Abstract
The CATERPILLER (CLR/NLR) gene family encodes a family of putative nucleotide-binding proteins important for host defense. Although nucleotide binding is thought to be central to this family, this aspect is largely unstudied. The CATERPILLER protein cryopyrin/NALP3 regulates IL-1β processing by assembling the multimeric inflammasome complex. Mutations within the exon encoding the nucleotide-binding domain are associated with hereditary periodic fevers characterized by constitutive IL-1β production. We demonstrate that purified cryopyrin binds ATP, dATP, and ATP-agarose, but not CTP, GTP, or UTP, and exhibits ATPase activity. Mutation of the nucleotide-binding domain reduces ATP binding, caspase-1 activation, IL-1β production, cell death, macromolecular complex formation, self-association, and association with the inflammasome component ASC. Disruption of nucleotide binding abolishes the constitutive activation of disease-associated mutants, identifying nucleotide binding by cryopyrin as a potential target for antiinflammatory pharmacologic intervention.
Keywords: inflammation, IL-1β
Mammalian genes encoding proteins with a conserved nucleotide-binding domain (NBD) and a leucine-rich region (LRR) have received much attention of late. This gene family known as CATERPILLER [for CARD, transcription enhancer, R(purine)-binding, pyrin, lots of leucine repeats, also CLR], NOD-LRR (for nucleotide-oligomerization domain), or NLR (for NACHT-LRR, soon to be renamed NBD-LRR) consists of >20 members (1–3). These proteins share structural similarity with a subfamily of plant disease resistance (R) proteins that mediate plant immunity to a wide array of microbial pathogens and environmental insults (4). Several of these genes have strong genetic association with human diseases. Among these, the CIAS1 (for cold induced autoinflammatory syndrome 1) gene is associated with three dominantly inherited periodic fevers: familial cold autoinflammatory syndrome (FCAS), Muckle–Wells syndrome, and neonatal-onset multisystemic autoinflammatory disease (5). All three syndromes are characterized by increased serum IL-1β, and anti-IL-1β therapy is the mainstay of treatment (6). The majority of known disease-associated mutations reside in the NBD, although not in residues predicted to directly interact with nucleotide (7).
Recent studies using CIAS1−/− mice indicate that cryopyrin is pivotal to the elaboration of IL-1β by macrophages in response to bacterial RNA, certain Gram-positive bacteria, uric acid and calcium pyrophosphate crystals, cellular toxins, and some Toll-like receptor agonists (8–12). In vitro, cryopyrin assembles with two CARD domain-containing adapter proteins, ASC/TMS1 and Cardinal/TUCAN/CARD8, as well as caspase-1 (the protease that cleaves pro-IL-1β to its mature form) to form a large IL-1β-converting holoenzyme known as the inflammasome (13). Periodic fever-associated mutations in cryopyrin are thought to cause constitutive inflammasome assembly. Additionally, WT and disease-associated mutant cryopyrin induce cell death when overexpressed (14, 15).
Although cryopyrin contains a consensus NBD that is postulated to be crucial in the protein's activity, the identity of the bound nucleotide and the role of nucleotide binding in cryopyrin function remain unexplored. Apoptotic protease activating factor-1, Apaf-1, shares a similar but distinct NBD with cryopyrin. Apaf-1 binds several adenosine-based nucleotides in vitro, and recombinant Apaf-1 isolated from eukaryotic cells is bound to dATP in its basal state (16, 17). Although the in vitro caspase-3 activating activity of Apaf-1 requires dATP, there are few detailed studies of nucleotide binding by CATERPILLER family members. A bacterially expressed partially purified NBD of CARD12 (Ipaf/Clan) exhibits ATP-binding activity (18). CIITA, the founding member of the CATERPILLER family, is associated with GTP when the protein is enriched from mammalian cells (19). The analysis of nucleotide binding by purified, full-length CATERPILLER family members has been technically difficult and remains unexplored.
We now show the successful isolation of recombinant cryopyrin from eukaryotic cells. This full-length protein binds ATP/dATP and can be specifically eluted from ATP-agarose. Intact nucleotide-binding capacity of cryopyrin is required for an array of functions attributed to this protein.
Results
Recombinant Cryopyrin Can Be Purified to Homogeneity by Sequential Affinity Chromatography.
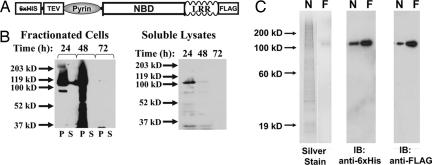
Studies of the nucleotide binding properties of cryopyrin (and other NBD-LRR proteins in plants and animals) have been hampered by the fact that overexpressed, full-length NBD-LRR proteins are largely insoluble. A dual-tagged recombinant cryopyrin with an N-terminal hexahistidine tag and a C-terminal FLAG tag was prepared in a baculoviral expression system (Fig. 1A). A portion of the cryopyrin expressed in baculovirus-infected Hi5 cells is found in the soluble fraction (Fig. 1B). Although this recombinant protein was significantly enriched with nickel and anion-exchange chromatography, a homogenous product was not achieved [supporting information (SI) Fig. 7A, WT lanes]. Ultimately, soluble recombinant cryopyrin was purified to homogeneity (with a yield of ≈10–20 μg per liter of cell culture) by immobilized nickel chromatography followed by an anti-FLAG immunoaffinity chromatography (Fig. 1C). The purified protein was immunoreactive with antibodies directed toward the C-terminal FLAG and N-terminal hexahistidine tags (Fig. 1C).
Fig. 1.
Recombinant cryopyrin expressed in insect cell culture can be purified to homogeneity by affinity chromatography. (A) Schematic of the recombinant tags on dual-tagged cryopyrin. N-terminal hexahistidine tag and TEV protease site and C-terminal FLAG tag are indicated. The Pyrin domain, NBD, and LRR domains indicate domains of the native protein. (B) Dual-tagged cryopyrin was expressed in Hi5 Insect cells by infection with recombinant baculovirus (multiplicity of infection = 0.5). After the indicated times, the cells were harvested, flash-frozen, resuspended in lysis buffer, and sonicated. After centrifugation at 15,000 × g for 1 h, the supernatants (S, soluble fraction) were removed and the pellets (P, insoluble fraction) were resuspended in an equal volume of 1× SDS/PAGE loading buffer. The soluble (S, supernatant) and insoluble (P, pellet) fractions (Left) were subjected to immunoblot analysis with anti-FLAG antibody. The soluble lysates from the infected cells fractions are run alone (Right) and demonstrate that immunoreactive material in the second lane in Left is not bleed-over from a neighboring lane. (C) Recombinant cryopyrin was purified by sequential affinity chromatography. Protein eluted from the nickel (N, 200 ng) followed by the anti-FLAG (F, 50 ng) columns were subjected to silver staining (Left) or immunoblot with indicated antibodies (Center and Right).
Cryopyrin Binds ATP and Exhibits ATPase Activity.
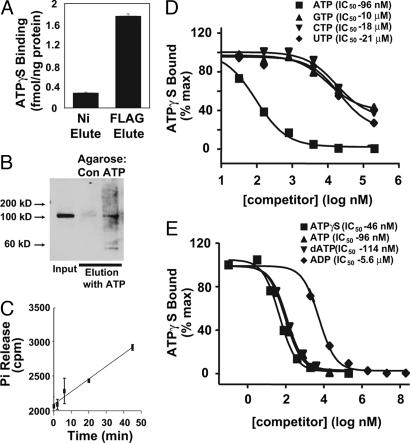
Protein eluted from immobilized nickel and anti-FLAG agarose each contained binding activity for ATPγS, a nonhydrolyzable ATP analog. There was nucleotide-binding activity detected in preparations of cryopyrin isolated by immobilized nickel chromatography. This activity was further enriched 6-fold by anti-FLAG affinity chromatography (Fig. 2A). Based on the amount of ATPγ S bound, the stoichiometry of ATPγ S to purified cryopyrin under these conditions is ≈1:3 (Fig. 2A). Although this stoichiometry is somewhat low, there are no other proteins observed in the preparations with sufficient abundance to account for this quantity of nucleotide binding. It is possible that purified cryopyrin requires a nucleotide exchange factor to facilitate stoichiometric ATP binding or that the some of the cryopyrin in the preparation is soluble but inactive.
Fig. 2.
Recombinant cryopyrin is an ATP-binding protein. (A) ATPγ S binding activity of the eluted fractions was assayed by filter binding assay as described in SI Materials and Methods using 200 nM ATPγ S. (B) WT cryopyrin eluted from NiNTA agarose was incubated with control or ATP-conjugated agarose. Specific ATP-binding proteins were eluted by incubation with ATP-containing buffer and analyzed by immunoblot with anti-FLAG antibody. (C) PO4 release from [γ-32P]ATP in the presence of highly purified cryopyrin (see Fig. 1C) was measured as described in SI Materials and Methods. Error bars indicate standard deviation of triplicate samples. (D and E) NiNTA and anion-exchange chromatography-enriched cryopyrin was used to assess the specificity of cryopyrin-associated nucleotide-binding activity. Cryopyrin was incubated with 2 nM 35S-ATPγ S (≈10,000 cpm/pmol) and the indicated concentration of unlabeled competitor nucleotides at 4°C for 30 min (at which time bound nucleotide was increasing linearly with increasing time), and bound ATPγ S was assayed. Curves represent nonlinear regression fit to single-site competition model (Y = 100/(1 + 10(X-Log(EC50)))).
The ATP-binding activity of cryopyrin was further demonstrated by association of cryopyrin with ATP-agarose or agarose and elution of the protein with ATP-containing buffer (Fig. 2B). The combination of measurable ATP binding in the preparation of purified cryopyrin and preferential binding of the protein to ATP-agarose indicates that cryopyrin is indeed an ATP-binding protein. Purified cryopyrin also exhibited ATPase activity, as shown by the release of 32P-PO4 from [γ-32P]ATP incubated with cryopyrin (Fig. 2C). Thus, highly purified, recombinant cryopyrin has both ATP-binding and hydrolysis activity.
The nucleotide-binding specificity of cryopyrin was assessed by competition for ATPγ S binding activity by recombinant cryopyrin with various unlabeled nucleotides. ATP competed for ATPγ S binding by partially purified cryopyrin with an IC50 of 96 nM, ≈1,000-fold lower concentrations than the IC50 for GTP, CTP, or UTP (Fig. 2D). Additionally, cryopyrin had a marked preference for ATP and ATPγ S over ADP but showed no significant difference between ATP and dATP (Fig. 2E). These studies conclusively demonstrate that cryopyrin is an adenine nucleotide-binding protein.
Mutation of the Walker A Motif of Cryopyrin Disrupts ATP Binding.
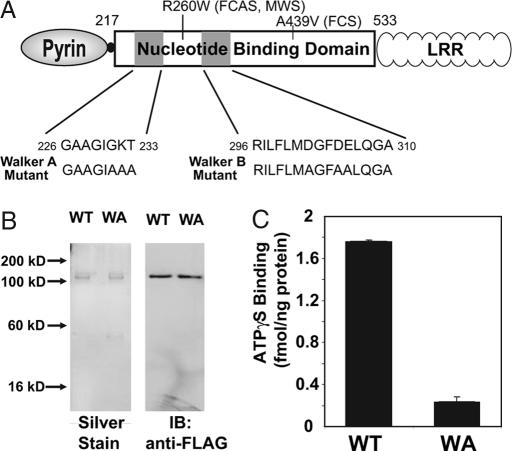
All CLR proteins are characterized by a predicted NBD contained within an expanded region of 12 conserved elements (1). Among these, the consensus Walker A motif contains the sequence GKS/T, which correlates to G231, K232, and T233 of cryopyrin. The lysine is involved in coordinating the γ-phosphate of ATP in many proteins with this motif (20). All three of these residues were mutated to alanine to generate the Walker A mutant (Fig. 3A). WT and Walker A mutant cryopyrin were expressed and partially purified by using NiNTA agarose and anion-exchange chromatography (SI Fig. 7A) or purified to homogeneity by using sequential affinity chromatography as described earlier (Fig. 3B). WT and Walker A mutant cryopyrin were assayed for ATPγ S binding at various stages of purification. Although the Walker A mutant exhibited nearly identical solubility and chromatographic properties, the detectable ATPγ S binding activity of a partially purified preparation of cryopyrin (SI Fig. 7B) or of a purified preparation of cryopyrin was extinguished by the mutation (Fig. 3C).
Fig. 3.
Mutation of the Walker A motif of cryopyrin abrogates its ATP-binding activity. (A) WT and Walker A mutant variants of cryopyrin. Mutated Walker A sequence is shown. Walker B mutation, used in SI Table 1, is noted. Disease-associated mutations, R260W and A439V, used in Fig. 5, are also noted. (B) WT and Walker A mutant (WA) cryopyrin were purified by sequential affinity chromatography as described in Fig. 1A. Silver staining (Left) and immunoblot analysis (Right) of 50 ng of each of the purified proteins is shown. (C) ATPγ S binding activity of the indicated proteins was assayed at 60 min in the presence of 200 nM ATPγ S. Error bars represent standard deviation of ATP-binding measurements determined in triplicate.
An Intact NBD Is Required for Cryopyrin-Induced IL-1β Production.
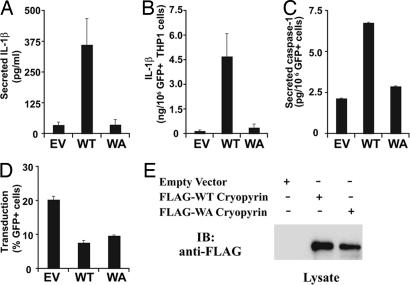
To determine whether ATP binding is required for cryopyrin-mediated IL-1β secretion in a biologically relevant cell type, the human myeloid–monocytic cell line THP1 was transduced with recombinant retrovirus expressing WT and Walker A mutant cryopyrin. The recombinant retrovirus also expressed the gene encoding GFP, which was used to identify transduced cells (21). Cells transduced with WT cryopyrin demonstrated increased IL-1β secretion when compared with retrovirus expressing only GFP (empty vector) or the Walker A mutant (WA) (Fig. 4A). When IL-1β production was corrected for the number of cells that expressed GFP (Fig. 4B), the same trend was observed. Levels of caspase-1 activation in retrovirally transduced cells mirrored the findings with IL-1β, suggesting the reduced levels of IL-1β observed with expression of the Walker A mutant cryopyrin were the result of a failure in caspase-1 activation by that protein (Fig. 4C). Differences between the effects of expression of WT and Walker A mutant cryopyrin are not attributable to lower viral transduction efficiency (assayed by flow cytometry) or cryopyrin expression (assayed by immunoblot analysis) (Fig. 4 D and E). Results from our current study differ from other published studies in which THP-1 cells transfected with plasmids that express WT cryopyrin do not spontaneously secrete IL-1β (22). Our results may reflect either higher levels of expression or greater efficiency of transduction associated with the use of spinoculation with recombinant retrovirus. Alternatively, spinoculation with recombinant virus may serve as a weak inflammasome-assembly triggering stimulus that can activate cells overexpressing cryopyrin but not those transduced with empty virus. We have observed a lack of spontaneous IL-1β secretion after transfection of plasmids encoding WT cryopyrin (Fig. 5C) just as reported by other groups (22). We have also used recombinant retrovirus to transduce CIAS1−/− mouse macrophages, although with a much lower efficiency of transduction. In these experiments, WT cryopyrin expression restored both LPS- and R838-induced IL-1β secretion whereas expression of the Walker A mutant did not (data not shown). Thus, the Walker A mutation eliminates spontaneous IL-1β production associated with high levels of cryopyrin overexpression and regulated IL-1β production. These findings indicate that nucleotide binding is an important step in cryopyrin-mediated caspase-1 and IL-1β production.
Fig. 4.
ATP binding by cryopyrin is essential for cryopyrin-induced IL-1β production. THP1 cells were transduced with recombinant retrovirus expressing the following proteins: empty vector (EV), no cryopyrin; WT cryopyrin; and Walker A mutant (WA) cryopyrin. (A) IL-1β secretion was measured 24 h after transduction by ELISA. (B) IL-1β measured in A was corrected for the number of GFP+ cells in the culture. (C) Secreted caspase-1 was assayed by ELISA. Error bars represent standard deviation between transductions performed in triplicate. (D) Retroviral transduction efficiency was determined by monitoring GFP expression with flow cytometry. (A–D) Error bars represent standard deviation between transductions performed in triplicate. (E) Expression levels of recombinant cryopyrin were assessed by immunoblot analysis with anti-FLAG antibody.
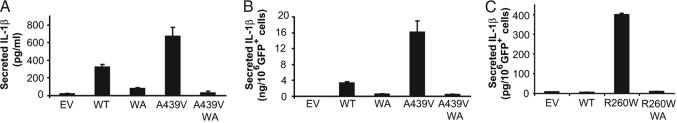
Fig. 5.
Constitutive activity of disease-associated mutant cryopyrin requires intact ATP-binding activity. (A and B) THP1 cells were transduced with recombinant retrovirus expressing the following proteins: empty vector (EV), no cryopyrin; WT cryopyrin; Walker A mutant (WA) cryopyrin; FCAS mutant cryopyrin (A439V); and FCAS mutant cryopyrin with Walker A mutation (A439V/WA). (A) IL-1β secretion was measured 12 h after transduction by ELISA. (B) IL-1β measured in A was corrected for the number of GFP+ cells in the culture. Error bars represent standard deviation between transductions performed in triplicate. (C) THP1 cells were transfected with plasmids expressing GFP and the following proteins: empty vector (EV), no additional protein; WT cryopyrin; FCAS/Muckle–Wells syndrome mutant cryopyrin (R260W); and R260W with Walker A mutation (R260W/WA). Secreted IL-1β was assayed by ELISA 24 h after transfection. Error bars represent standard error from duplicate measurements.
Gain of Function by Cryopyrin Disease-Associated Mutants Depends on ATP Binding.
CIAS1-associated fever syndromes are autosomal dominant in inheritance, and disease-related cryopyrin mutants lead to constitutive IL-1β production, suggesting that these mutations lead to a constitutively activated cryopyrin molecule. Disease-associated CIAS1 mutations are concentrated in the exon that encodes the NBD. It was unknown whether such mutations promote constitutive activation of cryopyrin by circumventing the requirement for nucleotide binding. We tested this by assessing the role of nucleotide binding in the in vivo function of disease-associated cryopyrin. The FCAS-associated A439V mutation was introduced into the Walker A mutant cDNA and designated as the A439V/WA double mutant (Fig. 5A). The A439V disease-associated mutant induced significantly more IL-1β secretion when compared with the WT control, consistent with previous reports (Fig. 5A) (13, 22). However, IL-1β production was absent in cells expressing the A439V/Walker A double-mutant molecules (Fig. 5A). Correction for the number of GFP-expressing cells in each culture yielded the same findings (Fig. 5B). This establishes that a disease-associated form of cryopyrin still requires nucleotide binding for its activation. To determine whether this finding could be extended to other disease-associated cryopyrin mutants, we tested the FCAS/Muckle–Wells syndrome-associated R260W mutant. THP1 cells transfected with plasmids encoding this mutant produced more IL-1β than those expressing the WT control, but mutations in the Walker A (labeled R260W/WA; see Fig. 3A for mutations) ablated IL-1β production (Fig. 5C). Combined, these findings suggest that therapies directed at nucleotide binding by cryopyrin represent a viable strategy to modulate the function of periodic fever-associated mutant proteins. This may be especially appealing for long-term control of these diseases where an oral agent might be preferable to current injectable anti-IL-1 therapies.
Overexpression of WT cryopyrin has been shown to cause an ASC1-dependent cell death (14). More recently, periodic fever-associated mutant cryopyrin expression has been shown to induce necrotic cell death (15). A reduction in viable cells was also observed when THP1 cells were transduced with retrovirus or adenovirus expressing the disease-associated mutant cryopyrins (SI Fig. 8 B and C). These constructs caused more IL-1β secretion as expected (SI Fig. 8A). The cell death associated with two disease-associated cryopyrin mutants also depends on functional ATP-binding activity (SI Fig. 8 B and C). The physiologic significance of cryopyrin-induced cell death remains to be determined; however, our data indicate that both the cell death-inducing and IL-1β-producing activities of cryopyrin require intact ATP binding by the protein.
Intact Nucleotide Binding Is Required for the Self-Association of Cryopyrin.
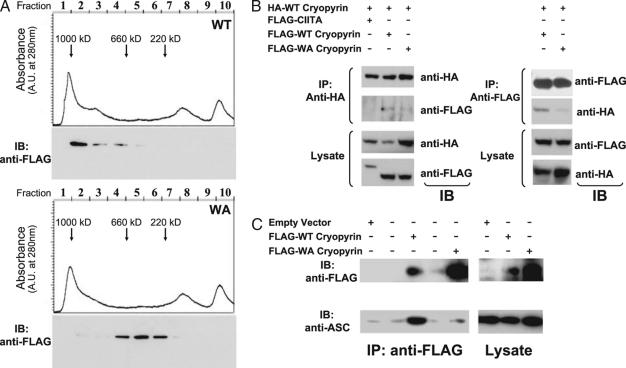
Binding of dATP by Apaf-1 is a critical step in the assembly of the apoptosome (23). It has been hypothesized that CATERPILLER proteins require nucleotide binding to form a macromolecular complex and activate their respective signaling pathways. To determine whether ATP binding by cryopyrin affected complex formation, soluble fractions from HEK293 cells expressing either WT cryopyrin or the Walker A mutant were subjected to gel filtration chromatography. These cells were used initially because protein expression in THP1 cells was low. Immunoblot analysis revealed that the WT cryopyrin protein was found predominantly in fractions approaching 1,000 kDa (Fig. 6A Upper), while the mutant protein ran between 200 and 600 kDa (Fig. 6A Lower). This size difference could reflect either self-association or assembly with other proteins by the WT protein. Because HEK293 cells do not secrete IL-1β and lack other known inflammasome components, it is more likely that the Walker A mutation blocked both ATP binding and self-association of the protein. To test this hypothesis, a bidirectional self-association analysis was performed by transfection of HA-tagged and FLAG-tagged cryopyrin followed by immunoprecipitation with either anti-HA or anti-FLAG antibodies and immunoblot analysis directed against the other tag. Whereas the WT cryopyrin protein was found to self-associate, the Walker A mutant protein associated less efficiently with coexpressed WT cryopyrin (Fig. 6B). CIITA, an NBD-LRR containing protein, did not associate with cryopyrin. Thus, an intact NBD is necessary for the specific self-association of cryopyrin.
Fig. 6.
Cryopyrin self-association and association with ASC require ATP binding. (A) Extracts from HEK293 cells transfected with plasmids encoding FLAG-tagged WT (Upper) or Walker A mutant (Lower) versions of cryopyrin were subjected to size-exclusion chromatography (as detailed in Materials and Methods). The chromatograms show UV absorbance plotted against volume of elution. Arrows indicate volume of elution for standards of the indicated molecular mass. Cryopyrin content (shown below the chromatographs) is determined by immunoblot analysis with anti-FLAG antibodies. (B) HEK293 cells were transfected with indicated plasmids. Lysates from the cells were subjected to immunoprecipitation with anti-HA antibodies (Left) and anti-FLAG antibodies (Right), and samples were analyzed by immunoblot analysis with the indicated antibodies. (C) THP1 cells were transduced with recombinant retrovirus expressing the following proteins: empty vector, no cryopyrin; FLAG-tagged cryopyrin; or Walker A mutant (WA) FLAG-tagged cryopyrin. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody and analyzed by immunoblot with anti-FLAG and anti-ASC. The second and fourth lanes in Left have no sample.
Intact Nucleotide Binding Regulates the Association of Cryopyrin with ASC.
Cryopyrin has been proposed to form a caspase-1 activating structure called the inflammasome, which contains cryopyrin, ASC, and CARDINAL (13). We sought to determine whether assembly of the cryopyrin-containing inflammasome depends on nucleotide binding by cryopyrin in THP1 cells, which express endogenous cryopyrin. Cryopyrin was isolated by immunoprecipitation with anti-FLAG antibody from cytosolic extracts of THP1 cells expressing the WT or Walker A mutant cryopyrin molecules and analyzed for their association with endogenous ASC. Coimmunoprecipitation of ASC with cryopyrin was drastically reduced with the Walker A mutant when compared with the WT protein (Fig. 6C), suggesting that cryopyrin requires binding of ATP to associate with ASC.
Discussion
This work shows that highly purified, recombinant full-length cryopyrin isolated from eukaryotic cells binds to and hydrolyzes ATP. Nucleotide binding is further demonstrated by mutagenesis, nucleotide competition, binding to ATP-agarose, and elution by ATP. Analysis of mutant cryopyrin lacking this activity indicates that ATP binding by cryopyrin is essential to a host of cryopyrin-mediated functions. Similar elimination of biological function of other CLR genes and plant R genes has been reported with mutation of the Walker A domain in the respective systems (18, 24–26). Our current data cannot distinguish between the possibility that nucleotide binding by cryopyrin is a regulated process controlling signal transduction by the protein or that nucleotide binding is required for proper folding or to stabilize a signaling competent protein but is not regulated during inflammasome assembly by the protein.
Studies of the mechanism of Apaf-1 and apoptosome assembly and the function of some plant R proteins suggest that nucleotide binding by cryopyrin is likely to be a regulated process controlling inflammasome assembly. Although Apaf-1-stimulated caspase-3 activation is not supported by nonhydrolyzable ATP or dATP analogs, suggesting a role for ATP hydrolysis in this process, the role of ATP hydrolysis in inflammasome assembly or disassembly remains to be characterized. Mutations in the plant R protein i-2, which confer constitutive activation of its signaling pathway, have been found to not hydrolyze ATP, suggesting that some human CATEPILLER proteins may also be bound to ATP during their active stage (25). Studies of inflammasome assembly using purified components, including cryopyrin, and defined nucleotides, including nonhydrolyzable analogs, are obvious extensions of this work to examine this problem.
We have also demonstrated that the disease-associated phenotype of mutant cryopyrin requires ATP binding. Using the model of regulated nucleotide binding controlling inflammasome assembly, our data imply that these mutations of cryopyrin are likely to promote ADP/ATP exchange, inhibit ATPase activity of the protein, or otherwise facilitate ATP binding and activation of the protein. Mutations that lead to constitutive activation of guanine nucleotide-binding proteins (G proteins) have been associated with loss of GTPase activity (locking the protein in GTP bound state) or with decreased GDP affinity (leading to increased GTP for GDP nucleotide exchange) (27, 28). Before this work, the enhanced activity of disease-associated mutants could have been attributed to an unregulated, ATP-independent activation of the protein or to a novel activity of the mutant protein not ascribable to the WT cryopyrin. This work indicates that the function of disease-associated cryopyrin proteins is still ATP-dependent. Determination of the molecular mechanism underlying the constitutive activity of these mutant proteins will rely on detailed biochemical analysis of their ATP-binding properties. However, disease-associated mutants are even more difficult to enrich and purify (data not shown); thus, improvements in the expression system will need to be made to undertake these studies.
This study serves as a proof-of-principle that disease-associated activation of cryopyrin signaling can be modulated by interference with the nucleotide binding of this protein. Nucleotide analogs with specificity for nucleotide using enzymes associated with specific diseases are currently used in a number of clinical settings. A prime example is the use of nucleotide analogs for the treatment of HIV (29). A pyrimidine-derived inhibitor of the BCR-ABL kinase, imatinib, has also proven successful for treating chronic myelogenous leukemia (30). In the future, cryopyrin-binding nucleotide analogs might have use in the treatment of hereditary inflammatory syndromes as well as other inflammatory conditions that use cryopyrin-mediated signaling.
Materials and Methods
Construction of CIAS1 Expression Plasmids.
Standard techniques of DNA manipulation were used (31). Details of cloning strategies and oligonucleotide sequences used are provided in SI Materials and Methods.
Cell Culture and Transfection.
THP1 and HEK293 (American Type Culture Collection, Manassas, VA) were cultured under recommended conditions. FuGENE 6 transfection reagent (Roche, Indianapolis, IN) was used for transfection of HEK293 cells.
Immunoprecipitation and Immunoblot Analysis.
Immunoprecipitations were carried out in 50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1 mM MgCl2, and 0.5% Nonidet P-40 supplemented with complete protease inhibitor mixture (Roche) and 1 mM ATP. Lysates from 5 × 107 cells were incubated for 30 min at 30°C before immunoprecipitation with agarose-conjugated anti-FLAG (M2) or anti-HA (12CA5) antibodies (Sigma–Aldrich, St. Louis, MO) followed by SDS/PAGE. Anti-hexahistidine antibody (His Probe, H3) was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-ASC antibody (AL177) was from Alexis Biochemicals (Lausen, Switzerland).
Expression of Recombinant Cryopyrins in THP1 Cells.
Recombinant retrovirus expressing cryopyrin and mutant variants was generated as described by Coffield et al. (21). Recombinant adenovirus was generated by using the AdenoX system (BD Biosystems, Franklin Lakes, NJ). THP1 cells were virally transduced by using a “spinoculation” protocol. Details are provided in SI Materials and Methods. Transfection of plasmids encoding cryopyrin and mutant variants into THP1 cells was carried out by using the Nucleofector V kit (Amaxa, Gaithersburg, MD) according to the manufacturer's protocol.
Flow Cytometry and ELISA.
Annexin V-PE (BD Biosciences) was used according to the manufacturer's protocols. Flow cytometry was performed on a FACScan instrument (BD Biosciences). An OptEIA human IL-1β ELISA kit (BD Biosciences) was used for secreted IL-1β quantitation. Secreted caspase-1 was detected by using a caspase-1 ELISA kit (Alexis Biochemicals).
Baculovirus and Insect Cell Culture.
Recombinant baculovirus expressing cryopyrin was generated by using the Bac to Bac system (Invitrogen, Carlsbad, CA). Insect cell culture and baculovirus infection were performed according to the manufacturer.
Purification of Recombinant Cryopyrin.
Frozen Hi5 cells were lysed by sonication, and recombinant proteins were purified by using NiNTA agarose (Qiagen, Valencia, CA) and agarose-conjugated anti-FLAG (M2) antibody (Sigma–Aldrich) using modified manufacturers' protocols. Anion-exchange chromatography was performed by using a Duoflow chromatography system (Bio-Rad, Hercules, CA). Details are provided in SI Materials and Methods.
ATPγ S Binding and ATP Hydrolysis Assays.
Nucleotide binding was assayed by using a nitrocellulose filter binding assay (32). Nucleotide hydrolysis assays were carried out by using a modified acidified charcoal precipitation (33). Further details are provided in SI Materials and Methods.
Gel Filtration Chromatography.
Soluble lysates from cultured cells were prepared by hypotonic lysis (buffer is described in SI Materials and Methods) and shearing through a 27-gauge needle. Soluble lysate (0.5 mg of total protein) was run on a Biosil 400 gel filtration column using a Bio-Rad Duoflow chromatography system in PBS with 2 mM DTT.
Acknowledgments
This work was supported by the Sandlers Program in Asthma Research and National Institutes of Health Grants R01AI057157, R01AI063031, and R01DE16326 (to J.P.-Y.T.). J.A.D. is supported by the Pfizer Fellowship in Infectious Diseases and National Institutes of Health Career Development Award K12RR023248. A.G.Z. is supported by National Institutes of Health Postdoctoral Training Fellowship T32AI007151.
Abbreviations
- LRR
leucine-rich region
- NBD
nucleotide-binding domain
- FCAS
familial cold autoinflammatory syndrome.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611496104/DC1.
References
- 1.Harton JA, Linhoff MW, Zhang J, Ting JP. J Immunol. 2002;169:4088–4093. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 2.Inohara N, Nunez G. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 3.Martinon F, Tschopp J. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Belkhadir Y, Subramaniam R, Dangl JL. Curr Opin Plant Biol. 2004;7:391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Nat Genet. 2001;29:301–305. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, Rubin BI, Kim HJ, Brewer C, Zalewski C, Wiggs E, et al. N Engl J Med. 2006;355:581–592. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht M, Domingues FS, Schreiber S, Lengauer T. FEBS Lett. 2003;554:520–528. doi: 10.1016/s0014-5793(03)01222-5. [DOI] [PubMed] [Google Scholar]
- 8.Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 10.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 11.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, Flavell RA. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Lich JD, Arthur JC, Ting JP. Immunity. 2006;24:241–243. doi: 10.1016/j.immuni.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 14.Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Nunez G. Biochem Biophys Res Commun. 2003;302:575–580. doi: 10.1016/s0006-291x(03)00221-3. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa A, Kambe N, Saito M, Nishikomori R, Tanizaki H, Kanazawa N, Adachi S, Heike T, Sagara J, Suda T, et al. Blood. 2007;109:2903–2911. doi: 10.1182/blood-2006-07-033597. [DOI] [PubMed] [Google Scholar]
- 16.Zou H, Li Y, Liu X, Wang X. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 17.Kim HE, Du F, Fang M, Wang X. Proc Natl Acad Sci USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C, Wang A, Wang L, Dorsch M, Ocain TD, Xu Y. Biochem Biophys Res Commun. 2005;331:1114–1119. doi: 10.1016/j.bbrc.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 19.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 20.Traut TW. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 21.Coffield VM, Jiang Q, Su L. Nat Biotechnol. 2003;21:1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowds TA, Masumoto J, Zhu L, Inohara N, Nunez G. J Biol Chem. 2004;279:21924–21928. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 24.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL. Plant Physiol. 2006;140:1233–1245. doi: 10.1104/pp.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 28.Der CJ, Finkel T, Cooper GM. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- 29.Pomerantz RJ, Horn DL. Nat Med. 2003;9:867–873. doi: 10.1038/nm0703-867. [DOI] [PubMed] [Google Scholar]
- 30.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 31.Silverman N, Maniatis T. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 32.Northup JK, Smigel MD, Gilman AG. J Biol Chem. 1982;257:11416–11423. [PubMed] [Google Scholar]
- 33.Brandt DR, Asano T, Pedersen SE, Ross EM. Biochemistry. 1983;22:4357–4362. doi: 10.1021/bi00288a002. [DOI] [PubMed] [Google Scholar]