1 | 130 | 2 | 1.5 (0.3, 5.6) | 13 (2–11) | 6.5 (2.0, 6.5) | 9.6 |
| Simbock 2003 | 397 | 9 | 2.3 (1.1, 4.2) | 69 (1–15) | 7.7 (3.9, 11.0) | 11.4 | 336 | 18 | 5.4 (3.4, 8.3) | 71 (1–12) | 3.9 (2.6, 5.4) | 6.2 |
| Simbock 2004 | 961 | 13 | 1.4 (0.8, 2.3) | 79 (1–25) | 6.1 (3.1, 11.7) | 14.6 | 275 | 14 | 5.1 (3.0, 8.5) | 37 (–8) | 2.6 (1.7, 3.9) | 4.1 |
| Tibati 2003 | 3,677 | 18 | 0.5 (0.3, 0.8) | 189 (1–40) | 10.5 (6.1, 17.3) | 23.7 | 1,127 | 15 | 1.3 (0.8, 2.2) | 132 (1–40) | 8.8 (5.1, 16.3) | 20.4 |
| Rota 2002 | 743 | 20 | 2.7 (1.7, 4.1) | 117 (1–30) | 5.8 (4.0, 9.8) | 12.5 | 608 | 19 | 3.1 (1.9, 4.8) | 189 (1–54) | 9.9 (6.1, 18.2) | 25.6 |
| Rota 2003 | 689 | 51 | 7.4 (5.6, 9.6) | 334 (1–44) | 6.5 (4.5, 9.5) | 18.6 | 486 | 25 | 5.1 (2.4, 7.6) | 104 (1–25) | 4.2 (2.4, 7.6) | 13.2 |
| Total | 7,334 | 128 | 1.7 (1.5, 2.1) | 873 (1–44) | 6.8 (5.6, 8.6) | 17.7 | 2,962 | 93 | 3.1 (2.6, 3.8) | 546 (1–54) | 5.9 (4.5, 7.9) | 17.6 |
Genetic Polymorphism.
We assayed seven previously described microsatellite loci (13, 19, 22) located on five chromosomes (chr.): POLYa (chr. 4), TA60 (chr. 13), ARA2 (chr. 11), Pfg377 and PfPK2 (chr. 12), and TA87 and TA109 (chr. 6). The analyses were based on 746 oocysts successfully isolated from 183 mosquito guts: seven loci amplified from 678 oocysts, plus six and five loci genotyped from additional 48 and 20 oocysts, respectively. The allele frequencies are given in supporting information (SI) Table 3.
All loci were highly polymorphic, showing an average (±SE) of 15.9 (±2.2) alleles per locus, ranging from 7 in Pfg377 to 23 in TA109. The mean allelic richness of parasite subpopulations within mosquitoes across sites and samples was identical in the two vectors (1.79 ± 0.05 for A. gambiae and 1.79 ± 0.04 for A. funestus) and did not differ significantly within each site between the two species (Wilcoxon two-sample test, P = 0.7).
The genotypic frequencies suggested considerable inbreeding in the P. falciparum populations of either vector. The total observed and expected frequencies of heterozygotes were, respectively, HO = 0.42 ± 0.03 and HE = 0.79 ± 0.05 in A. gambiae and HO = 0.32 ± 0.03 and HE = 0.79 ± 0.04 in A. funestus. The two species did not differ in the mean expected heterozygosities of their parasite populations (SI Table 3) within each site or within the total sample (Wilcoxon two-sample tests, P > 0.3).
The discrepancy between observed and expected heterozygosities remained high and significant for each locus and within each mosquito species when we considered only mosquito guts with 10 or more oocysts per gut (HO = 0.38 ± 0.02 and HE = 0.79 ± 0.04 in A. gambiae and HO = 0.35 ± 0.05 and HE = 0.70 ± 0.05 in A. funestus).
Population Structure.
We used F statistics to explore the pattern and distribution of heterozygote deficits. For the whole population of oocysts distributed in their respective mosquito hosts (subpopulations), the total heterozygote deficit FIT, which measures the overall departure from panmixia, was FIT = 0.53 (C.I. = 0.49–0.56, P < 10−4). For oocyst subpopulations from the two species separately, FIT = 0.47 (C.I. = 0.43–0.51, P < 10−4) for A. gambiae and FIT = 0.60 (C.I. = 0.57–0.64, P < 10−4) for A. funestus. This difference was due to the fraction of the total allelic variance of P. falciparum oocysts that was distributed among mosquitoes in the two vector species (see section titled FST between mosquitoes). The total heterozygote deficit FIT can be partitioned into two components: FIS, which measures the deficit due to the deviation from panmixia within a mosquito gut, i.e., the deviation from random union of gametes within each oocyst relative to gametes from different oocysts within the same mosquito; and FST, which measures the heterozygote deficit due to the partitioning of the total allelic variance among populations. This latter measure can be further split into a component that measures heterozygote deficit due to differences between subpopulations (oocysts within mosquito guts) within a sample (FST between mosquitoes) and one that measures heterozygote deficit due to the three additional parameters that contribute to population structure: sampling date, site, and Anopheles species.
FIS within mosquitoes.
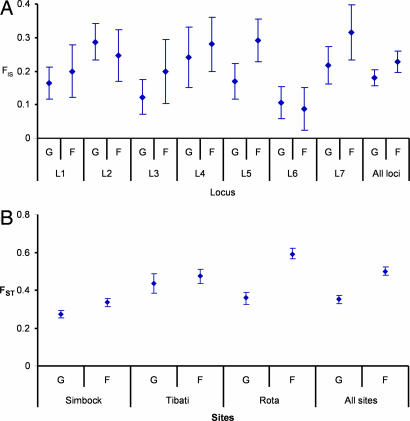
FIS measures departure from panmixia due to the nonrandom association of alleles within oocysts in mosquitoes (23). FIS was calculated for the 112 mosquitoes (subpopulations) with at least two oocysts. The average FIS was 0.18 (C.I. = 0.14–0.23, P < 10−4) for A. gambiae (68 subpopulations) and 0.23 (C.I. = 0.17–0.28, P < 10−4) for A. funestus (44 subpopulations). Single-locus FIS estimates were also positive and significant (Fig. 1A) and did not differ between species when all loci were combined (Wilcoxon two-sample test, P = 0.3) or treated separately (see C.I. 95% of Fig. 1 A).
Fig. 1.
Inbreeding coefficient statistics for P. falciparum oocysts collected from A. gambiae (G) and A. funestus (F) mosquitoes in Simbock and Tibati (Cameroon) and in Rota (Kenya). (A) FIS measures the deficiency of heterozygotes resulting from nonrandom union of gametocytes within individual mosquito guts; bars indicate the 95% C.I. for each FIS value. (B) FST measures the deficiency of heterozygotes resulting from nonrandom distribution of oocyst genotypes among mosquito guts. L1 to L7 are the following seven microsatellite loci, respectively: POLYa, TA60, ARA2, Pfg377, PfPK2, TA87, and TA109 (ref. 19).
We estimated the selfing rate (s) from the relationship FIS = s/(2 − s), assuming the FIS within mosquitoes only resulted from self-fertilization. The mean selfing rate for the whole sample was s = 0.33 (C.I. = 0.24–0.42); for each vector species, s = 0.30 (C.I. = 0.24–0.37) for A. gambiae and 0.37 (C.I. = 0.29–0.45) for A. funestus.
FST between mosquitoes.
A nonrandom distribution of genotypes among mosquitoes accounted for the largest component of the departure from panmixia. For the total population, FST = 0.41 (C.I. = 0.39–0.43, P < 10−4); for A. gambiae, FST = 0.35 (C.I. = 0.33–0.37, P < 10−4); and for A. funestus, FST = 0.50 (C.I. = 0.48–0.52, P < 10−4). The variation among samples ranges from FST = 0.27 (C.I. = 0.25–0.29) for A. gambiae in Simbock to FST = 0.59 (C.I. = 0.57–0.62) for A. funestus in Rota (Fig. 1B). The FST values observed between mosquitoes were similar to those previously observed for populations of P. falciparum from Papua New Guinea (13) or from Kenya (for example, average FST = 0.36 for 11 localities in Kenya; see ref. 19).
FST between sites.
There were no significant differences among sampling sites: FST ranged from 0.004 to 0.02 (P > 0.1) using HIERFSTAT (see Materials and Methods). We have also used FSTAT version 2.9.4 to assess differentiation among sites because the values obtained can be compared with published data on genetic differentiation of oocysts. The estimates of differentiation between sites ranged from 0.02 to 0.12, indicating weak but significant spatial differentiation in all cases (Table 2).
Table 2.
Geographical genetic differentiation between parasite populations from three African sites within and between Anopheles species
| Species |
A. gambiae |
A. funestus |
||
|---|---|---|---|---|
| Tibati | Rota | Tibati | Rota | |
| A. gambiae | ||||
| Simbock | 0.04 (0.02–0.07)* | 0.02 (0.01–0.02)* | 0.03 (0.02–0.04) | 0.07 (0.04–0.10)* |
| Tibati | 0.04 (0.02–0.07)* | 0.12 (0.07–0.18)* | ||
| A. funestus | ||||
| Simbock | 0.05 (0.03–0.08)* | 0.02 (0.01–0.04)* | 0.03 (0.02–0.04)* | 0.08 (0.04–0.11)* |
| Tibati | 0.03 (0.02–0.04)* | 0.10 (0.08–0.13)* | ||
FST and 95% C.I. (15,000 bootstrap simulations over loci) are shown. Departure of FST from zero was tested by using the Bonferroni-corrected P value as implemented in FSTAT version 2.9.4.
*Statistically significant (P < 0.001).
FST between sampling dates.
Simbock was sampled in 2002, 2003, and 2004; Rota was sampled in 2002 and 2003. The allele frequency distributions were similar at different times (for all HIERFSTAT tests, P > 0.05). We therefore combined the samples from different times at these two sites.
Genetic differentiation between Anopheles species.
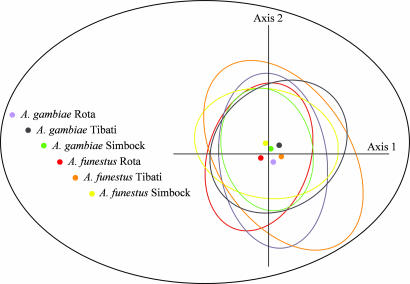
We used canonical correspondence analysis (24) to determine the relative contribution of the vector species to the global genetic structure of the P. falciparum populations. A graphic representation of the results shows largely overlapping distributions (centroids and ellipses of the 95% C.I. in Fig. 2) of the genetic variability of P. falciparum between vector species and among sites. Thus, the vector species did not contribute to the genetic population structure of the parasite.
Fig. 2.
Results of the canonical correspondence analysis. Relative contribution of the variables “site” and “Anopheles species” to the genetic structure of P. falciparum populations infecting the two Anopheles species in the three sites. Oocyst populations were projected on the first two axes of the canonical analyses. Centroids (dots) of each parasite population are surrounded by the 95% C.I.s (ellipses).
A Monte Carlo permutation test on the first four canonical correspondence analysis axes that combines species and sites is statistically significant (P = 0.001). This suggests some degree of geographic differentiation (see FST between sites; Table 2), although not between species. This interpretation was corroborated by FSTAT version 2.9.4 analysis, performing 20 random samplings of two sets of oocyst subpopulations within each species at each site. The interspecies FST values obtained were all included inside the range of intraspecies FST values.
Linkage Disequilibrium.
Linkage disequilibrium was significant for each of the 21 pairs of loci in A. gambiae, with R = 0.60–0.69 (P < 10−3 with Bonferroni correction), and for 15 of 21 pairs in A. funestus, R = 0.53–0.79 (P < 10−3). The difference between the two species is likely due to the smaller number of oocyst subpopulations (i.e., infected mosquitoes with more than one oocyst per gut) in A. funestus (44) than in A. gambiae (68). In Simbock, 20 of 21 pairs display significant linkage disequilibrium in 2002, 19 of 21 pairs in 2003, and 19 of 21 pairs in 2004. In Rota, all 21 pairs were in linkage disequilibrium in 2002 and 2003. Thus, linkage disequilibrium is stable over years in each site, corroborating that the “clonal” population structure persists over the generations.
The seven microsatellite loci are distributed among five different chromosomes; therefore, only 2 of 21 pairwise comparisons involved loci on the same chromosome (19).
Discussion
Our results address three issues with important epidemiological consequences: the degree of inbreeding and the extent to which the population structure of P. falciparum is closer to clonal rather to panmictic; the degree of genetic differentiation among geographically separated populations of P. falciparum; and the distribution of the Plasmodium oocyst pool within its major mosquito vector species, A. gambiae and A. funestus, which may be indicative of a differential effectiveness in malaria transmission between the two species.
The “clonal theory” of parasitic protozoa (25) proposed that the population structure of many parasitic protozoa, and in particular malaria, appears to be a result of clonal propagation rather than of randomly mating parasites, although the relative importance of both reproductive mechanisms can vary within and among parasitic species in different regions and epidemiological settings. Plasmodium was considered a likely exception because this idea seems to be in conflict with the biology of the species: The transmission of the parasite from a human host to another necessarily requires a sexual stage in the mosquito vector and, thus, the formation of gametes, fertilization, and meiosis. However, although the sexual reproduction in the mosquito host is obligatory, it does not necessarily result in recombination between genetically distinct gametes. If the combining gametes both derive from the same clonal population of haploid ancestors, fertilization will occur between genetically identical male and female gametocytes, and thus physiological sexuality does not necessarily imply genetic sexuality. The issue remains of the extent to which fertilization will imply genetically identical or different gametes, in natural populations of P. falciparum sampled from different vector species, and/or in different epidemiological settings.
The extent of cross-fertilization and panmixia relative to clonal reproduction in P. falciparum has indeed been the subject of considerable debate during the last decade (13, 19, 26–30). The current consensus is that this parasite species displays a range of population structures associated with the levels of transmission intensity (13). Regions of high P. falciparum transmission intensity correlate with high prevalence (31–32) and high genetic diversity of the parasite (13, 15, 33), which result in a high proportion of (genetically) mixed infections in humans. Increased genetic diversity in the human host increases the probability that the mosquito will obtain genetically distinct gametocytes with its blood meal. This, in turn, increases the probability of parasite outcrossing within the mosquito gut (34–35). Conversely, higher levels of inbreeding and clonal population structure are expected in regions with lower transmission rates (13, 36).
We collected our mosquitoes in three different African regions of high-transmission intensity, with an annual entomologic inoculation rate ranging between 100 and 300 infective bites per person per year (2, 37). Accordingly, we observed high genetic diversity of P. falciparum in each of the two vector species: HE = 0.79 ± 0.05 for A. gambiae and HE = 0.79 ± 0.04 for A. funestus. These values are similar to those obtained in a recent study using the same microsatellite loci to explore the level of genetic diversity in P. falciparum oocyst stages in Kenya (HE = 0.76–0.88; ref. 19). Our results are also consistent with several investigations conducted on haploid blood stages in highly endemic African populations (12, 13, 15, 38, 39).
In agreement with the clonal theory and despite the high endemicity, we found strong and highly significant heterozygote deficits at the subpopulation (mosquito) level: In both A. gambiae and A. funestus, the observed heterozygosities were only approximately one-half of the expected values under the hypothesis of random union of the gametes. This led to estimates of the self-fertilization index (s) of 0.30 and 0.37, respectively, for A. gambiae and A. funestus, i.e., that approximately one gametocyte of every three or four is involved in selfing. These selfing values are similar to those previously reported for the oocyst populations of A. gambiae in Kenya: s = 0.25 (FIS = 0.15) (19).
Read et al. (40) estimated from observations of gametocyte sex ratios in the peripheral blood of patients from Madang in Papua New Guinea that 62% of all P. falciparum matings are between genetically identical gametes. Based on evidence from oocyst genotyping using antigen-coding loci, overall inbreeding coefficients were estimated to be 0.90 in Papua New Guinea (18) but 0.34 in Tanzania (17). This difference was attributed to the lower levels of malaria transmission in Papua New Guinea as compared with Tanzania (18). However, in an earlier study (19), we observed that selfing in P. falciparum is not a local strategy that might occur in some but not other localities or occasions, but rather that it is a generalized life history trait of P. falciparum, which occurs even in high-transmission regions. Moreover, the observation made earlier (19) that there is in P. falciparum a considerable proportion of additional inbreeding due to nonrandom distribution of oocyst genotypes among mosquito guts (average FST = 0.35 for 11 P. falciparum populations from Kenya infecting A. gambiae) was confirmed in the present study for the two vectors, A. gambiae and A. funestus (average FST = 0.35 and 0.50, respectively).
The evidence of considerable inbreeding derived from heterozygote deficits was confirmed by the high level of linkage disequilibrium between loci observed in all three localities and in either vector species. High linkage disequilibrium can be due to the high level of inbreeding arising both from selfing and from the consequent high relatedness among oocysts within each mosquito, so that there is little opportunity for recombination. Similar results have been obtained with parasitized blood samples (13, 17, 19, 39, 41) from different African regions and different years or seasons displaying a very wide variety of transmission intensities. The conclusion is that the population structure of P. falciparum is predominantly clonal and that this is the case for parasites obtained from both of the two major vectors of P. falciparum in Africa, A. gambiae and A. funestus. Thus, P. falciparum cannot be considered a panmictic species even in African regions with the highest endemicity. This has important epidemiological consequences: nonrandom mating and “clonality” in highly endemic regions can facilitate the spread of antimalarial drug resistance (42) and thus rapidly impair the effectiveness of measures for malaria control if monitoring lags behind.
How much genetic differentiation occurs among different regions or among vector species also has considerable epidemiological consequences, one of the most important being the understanding of the spread of the parasite resistance to drugs and vaccine (42). The degree of geographic differentiation between populations separated by ≈320 km (Simbock to Tibati) or >2,000 km (Cameroon to Kenya) is significant but quite low, FST = 0.02 to 0.12, by one measure (FSTAT version 2.9.4.) and not significant, FST = 0.004 to 0.02, by a different statistical method (HIERFSTAT; see Results). Bogreau et al. (39) have observed in blood samples a substantial degree of differentiation between urban and rural sites at the scale of the African continent (FST = 0.17–0.24). However, microsatellite investigations of blood stages, conducted in highly endemic regions, are consistent with those reported in the present paper, showing low levels of geographic differentiation even over considerable distances. Geographic differentiation is weak among four Sudan sites separated by 420 km (FST = 0.006–0.11; ref. 12). Similarly, the genetic differentiation among microsatellite loci from three different countries is very low (FST = 0.04; ref. 27). From human bloodstages, Anderson et al. (13) noted that the genetic variation of P. falciparum at the African scale is distributed mostly within localities and not at all or very little among them (FST = 0.003–0.012), despite distances >2,000 km. We found the same feature of the genetic variation distribution in Africa at the oocyst level by sampling the parasite within the mosquito definitive host.
Finally, our study addresses the role of the mosquito vector in shaping P. falciparum population structure. Approximately 10 Anopheles species are vectors of P. falciparum in Africa (3, 4). The most important ones are A. gambiae and A. funestus because they are widely distributed throughout the continent, are highly anthropophilic and endophilic, and exhibit high survival rates (6, 7). In Kenya, the incidence of infected mosquitoes was not significantly different between the two vector species A. gambiae and A. funestus, but in the two Cameroon populations, the proportion of infected mosquitoes was significantly higher in A. funestus than in A. gambiae and as it was for all samples combined (3.1% vs. 1.7%, Table 1).
For large-scale malaria vector control, it is critical to understand whether ecological and other biological differences between the two most important vectors affect the population structure of the parasite with possible consequences on their efficiency in malaria transmission. One obvious major concern is the possibility for the P. falciparum parasite to evolve and adapt differentially to the different Anopheles species present in a malaria transmission area. If this is the case, implementation of a strategy aiming at neutralizing one vector species by introgression of a transgenic resistance trait may buffer the advantage created by the introduction of the transgene because it may liberate a niche for a rare parasite strain adapted to a different vector species to exploit, with major consequences on the efficiency of vector control strategies. Despite its potentially great significance, we know of no study, other than the present one, that has addressed this issue. Our results demonstrate a lack of evidence for any significant influence of malaria vector species diversity on the population structure of the transmitted pathogen, both A. gambiae and A. funestus being equally prone to be infected by any P. falciparum genotype. The population structure of P. falciparum is thus similar for the two species, with no segregation of particular P. falciparum genotypes within any of the two major African vector species.
Materials and Methods
Sampling Sites.
Mosquitoes were collected in two sites from two ecoclimatic areas of Cameroon, Central Africa, Simbock (lat 3°51′N, long 11°30′E) and Tibati (lat 6°28′N, long 12°37′E), and from Rota (lat 0°06.3′S, long 34°40.3′E) in Western Kenya, East Africa. Simbock is a suburb of Yaoundé, the capital of Cameroon, within the equatorial climate zone. Annual rainfall is ≈1,500 mm spread throughout the year. There are two rainy seasons, one from September to November and a shorter one from March to June. Tibati is in a rural setting ≈320 km north of Simbock, within the highland tropical climatic area of the Adamawa Plateau, with annual rainfall >1,500 mm and mean annual temperature of ≈22°C. Vegetation is of the Sudan–Guinean type. The single rainy season extends from March to October (43). Rota is in a rural setting, on the marshes of Lake Victoria in Western Kenya, with a tropical climate and annual rainfall >1,000 mm. In all three highly endemic malaria localities, transmission is perennial, with inoculation rates of ≈200 (Tibati), ≈300 (Simbock), and 100–300 (Rota) infective bites per human per year (2, 3, 37).
Mosquito Collections and Field Processing.
Anopheles mosquitoes were collected in Simbock in November 2002, March 2003, and November 2004; in Tibati in September 2003; and in Rota from February to May in 2002 and 2003 (two replicate samples). In Cameroon, mosquitoes were captured between 2000 and 0600, indoors and outdoors, before feeding, from 10 houses ≈1 km or less apart in Simbock. In Tibati, mosquitoes were captured from seven houses, <5 km from each other. Specimens from Rota were aspirated from the walls of 10 huts in the morning and reared for 1 week in the laboratory. A. gambiae s.s. and A. funestus females were found in all three sites surveyed (along with other P. falciparum-transmitting Anopheles species; ref. 3). Once identified by means of morphological keys (6, 7), the mosquitoes were dissected in a saline solution on the morning of their collection in Cameroon and 1 week after collection in Rota to allow for the development of oocysts in infected females. Dissected midguts were checked for the presence of oocysts under a compound microscope. Oocyst-infected guts were stored individually, along with the remainder of the mosquito body, in 1.5-ml microtubes with 80% ethanol. Samples were transported to the Montpellier laboratory and stored at 4°C. Infected females belonging to the A. gambiae complex and A. funestus group were identified to species by PCR diagnostic assays (44, 45).
Oocyst Dissection.
Oocysts were dissected from each gut after gradual rehydration of the specimens in distilled water by using a Leica (Deerfield, IL) inverted DMIRB microscope. Individual oocysts were teased from the basal lamina of the midgut epithelium and transferred to individual 0.5-ml tubes with 10 μl of distilled water. Tips and dissecting tools were washed in Javel water and 70% ethanol and changed regularly to avoid contamination. Parasite DNA from each oocyst was extracted by using the DNeasy Tissue Kit of Qiagen (Valencia, CA).
Microsatellite Genotyping.
Seven microsatellite loci were selected among the 12 P. falciparum loci used by Anderson et al. (13), initially described by Su and Wellems (22). The seven microsatellite loci with their chromosome location and GenBank accession number are as follows: POLYa (chr. 4, G37809), TA60 (chr. 13, G38876), ARA2 (chr. 11, G37848), Pfg377 (chr. 12, G37851), PfPK2 (chr. 12, G37852), TA87 (chr. 6, G38838), and TA109 (chr. 6, G38842). We followed a two-step strategy for amplification (19). The microsatellite PCR products were sized relatively to an internal standard and resolved by using GENESCAN software version 3.1 on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Vector Infection Rate and Parasite Distribution.
Rates of mosquito infection, number of oocysts per mosquito, mean intensity, indices of parasite distribution within mosquitoes (variance-to-mean ratio and index of discrepancy; ref. 46), and comparisons between Anopheles species were computed by using the QUANTITATIVE PARASITOLOGY 3.0 computer program (47).
Genetic Analysis.
Genetic variability was quantified as allelic richness by using FSTAT version 2.9.4 (48, 49), as well as observed (HO) and expected heterozygosity under panmixia (HE), by using the unbiased estimator of Nei (50), correcting for small samples. Comparisons between populations were performed by using nonparametric tests on two or more samples (Wilcoxon two-sample and Kruskal–Wallis tests) by using the R software (51, 52).
We used FSTAT version 2.9.4 (49) to calculate F statistics. For a two-level population structure (individual and subpopulation levels), the total FIT, measuring the inbreeding coefficient of an individual (I) relative to the total (T), can be partitioned into FIS, inbreeding coefficient of an individual (I) relative to the subpopulation (S), which measures departure from Hardy–Weinberg expectations under the hypothesis of panmixia within each subpopulation; and FST, which measures genetic differentiation between subpopulations (S) in the total population (T): FIT = 1 − (1 − FIS) (1 − FST). The data set was organized into three levels: (i) oocysts within the mosquito (subpopulation or the mosquito level), (ii) oocysts infecting mosquitoes within a site (population or site level), and (iii) all oocysts infecting mosquitoes in the whole geographic area sampled (metapopulation or geographic area level).
FIS was estimated and tested by randomizing alleles between oocysts within each of the 112 oocyst subpopulations (112 infected mosquitoes with more than one oocyst). FST between subpopulations (the fraction of the total allelic variance due to clustering of mosquitoes within geographical sites) was estimated and tested by randomizing oocyst genotypes between subpopulations (mosquitoes) within each population (site). Because FSTAT version 2.9.4 is designed for a two-level model and cannot take into account the three-level structure of our data set, differentiation between sites was estimated in two ways. First, the fixation index FST was tested by using FSTAT version 2.9.4 on all oocysts sampled in each locality by randomizing oocyst genotypes between populations relative to total. Second, because the highly significant differentiation observed between subpopulations strongly affects population differentiation computed between oocyst populations from different vectors, sites, or years (53), the fixation index was also estimated by using HIERFSTAT version 0.04-2 (54), which computes F statistics from any number of hierarchical levels, enabling us to take into account the highly significant effect of the mosquito level on parasite population structure to estimate spatial differentiation between populations. Significance of the hierarchical F statistics was assessed by 1,000 permutations of subpopulations (mosquitoes) among populations (sites). The crossed factor “date of sampling” was also estimated, and its significance was assessed by using HIERFSTAT version 0.04-2.
The Anopheles species cannot be considered as a hierarchical level because mosquitoes from different species can feed on the same human host. To measure and illustrate levels of genetic differentiation between P. falciparum harbored by the two vectors in the three sites, a canonical correspondence analysis was first carried out by using CANOCO software. Canonical correspondence analysis searches for multivariate relationships between two data sets (e.g., genetic data and species environmental data) (55). Only oocysts for which all seven microsatellite loci could be scored were considered. The significance of the canonical axes was tested with a Monte Carlo permutation test (24), which allows estimation of the 95% C.I.s of the centroids of each population. A second analysis used FSTAT version 2.9.4., performing 20 random samplings of two equal sets of oocyst subpopulations within each Anopheles species in each site, to estimate and assess the significance of 20 FST pair values for each species. These intraspecies FST values were compared with the interspecies FST values of the oocyst subpopulations of the two species in each site.
Linkage disequilibrium between pairs of loci was measured with the correlation coefficient R generalized for the multiallelic case. Statistical significance was assessed by a permutation test: Genotypes at two loci are associated at random a number of times, and the log-likelihood G test statistic is recalculated on the randomized data. We applied a Bonferroni correction to the P values (P value times the number of tests), as implemented in FSTAT version 2.9.4.
Acknowledgments
We thank Pierre Kengne for technical support on Anopheles species identification and Yannis Michalakis, Thierry de Meeûs, and Frank Prugnolle for helpful discussions. We are grateful to the whole teams of the Laboratoire de Recherche sur le Paludisme of the Organisation de Lutte contre les Endémies en Afrique Centrale (OCEAC) and the Kenya Medical Research Institute (KEMRI) for their help. This work was supported by the Institut de Recherche pour le Développement (IRD), the Centre National de la Recherche Scientifique (CNRS), the Recherche sur le VIH/sida et sur le Paludisme program, and a student fellowship from François Lacoste and the Fondation de France and Fondation pour la Recherche Médicale Grant FDT 20051205940 (to Z.A.).
Abbreviations
- C.I.
confidence interval
- chr.
chromosome.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702715104/DC1.
References
- 1.World Health Organization. World Malaria Report 2005. World Health Organization and United Nations Children's Fund; 2005. [Google Scholar]
- 2.Antonio-Nkondjio C, Awono-Ambene P, Toto JC, Meunier JY, Zebaze-Kemleu S, Nyambam R, Wondji CS, Tchuinkam T, Fontenille D. J Med Entomol. 2002;39:350–355. doi: 10.1603/0022-2585-39.2.350. [DOI] [PubMed] [Google Scholar]
- 3.Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkham T, Fontenille D. J Med Entomol. 2006;43:1215–1221. doi: 10.1603/0022-2585(2006)43[1215:cotmvs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Fontenille D, Simard F. Comp Immunol Microbiol Infect Dis. 2004;27:357–375. doi: 10.1016/j.cimid.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Coluzzi M. Parassitologia. 1999;41:277–283. [PubMed] [Google Scholar]
- 6.Gillies MT, Coetzee M. Publ S Afr Inst Med Res. 1987;55:1–143. [Google Scholar]
- 7.Gillies MT, De Meillon B. Publ S Afr Inst Med Res. 1968;54:1–343. [Google Scholar]
- 8.della Torre A, Tu Z, Petrarca V. Insect Biochem Mol Biol. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Dia I, Diop T, Rakotoarivony I, Kengne P, Fontenille D. J Med Entomol. 2003;40:279–283. doi: 10.1603/0022-2585-40.3.279. [DOI] [PubMed] [Google Scholar]
- 10.Minakawa N, Sonye G, Mogi M, Githeko A, Yan G. J Med Entomol. 2002;39:833–841. doi: 10.1603/0022-2585-39.6.833. [DOI] [PubMed] [Google Scholar]
- 11.Fontenille D, Lochouarn L, Diagne N, Sokhna C, Lemasson JJ, Diatta M, Konate L, Faye F, Rogier C, Trape JF. Am J Trop Med Hyg. 1997;56:247–253. doi: 10.4269/ajtmh.1997.56.247. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Muhsin AA, Mackinnon MJ, Awadalla P, Ali E, Suleiman S, Ahmed S, Walliker D, Babiker HA. Parasitology. 2003;126:391–400. doi: 10.1017/s0031182003003020. [DOI] [PubMed] [Google Scholar]
- 13.Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, et al. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 14.Conway DJ, Machado RL, Singh B, Dessert P, Mikes ZS, Povoa MM, Oduola AM, Roper C. Mol Biochem Parasitol. 2001;115:145–156. doi: 10.1016/s0166-6851(01)00278-x. [DOI] [PubMed] [Google Scholar]
- 15.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 16.Mu J, Duan J, Makova KD, Joy DA, Huynh CQ, Branch OH, Li WH, Su XZ. Nature. 2002;418:323–326. doi: 10.1038/nature00836. [DOI] [PubMed] [Google Scholar]
- 17.Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T, Walliker D. Parasitology. 1994;109:413–421. doi: 10.1017/s0031182000080665. [DOI] [PubMed] [Google Scholar]
- 18.Paul RE, Packer MJ, Walmsley M, Lagog M, Ranford-Cartwright LC, Paru R, Day KP. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 19.Razakandrainibe FG, Durand P, Koella JC, De Meeus T, Rousset F, Ayala FJ, Renaud F. Proc Natl Acad Sci USA. 2005;102:17388–17393. doi: 10.1073/pnas.0508871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medley GF, Sinden RE, Fleck S, Billingsley PF, Tirawanchai N, Rodriguez MH. Parasitology. 1993;106:441–449. doi: 10.1017/s0031182000076721. [DOI] [PubMed] [Google Scholar]
- 21.Pichon G, Robert V, Tchuinkam T, Mulder M, Verhave JP. Parasite. 1996;3:165–167. [Google Scholar]
- 22.Su X, Wellems TE. Genomics. 1996;33:430–444. doi: 10.1006/geno.1996.0218. [DOI] [PubMed] [Google Scholar]
- 23.Weir BS, Cockerham CC. Evolution (Lawrence, Kans) 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 24.Ter Braak CJF. CANOCO – Fortran program for canonical community ordination. Ithaca, NY: Microcomputer Power; 1987. [Google Scholar]
- 25.Tibayrenc M, Kjellberg F, Ayala FJ. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rich SM, Hudson RR, Ayala FJ. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conway DJ, Roper C, Oduola AM, Arnot DE, Kremsner PG, Grobusch MP, Curtis CF, Greenwood BM. Proc Natl Acad Sci USA. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abderrazak SB, Oury B, Lal AA, Bosseno M-F, Force-Barge P, Dujardin J-P, Fandeur T, Molez J-F, Kjellberg F, Ayala FJ, et al. Exp Parasitol. 1999;92:232–238. doi: 10.1006/expr.1999.4424. [DOI] [PubMed] [Google Scholar]
- 29.Urdaneta L, Lal A, Barnabé C, Oury B, Goldman I, Ayala FJ, Tibayrenc M. Proc Natl Acad Sci USA. 2001;98:6725–6729. doi: 10.1073/pnas.111144998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartl DL, Volkman SK, Nielsen KM, Barry AE, Day KP, Wirth DF, Winzeler EA. Trends Parasitol. 2002;18:266–272. doi: 10.1016/s1471-4922(02)02268-7. [DOI] [PubMed] [Google Scholar]
- 31.Beier JC, Killeen GF, Githure JI. Am J Trop Med Hyg. 1999;61:109–113. doi: 10.4269/ajtmh.1999.61.109. [DOI] [PubMed] [Google Scholar]
- 32.Smith DL, Dushoff J, Snow RW, Hay SI. Nature. 2005;438:492–495. doi: 10.1038/nature04024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, Johnson JR, Le Roch KG, Sarr O, Ndir O, et al. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dye C, Williams BG. Proc Biol Sci. 1997;264:61–67. doi: 10.1098/rspb.1997.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill WG, Babiker HA, Ranford-Cartwright LC, Walliker D. Genet Res. 1995;65:53–61. doi: 10.1017/s0016672300033000. [DOI] [PubMed] [Google Scholar]
- 36.Smith JM, Feil EJ, Smith NH. Bioessays. 2000;22:1115–1122. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 37.Ndenga B, Githeko A, Omukunda E, Munyekenye G, Atieli H, Wamai P, Mbogo C, Minakawa N, Zhou GF, Yan GY. J Med Entomol. 2006;43:200–206. doi: 10.1603/0022-2585(2006)043[0200:pdomvi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 38.Pearce R, Malisa A, Kachur SP, Barnes K, Sharp B, Roper C. Mol Biol Evol. 2005;22:1834–1844. doi: 10.1093/molbev/msi177. [DOI] [PubMed] [Google Scholar]
- 39.Bogreau H, Renaud F, Bouchiba H, Durand P, Assi SB, Henry MC, Garnotel E, Pradines B, Fusai T, Wade B, et al. Am J Trop Med Hyg. 2006;74:953–959. [PubMed] [Google Scholar]
- 40.Read AF, Narara A, Nee S, Keymer AE, Day KP. Parasitology. 1992;104:387–395. doi: 10.1017/s0031182000063630. [DOI] [PubMed] [Google Scholar]
- 41.Durand P, Michalakis Y, Cestier S, Oury B, Leclerc MC, Tibayrenc M, Renaud F. Am J Trop Med Hyg. 2003;68:345–349. [PubMed] [Google Scholar]
- 42.Hastings IM. Parasitology. 2006;132:615–624. doi: 10.1017/S0031182005009790. [DOI] [PubMed] [Google Scholar]
- 43.Olivry JC. Fleuves et Rivières du Cameroun. Bondy, France: Orstom; 1986. [Google Scholar]
- 44.Koekemoer LL, Kamau L, Hunt RH, Coetzee M. Am J Trop Med Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- 45.Scott JA, Brogdon WG, Collins FH. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 46.Poulin R. Int J Parasitol. 1993;23:937–944. doi: 10.1016/0020-7519(93)90060-c. [DOI] [PubMed] [Google Scholar]
- 47.Rozsa L, Reiczigel J, Majoros G. J Parasitol. 2000;86:228–232. doi: 10.1645/0022-3395(2000)086[0228:QPISOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 48.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. [Google Scholar]
- 49.Goudet J. J Heredity. 1995;86:485–486. [Google Scholar]
- 50.Nei M. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Team RDC. R Foundation for Statistical Computing. Austria: Vienna; 2005. [Google Scholar]
- 52.Sokal RR, Rohlf RJ. Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman; 1995. [Google Scholar]
- 53.Yang RC. Evolution (Lawrence, Kans) 1998;52:950–956. doi: 10.1111/j.1558-5646.1998.tb01824.x. [DOI] [PubMed] [Google Scholar]
- 54.Goudet J. Mol Ecol Notes. 2005;5:184–186. [Google Scholar]
- 55.Ter Braak CJF. Ecology. 1984;67:1167–1179. [Google Scholar]