Abstract
At least 5% of the human genome predating the mammalian radiation is thought to have evolved under purifying selection, yet protein-coding and related untranslated exons occupy at most 2% of the genome. Thus, the majority of conserved and, by extension, functional sequence in the human genome seems to be nonexonic. Recent work has highlighted a handful of cases where mobile element insertions have resulted in the introduction of novel conserved nonexonic elements. Here, we present a genome-wide survey of 10,402 constrained nonexonic elements in the human genome that have all been deposited by characterized mobile elements. These repeat instances have been under strong purifying selection since at least the boreoeutherian ancestor (100 Mya). They are most often located in gene deserts and show a strong preference for residing closest to genes involved in development and transcription regulation. In particular, constrained nonexonic elements with clear repetitive origins are located near genes involved in cell adhesion, including all characterized cellular members of the reelin-signaling pathway. Overall, we find that mobile elements have contributed at least 5.5% of all constrained nonexonic elements unique to mammals, suggesting that mobile elements may have played a larger role than previously recognized in shaping and specializing the landscape of gene regulation during mammalian evolution.
Keywords: exaptation, genome evolution, transposon, vertebrate cis-regulation
Comparative analysis of mammalian genomes has recently revealed that at least 5% of the human genome evolves under purifying selection (1). Protein-coding exons are the most studied class of these conserved elements, yet they constitute only a third of this set, slightly more if related untranslated regions are included (2). Thus, the majority of conserved bases in the human genome do not appear in mature mRNA transcripts (reviewed in ref. 3).
Complex metazoans seem to harbor significantly more conserved non-protein-coding sequence than simpler organisms (4). In vertebrates, many of these regions seem to serve as regulatory elements controlling the transcription of nearby genes (5–8). The evolution of regulatory regions is believed to be a major force behind the observed morphological diversity within the vertebrate lineage (9, 10), yet how this additional regulatory sequence was created is currently far from understood.
More than 50 years ago, when transposable elements were first discovered, B. McClintock (11) termed them “controlling elements” because of how they affect the expression of neighboring genes. Fifteen years later, Britten and Davidson (12) expanded this idea by hypothesizing that repetitive elements can act to distribute regulatory sequences throughout the genome and, in doing so, enriching, possibly even creating, whole pathways.
First glimpses of this phenomenon were explored in the pregenomic era and compiled into a hand-curated list of cases where researchers had come across individual mobile element instances that acquired a cellular role (13), a process termed “exaptation” (as opposed to adaptation) by Gould and Vrba (14). In the early genomic era, 1 Mb of the human and mouse genomes was examined for exaptation of mobile elements (15). A later analysis of 1.9 Mb of the human genome sequenced in 28 additional mammals came up with another handful of ancestral repeats evolving under strong purifying selection (16). More recent works, focusing on large families of constrained paralogous non-protein-coding sequences (17, 18), were able in two cases to explicitly implicate these families as originating from mobile elements (19, 20). Recent work has also elucidated that some mobile elements may be rich in transcription factor-binding sites (21). Combined, these observations suggest that the ideas of McClintock, Britten, and Davidson should be revisited on a genomic scale.
Here, we perform a genome-wide scan for mobile element instances exapted into putative cis-regulatory roles, by analyzing a large set of constrained nonexonic sequences with clear repetitive origins. We find this set by looking for repetitive origins in a conservative set of putative cis-regulatory regions, which covers <1.5% of the human genome and has been under strong purifying selection since the boreoeutherian ancestor (100 Mya), predating the human–dog split. We show that even by these conservative measures, thousands of constrained nonexonic elements (CNE), totaling over one million bases, including >5% of all CNEs unique to mammals, were deposited by interspersed repeats. These elements are significantly enriched near genes associated with the regulation of transcription and development. We also show that particular repeat portions are preferentially exapted into nonexonic functions and examine the reelin pathway, where all known receptor-related genes have acquired similar putative regulatory regions by conserving a repeat instance of the same type.
Results
Constrained Nonexonic Elements from Transposable Origins.
To construct an initial set of highly conserved human elements, we combined three complementary approaches to detect purifying selection on the boreoeutherian subtree (see Methods for details): resistance to base substitutions (4), resistance to microinsertions and deletions (22), and a simple windowing method to calculate percent identity in a multiple alignment, combining resistance to both substitutions and in-dels. We applied these methods to a syntenic multiple alignment between human, chimp, rhesus (Macaque Genome Sequencing Consortium, personal communication), rat, mouse, and dog.
We used only the highest scoring elements from each method, and augmented these elements with clear syntenic alignments between human and chicken, frog, fugu, tetraodon, or zebrafish; no neutrally evolving DNA should be alignable at these distances (23). Combined, these regions cover 3.5% of the human genome, constituting a conservative set compared with the 5% or more believed to be under purifying selection (1).
To obtain a nonexonic subset, we filtered out all regions found in any known or reliably predicted mature transcript (see Methods). Remaining regions were then required to be within syntenic alignments between human and chimp, rhesus, rat, mouse, and dog, leaving us with 1.45% of the genome as constrained boreoeutherian nonexonic elements. Each of the four conservation measures uniquely contributes >8% of this set, attesting to the value of combining rather than arbitrating between them.
In each of these six species, we then intersected this set with mobile element subfamilies annotated by RepeatMasker (24, 25). We used only mobile element subfamilies that have a presence in primates, rodents, and dog. Because these subfamilies appear across the boreoeutherian subtree, we term them “pan-boreoeutherian” [supporting information (SI) Text, section S1, and SI Fig. 5]. The intersection of our conserved nonexonic elements with the pan-boreoeutherian repeat subfamilies resulted in a set of 10,402 highly constrained nonexonic elements with clear repetitive origins. All elements are at least 50 bp long, with a maximum of 489 bp and a mean of 100 bp. The set covers just over 1 Mb (0.04%) of the human genome.
Data Set Validation.
We used a second set of tools to reaffirm that these regions are indeed mobile element fragments evolving under purifying selection. First, we used Blastz (26) to realign all repeat consensus sequences to the human genome. Using sensitive thresholding, we were able to recover 98% of the constrained regions. Secondly, we validated that these regions are indeed evolving under purifying selection. The regions resisting insertions and deletions were previously shown to have a false discovery rate (FDR) of 1% (22). Using PhyloP (27) to compute the likelihood of a given multiple alignment under the species tree of neutral substitutions, all elements except 20 rejected the neutrality assumption at a FDR of 1%. The 20 exceptions all evolve less stringently within mammals, but each has a clear (>70%id) match to an orthologus region in a non-mammal and all were thus retained.
Constrained Regions Originate from All Walks of Transposon Life.
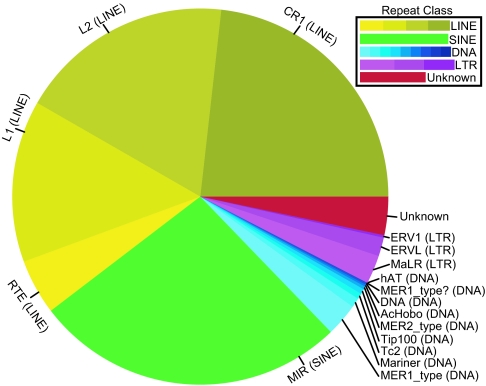
Fig. 1 shows the distribution of constrained nonexonic bases with respect to the progenitor mobile element. Strikingly, despite our stringent filtering, all four characterized classes of repeats are present, with long interspersed elements (LINEs) and short interspersed elements (SINEs) contributing the bulk of the constrained nonexonic sequence.
Fig. 1.
Contribution of repeat classes and families to the number of exapted bases surveyed. Autonomous LINEs and their SINE dependents dominate the set, yet all classes of mobile elements, including DNA transposons and LTRs, contribute nonnegligible amounts of novel functional DNA. The unknown class is made entirely of MER121 instances.
Comparing the distribution of CNEs from mobile elements to the overall abundance of each repeat in human (SI Tables 1–3), one can see a general trend where older repeats contribute proportionally more CNEs compared with their overall genome-wide abundance. This trend is partly a result of our strict screening. By focusing only on exaptations that predate our speciation from the carnivores (represented by dog) to support our functional claim, we bias against newer repeat subfamilies that may have undergone substantial proliferation after this split. Such is the case of the L1s and Alus that proliferated together as the L2/MIR pair was becoming less prevalent (28). In fact, the Alus that nowadays constitute >10% of the human genome are represented in our screen by a single subfamily, the “Fossil Alu Monomers” [FAM (29)], of which only a single instance is annotated in the dog genome. More ancient repeats likely have a higher ratio of exapted to genomic bases because, as a mobile element loses its ability to proliferate, all nonexapted copies continue to decay at a neutral rate, eventually mutating beyond our ability to identify their ancestry. After enough time, only exapted copies remain recognizable. Such seems to be the case of MER121, a paralog family of a thousand copies in the human genome whose evolutionary origins can now only be speculated to originate from an interspersed repeat (18, 25). Appropriately, this family makes up the “unknown” category in Fig. 1 and SI Table 1, and has the highest ratio of exapted to genomic copies.
Specific Parts of Mobile Elements Tend to Be Exapted.
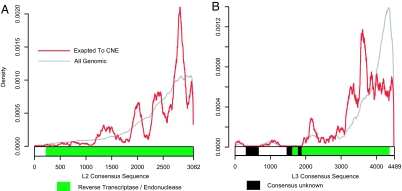
In the vast majority of instances, only a portion of the mobile element, rather than its entire length, exhibits extreme conservation. Truncation is a well known phenomenon in LINE repeats, where newly integrated copies are often truncated to varying degrees at their 5′ end (30). This phenomenon is apparent in a histogram showing how many times each base in the LINE consensus appears in the human genome (Fig. 2 A and B). Yet, a similar histogram of only exapted consensus regions departs markedly from this background, peaking at very different regions for both the L2 and L3 elements. This difference is suggestive not only of exaptation per se, but of one that depends on the sequence content of the LINE elements themselves. It could be that these sections of the LINEs are functional upon insertion, or become so after a few fortuitous mutations, and are therefore more likely to be exapted [as was previously observed for exonic exaptations (19, 31)]. SI Fig. 6 A and B gives two additional examples for other classes of repeats.
Fig. 2.
Preferential exaptation of specific portions of mobile elements. For each base in the mobile element consensus (x axis), the relative abundance is plotted (y axis). The abundance throughout the entire genome is shown in gray, and the abundance that has come under strong purifying selection for a nonexonic function is in red. The L2 (A) and L3 (B) genomic overabundance of 3′-end bases reflects the well known phenomenon of 5′ truncation in LINEs (30). In contrast, the distinct red peaks in each graph may well delineate the locations of gene regulatory elements overlapping the coding region of these two LINEs. Only the core consensus sequences of the L2 and L3, as defined by the RepeatMasker libraries, are shown (24).
Constrained Repetitive Elements Cluster Distally Around Developmental Genes and Transcription Regulators.
To obtain clues as to the putative functions of the exapted CNEs, we examined their relative abundance near functionally annotated genes. Distal enhancers can affect the transcription of a neighboring gene from a distance of as much as 1 Mb of genomic sequence (32). For this reason, we assigned exapted elements to the gene with the closest transcriptional start site (TSS), if one existed within 1 Mb. Our statistical test compares the distribution of exapted elements with a uniform distribution over all bases in the genome. For each term in the Gene Ontology [GO (33)], we calculated the P value for how surprising it is to see the number of exaptations assigned to genes with the given GO term, compared with the fraction of bases in the genome that will result in assignment to a gene carrying the same GO term.
This model is not biased by gene length because it uses only the location of the TSS. Nor is the model biased by genes residing next to gene deserts vs. those found in tight gene clusters, because larger basins of attraction result in a more probable null assignment. The overall top scoring GO term for this set is “development” (uncorrected P < 2 × 10−75). Many of its subterms are highly enriched as well, such as “system development” (4 × 10−55) and “nervous system development” (4 × 10−53). The second best scoring term is “transcription regulator activity” (2 × 10−72) along with many of its subterms. Of the more specific terms, we find particular enrichment for “cell recognition” (4 × 10−23) and related terms such as “neuron recognition,” “transmembrane receptor protein tyrosine kinase signaling,” “GPI anchor binding,” and “cell adhesion” (3 × 10−14). SI Table 4 shows the top scoring GO terms for this test.
The exapted regions tend to cluster around individual genes, often 20–30 instances within 1 Mb. To ensure that large clusters around a handful of genes are not the sole cause for GO term enrichment, we used the same association rule to assign exaptations to genes, but instead of using a uniform null distribution over all bases in the genome, we used a hypergeometric distribution over genes, now allowing each gene to be selected only once. The GO categories of “development” and “transcription regulator activity” were again at the top of the list with somewhat diminished but still very significant P values (8 × 10−24 and 6 × 10−19, respectively). “Cell adhesion” (6 × 10−11) and related terms also featured prominently. SI Table 5 shows all of the top scoring GO terms for this second test.
One may also suspect that mobile elements in general congregate near genes enriched for the observed GO terms, perhaps because the chromatin surrounding these genes is more accessible during germline transposition. This localization would cause any random subset of mobile elements to appear highly enriched for the same GO terms. To eliminate this possibility, we conducted a third, hypergeometric test where we assigned GO terms to the set of all mobile element instances from pan-boreoeutherian subfamilies, according to the above association rule of closest TSS within 1 Mb. We then computed the likelihood that the observed GO terms for genes near exapted CNEs can be obtained by selecting a random subset of repeats. This test resulted in extreme significance for the same terms, including “transcription regulator activity,” “development,” and “cell adhesion” (3 × 10−64, 3 × 10−60, and 2 × 10−15, respectively). The results of this third statistical test demonstrate that the enrichment we see is not due to an insertion bias of mobile elements, because the distribution of mobile elements exapted as CNE sequence is significantly different from the distribution of all mobile elements from these same families. Landing near a gene associated with development or transcriptional regulation makes an interspersed repeat much more likely to be exapted as a CNE. SI Table 6 shows all of the top scoring GO terms for this test, and SI Text, section S2, gives formal definitions of all three tests.
In search of subsets of exaptations whose annotation would suggest specific specialization, we also investigated GO enrichments at the different taxonomic levels of interspersed repeats (classes, families, and subfamilies), as well as for smaller sets of exaptations of the same consensus portion, or of smaller subsets of the most sequence similar exapted CNEs. Overall, these smaller categories were found enriched for the same functional annotation as the all-inclusive large set (SI Text, sections S3, S7, and S8, and SI Tables referenced therein).
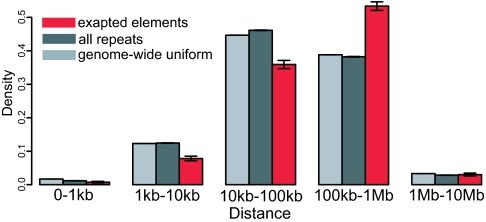
We compared a histogram of exapted CNEs distance from the nearest TSS with that of all bases in the human genome, as well as that of all repeat instances from pan-boreoeutherian subfamilies. As Fig. 3 shows, whereas both background distributions are similar, the exapted CNE set is clearly enriched for lying 0.1–1 Mb away from the nearest gene, at the expense of closer localizations. This enrichment suggests that these CNE are preferentially involved in distal cis-regulation.
Fig. 3.
Distribution of genomic distances to the closest known gene start site. We calculated the distribution of the distances to the closest TSS for three sets: all exapted CNE elements (red), all mobile element subfamilies predating the human–dog split of which the exapted CNEs are a subset (dark gray), and the set of all sequenced bases in the human genome (light gray). Although the set of all repeats is seen to be roughly distributed uniformly across the genome, the functional set departs from this null distribution by exhibiting a substantial overabundance at the distal range of 100 kb–1 Mb from the TSS. Error bars show the 95% confidence intervals for all three sets, but only the exapted elements have confidence intervals large enough to visualize. We calculated the confidence intervals by treating these data sets as samples from a multinomial distribution, and the confidence intervals are representative of the true proportions from which we were sampling.
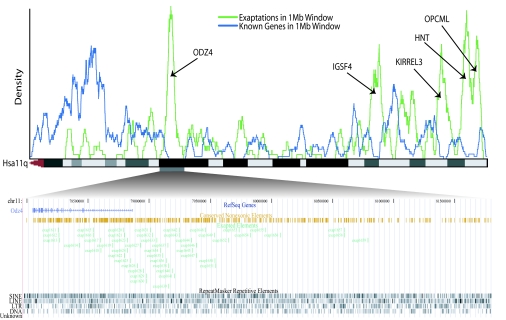
To further investigate the spatial congregation of exapted CNEs, we plotted the density of exaptations genome-wide, observing a very strong anti-correlation with gene density (Fig. 4). Indeed, the densest clusters are found in gene deserts most often flanked by genes involved in neuronal development, including cell adhesion (SI Table 16).
Fig. 4.
Spatial clustering and anti-correlation of exaptation events with gene density along human chromosome arm 11q. (Upper) The density of surveyed CNE exaptation events is plotted (green) and shown to anti-correlate with that of all protein coding genes (blue) along the ideogram of Hsa 11q. Exaptation clusters are clearly most often found in large gene deserts, suggesting that these clusters play cis-regulatory roles. We annotate the densest clusters (including the genome-wide top two from SI Table 16) with the name of a nearby gene that we speculate they regulate. (Lower) A zoom-in of nearly 4 Mb of the human genome holding the largest exaptation cluster (green), shown along with the Odz4 signaling and transcription regulatory gene (blue). The cluster is clearly centered around the transcription start site of the gene and mostly resides in its flanking gene desert. Also shown are the conserved nonexonic elements (gold) and RepeatMasker annotations (black), whose carefully controlled intersection resulted in the set of exapted CNEs.
Several additional avenues of investigation were inconclusive. The composition of local exaptation clusters appears diverse, overlapping documented enhancers were much longer than our exaptation events, and overlap with novel putative transcription start sites was inconclusive (SI Text, sections S4–S6).
Exaptations in the Reelin-Signaling Pathway.
Spurred by Britten and Davidson's early hypothesis (12), we attempted to investigate whether exapted elements, as a whole or broken by taxonomic groups, are also enriched for in particular molecular pathways (SI Tables 17 and 18). Unfortunately, mammalian pathway annotation is currently in its infancy, with only a small fraction of pathways annotated. Nonetheless, our attention was drawn to the reelin-signaling pathway, which allows neurons to complete their migration in the developing brain. Both the L1 family of LINEs and the MIRb subfamily of SINEs have at least one exaptation near each of the four genes that are known to be involved in response to the extracellular RELN signal: VLDLR, LRP8 (ApoER2), DAB1, and FYN. VLDLR and LRP8 are transmembrane receptors that, when bound by RELN, cause the tyrosine phosphorylation of DAB1 by FYN (34). Both enrichments are equally unlikely against the background genomic distribution of L1s and MIRb (5 × 10−6 and 7 × 10−6, respectively). The pathway itself is not completely understood downstream of these four genes. Interestingly, it is thought that some of the downstream targets could be cell adhesion molecules (35, 34), matching our observation above for the enrichment of exaptation events near these genes.
The MIRb exaptations all originate from overlapping sections of the MIRb consensus. It is thus plausible that these instances add similar regulatory regions to each gene in the receptor pathway. To examine this hypothesis, we identified potential transcription factor-binding sites orthologously conserved between human, chimp, macaque, rat, mouse, and dog (see Methods). Each of the four genes has an instance near it that contains orthologously conserved sites for En-1, Oct-1, YY-1, SRY, and v-Myb. However, whereas the position of these binding sites is conserved between species for each gene separately (CNE orthologs), no single predicted binding site comes from the same bases of the progenitor MIRb for all four genes (CNE paralogs). In fact, even irrespective of known binding sites, there are almost no columns where a base is perfectly conserved across all four paralog subtrees. The MIRb copies near each gene seem to have diverged differently from the consensus. However, the progenitor consensus sequence often has multiple predicted binding sites for each of the five transcription factors. Some orthologs seem to have conserved one or more of the instances, whereas their paralogs have conserved different binding sites for the same factor, thus presumably retaining function while diverging in sequence (SI Fig. 7).
Flux of CNE Exaptation from Mobile Elements.
Our conservative set of 10,402 exapted CNEs implicates 4% of all CNEs (2.5% of CNE bases) predating the human–dog split as having clear origins in mobile element insertions. Some CNEs, however, are as old as the vertebrate lineage itself (36). Current estimates suggest that repeat families can be recognized only if they are younger than 200 million years (Myr) (37), implying that some observed CNEs may well have evolved from exaptation of repeat families that have since decayed to the point where they cannot be recognized as such. We can thus attempt to refine our estimate of CNEs from mobile element origins, by considering only the subset of CNEs born after the avian–mammal split, represented by all 180,954 CNEs not found in the chicken genome. Of the identified exapted regions, 9,903 have no clear syntenic ortholog in chicken, suggesting that at least 5.5% of all CNEs born on this branch are from mobile elements. This estimate should increase as closer outgroups to the carnivore split, such as platypus and opossum are published (see SI Text, section S9, and SI Fig. 8). Normalizing for the estimated branch length between the avian and carnivore splits, we obtain a lower bound on the rate of exaptation on this branch that is ≈22,000 mobile elements exapted as CNEs for every substitution per site of branch length. The number of sampled species and branch lengths on the primate tree is currently insufficient to identify elements that have come under purifying selection during this time frame. However, well established examples make it clear that the mechanism of interspersed repeat exaptation into gene regulatory roles persists in the primate lineage (38, 39). A hypothetical extrapolation from the above estimate using the branch length from the extant human genome back to the speciation of Galago, one of the most distantly related primates, suggests that a substantial set of 2,650 CNE elements under strong purifying selection have been exapted from mobile elements in humans since this early primate ancestor.
Discussion
Revisiting an Age-Old Hypothesis.
McClintock's discovery that mobile elements can influence the expression of nearby genes (11) has been validated dozens of times (13), most recently in the form of exapted distal cis-regulation from >480 kb downstream of the target gene (19). The current survey reaffirms the widespread nature of this phenomenon at the genomic scale. It also takes an important step toward understanding the fundamental nature of cis-regulatory exaptation, by clearly highlighting specific regions within each repeat that are most prone to it. One may speculate that some of these exapted regions already play a regulatory role in the progenitor repeat.
Britten and Davidson (12) hypothesized that the dispersion of repetitive sequences with strong exaptation potential throughout the genome could allow for a whole “battery” of genes to suddenly become coregulated, augmenting an existing pathway, or even creating one from scratch, especially in the context of development. Remarkably, our much more recent appreciation for the complexity and modularity of vertebrate gene regulation serves only to strengthen this early insight. Exapted elements are indeed extremely enriched for clustering near developmental genes, even when considering the background distribution of transposon insertions. In fact, transposons seem to be biased against inserting and remaining near genes involved in developmental regulation (40). Thus, our enrichment is not due to an insertional bias of transposons, but rather a bias in retention, suggesting that they may carry something that may affect the regulation of these genes either beneficially or detrimentally. Developmental functions also dominate the list of genes flanking the largest spatial clusters of exapted elements in the genome. By transposing into the chromatin-accessible region surrounding an active transcription start site, mobile elements may seed novel transcription factor-binding sites and, through both functional and nonfunctional insertions, repeatedly drive older cis-regulatory elements further away from their target genes. Thus, whereas Britten and Davidson (12) did not foresee distal cis-regulation at distances of a megabase 36 years ago, their theory of transposon-mediated regulatory network evolution indirectly predicts it, and our observations provide circumstantial support for this theory. However, to fully verify their hypothesis, we must understand how these exapted CNEs affect developmental gene expression in the context of their regulatory networks (41).
Reelin signaling is believed to result in the activation of genes involved in cell adhesion, a theme that has been seen often in recent papers exploring the evolution of regulatory elements. Exapted instances of the previously discovered LF-SINE, which is thought to have been most active at the base of the tetrapods, are enriched near genes involved in cell adhesion (19). The elements we explore here originated mostly along the mammalian branch and also show significant enrichment for being near cell adhesion genes. A recent work looking at rapidly evolving regions in the human lineage also reports a strong enrichment near genes involved in cell adhesion (7). These results suggest that cell adhesion genes (perhaps mostly those involved in brain wiring) have been constantly refining their expression patterns throughout the last 300 Myr and into the present day.
The majority of current whole-genome experimental and computational approaches to gene regulation, such as tiling arrays used in ChIP-chip experiments (42), and transcription factor-binding site prediction (43), choose to ignore repetitive regions, for pragmatic reasons, assuming that most if not all are inert. Our analysis, however, suggests that, whereas the fraction of repeat copies that have come under strong purifying selection is indeed small, as a fraction of all putative regulatory elements under the same selective pressures, they constitute a pronounced minority. Indeed, as our appreciation for the contributions of repeats to different aspects of genome evolution continues to grow (44), it now seems that these unwanted, and often ignored, children of the genome played multiple crucial roles during the evolution of the human lineage.
Methods
Sequence Data Sources.
The University of California, Santa Cruz (UCSC) assemblies and repeat masker libraries we used per species are as follows: human (Mar2006/hg18/RM051101), chimp (Mar2006/panTro2/RM060120), macaque (Jan2006/rheMac2/RM20060120), rat (Nov2004/rn4/RM060314), mouse (Feb2006/mm8/RM060120), dog (May2005/canFam2/RM20050305), chicken (May2006/galGal3), frog (Aug2005/xenTro2), tetraodon (Feb2004/tetNig1), zebrafish (Mar2006/danRer4), and fugu (Aug2002/Fr1).
Generation of Constrained Nonexonic Elements.
Three mammalian sources of conserved elements were used: top-scoring elements resisting insertion and deletions from ref. 22 covering 2% of the human genome; same-sized set of elements resistant mostly to substitutions, generated by running phastCons (4) on a syntenic multiple alignment of human, chimp, macaque, mouse, rat, and dog; and a same-sized set comprising all sliding windows along a syntenic alignment of human, mouse, and dog, where 42 of the 50 bases (84%) of alignment columns were identical, resisting both substitution and in-dels. All regions of the human genome that syntenically aligned to chicken, frog, tetraodon, zebrafish, or fugu at 70% identity or better over at least 50 bases were also added to our final set. We filtered the resulting set of constrained elements by removing bases overlapping any known or reliably predicted mature transcript: refSeq and UCSC known genes, any GenBank cDNA/mRNA reliably alignable to human from a variety of species, human spliced ESTs, known and predicted pseudo genes, RNA genes, micro RNAs, and all Ensembl and Exoniphy gene predictions (45). After filtering, our constrained nonexonic set totaled 1.45% of the human genome.
Comparison with Neutral Rate.
We used a model of neutral evolution computed by PhyloP (27) from 4-fold degenerate sites in the ENCODE regions (46).
Calculating GO Enrichment.
All UCSC hg18 Known Genes (45) splice variants were combined into human gene loci. Each locus was assigned a representative TSS and the union of all Gene Ontology (GO) annotations (33) assigned to its variants. All loci lacking meaningful GO annotation were removed, leaving a set of 14,277 annotated loci.
Identification of Potential Binding Sites.
We used the Transfac free matrices (version 6.0) and search tools to identify potential transcription factor-binding sites (47). All sites were found by the P-Match search tool while minimizing the sum of false-positive and false-negative hits.
Acknowledgments
We thank the Macaque Genome Sequencing Consortium, Robert Baertsch, and Rachel Harte for sharing unpublished data, as well as Mark Diekhans, Jim Kent, Andy Kern, Martina Koeva, Jacob Pedersen, Katie Pollard, Brian Raney, Sofie Salama, Adam Siepel, Daryl Thomas, and the University of California, Santa Cruz Computational and Functional Genomics Group for technical advice. We also thank Juergen Brosius and David Kingsley for discussion.
Abbreviations
- CNE
constrained nonexonic element
- GO
Gene Ontology
- TSS
transcriptional start site
- LINE
long interspersed element
- SINE
short interspersed element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611223104/DC1.
References
- 1.International Mouse Genome Sequencing Consortium. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Dermitzakis ET, Reymond A, Antonarakis SE. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- 4.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 6.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 9.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 10.Carroll SB. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClintock B. Cold Spring Harbor Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Britten RJ, Davidson EH. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 13.Brosius J. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 14.Gould SJ, Vrba ES. Paleobiology. 1982;8:4–15. [Google Scholar]
- 15.Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. Genet Res. 2003;82:1–18. doi: 10.1017/s0016672303006268. [DOI] [PubMed] [Google Scholar]
- 16.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bejerano G, Haussler D, Blanchette M. Bioinformatics. 2004;20(Suppl 1):I40–I48. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- 18.Kamal M, Xie X, Lander ES. Proc Natl Acad Sci USA. 2006;103:2740–2745. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 20.Nishihara H, Smit AFA, Okada N. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornburg BG, Gotea V, Makalowski W. Gene. 2006;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 22.Lunter G, Ponting CP, Hein J. PLoS Comput Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Chicken Genome Sequencing Consortium. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 24.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. Seattle, WA: Institute for Systems Biology; 1996–2004 www.repeatmasker.org. [Google Scholar]
- 25.Jurka J. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siepel AC, Pollard KS, Haussler D. Proceedings of the Tenth Annual International Conference on Research in Computational Molecular Biology (RECOMB 06); New York: Association for Computing Machinery; 2006. pp. 190–205. [Google Scholar]
- 28.Smit AF. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 29.Nishihara H, Terai Y, Okada N. Mol Biol Evol. 2002;19:1964–1972. doi: 10.1093/oxfordjournals.molbev.a004020. [DOI] [PubMed] [Google Scholar]
- 30.Deininger PL, Moran JV, Batzer MA, Kazazian HHJ. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Lev-Maor G, Sorek R, Shomron N, Ast G. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 32.Kleinjan DA, van-Heyningen V. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tissir F, Goffinet AM. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- 35.Hammond V, Howell B, Godinho L, Tan SS. J Neurosci. 2001;21:8798–8808. doi: 10.1523/JNEUROSCI.21-22-08798.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatesh B, Kirkness EF, Loh Y, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, et al. Science. 2006;314:1892. doi: 10.1126/science.1130708. [DOI] [PubMed] [Google Scholar]
- 37.Brosius J. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- 38.Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP. J Biol Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 40.Simons C, Pheasant M, Makunin IV, Mattick JS. Genome Res. 2006;16:164–172. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davidson EH, Erwin DH. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 42.Buck MJ, Lieb JD. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Tompa M, Li N, Bailey TL, Church GM, DeMoor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 44.Biemont C, Vieira C. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- 45.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. Nucleic Acids Res. 2006;34:590–598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ENCODE Project Consortium. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 47.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]