Abstract
Malignant astrocytomas are the most common and lethal adult primary brain tumor. Retroviral gene trapping of nontransformed neonatal astrocytes from a glial fibrillary acidic protein (GFAP):V12Ha-Ras murine astrocytoma model led to isolation of the transcription factor Gata6. Loss of Gata6 resulted in enhanced proliferation and transformation of astrocytes. Human malignant astrocytoma cell lines, explant xenografts, and operative specimens demonstrated loss of GATA6 expression. Loss-of-function GATA6 mutations with loss of heterozygosity of the GATA6 locus were found in human malignant astrocytoma specimens but not in lower-grade astrocytomas or normal adult astrocytes. Knockdown of Gata6 expression in V12Ha-Ras or p53−/− astrocytes, but not in parental murine or human astrocytes, led to acceleration of tumorgenesis. Knockin GATA6 expression in human malignant astrocytoma cells reduced their tumorgenic growth with decreased VEGF expression. Collectively, these data demonstrate that GATA6, isolated from a murine astrocytoma model, is a novel tumor suppressor gene that is a direct target of mutations during malignant progression of murine and human astrocytomas. This work also demonstrates the utility of random mutagenesis strategies, such as gene trapping, on murine cancer models toward discovery of novel genetic alterations in corresponding human cancers.
Glioblastoma multiforme (GBM) are the most common and malignant of all primary human brain tumors (1, 2). Current management includes radiation and concomitant chemotherapy; however, the median survival remains ≈1 year (3). Although pathologically indistinguishable, some GBM develop from a preexisting low-grade astrocytoma (LGA) (“secondary GBM”), whereas others appear de novo (“primary GBM”) (2, 4). We and others have demonstrated increased levels of activated p21-Ras in GBM without primary oncogenic Ras mutations (5–7). We used ES cell transgenesis to generate genetically engineered mouse (GEM) models of astrocytomas by expressing oncogenic Ha-Ras (V12Ha-Ras) under control of the astrocyte-specific human glial fibrillary acidic protein (GFAP) promoter (8). One strain of the GFAP:V12Ha-Ras GEM (RasB8) are alive to reproductive age but ultimately develop LGA and die from high-grade astrocytomas (HGA) by ≈3–4 months of age (8, 9). GFAP-positive astrocyte cultures established from newborn (B8-P0) or 3-month-old (B8–3mth) mice harboring HGA both express the V12Ha-Ras transgene with elevated levels of activated Ras [supporting information (SI) Figs. 7 and 8]; however, B8-P0, unlike B8–3mth, astrocytes do not grow in soft agarose or develop tumors in Nod-Scid mice (SI Table 1). This suggests that the V12Ha-Ras transgene does not suffice to transform astrocytes but promotes acquisition of additional transforming molecular alterations, several of which are known to occur in human astrocytomas (8, 9).
In this study the B8 model was used as a “gene-discovery” reagent. Using retroviral gene trapping to screen for genetic modifiers that accelerate transformation of B8-P0 astrocytes, we identified the transcription factor Gata6 as a relevant novel tumor suppressor gene in human astrocytomas. Loss of Gata6 expression, with mutations in its DNA binding domain and loss of heterozygosity (LOH), was present in B8 murine HGA and not LGA and also human GBM and not LGA. This suggests that loss of Gata6 transcriptional regulation contributes to astrocytoma progression rather than initiation. Furthermore, short hairpin RNA (shRNA) knockdown of GATA6 in V12Ha-Ras transfected murine and human astrocytes accelerated transformation, whereas inducible and constitutive replacement of GATA6-null human GBM cells inhibited their tumorigenic growth. Collectively, these data support GATA6, a genetic alteration discovered by using a GEM astrocytoma model, as a functionally relevant novel human astrocytoma tumor suppressor gene.
Results
A Retroviral Gene-Trapped V12Ha-Ras Astrocyte Library.
Primary astrocyte cultures were established from normal murine astrocytes (NMAs) from newborn CD1-ICR mice (NMA-P0), plus newborn (B8-P0) and 3-month-old (B8–3mth) B8 mice, the latter harboring malignant HGA. Purity of the derivative astrocyte cultures was >90% as determined by IFC for GFAP (SI Fig. 7). B8-P0 and B8–3mth astrocytes expressed β-gal, indicative of the V12Ha-Ras:IRES-LacZ (8) transgene, and had equivalent and elevated levels of Ras-GTP compared with NMA-P0 astrocytes (SI Fig. 8). A total of 1.2 × 106 NMA-P0 or B8-P0 plated astrocytes (up to 30 passages) did not form soft agar colonies or grow intracranial xenografts in Nod-Scid mice, whereas ≈5–10% of B8–3mth astrocytes grew soft agar colonies and developed intracranial tumors (SI Table 1 and Fig. 1B).
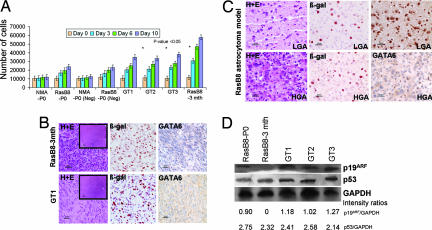
Fig. 1.
Proliferation and tumorigenic properties of gene-trapped astrocytes derived from the B8 GEM astrocytoma model. (A) MTT proliferation assays of NMAs from CD1-ICR newborn pups (NMA-P0), newborn B8 pups (B8-P0), both transduced with empty vector controls (Neg), gene-trapped B8-P0 astrocyte clones (GT1, GT2, and GT3), and B8–3mth astrocytoma cultures derived from a mouse harboring a HGA. Gene-trapped B8-P0 clones have a proliferation advantage compared with parental and Neg NMA-P0 and B8-P0 astrocytes, approaching the proliferation rate of the transformed B8–3mth astrocytoma cultures. (B Upper) Pathological features of invasive malignant astrocytoma in the frontal cortex of Nod-Scid mice 1 month after injection of 106 B8–3mth astrocytes (H&E). β-Gal staining confirms expression of the GFAP:V12HaRas-IRES-LacZ transgene. GATA6 expression is lost in the HGA. (Lower) Pathological features of invasive malignant astrocytoma in the frontal cortex of Nod-Scid mice that developed in ≈7% of B8-P0 gene-trapped astrocytes (GT1 clone shown; H&E), with β-gal expression from both the GFAP:V12HaRas-IRES-LacZ transgene and the gene-trap vector. Parental or empty gene-trap vector-negative NMA and B8-P0 controls did not grow in Nod-Scid mice up to 6 months of observation. The finding of loss of Gata6 expression in the HGA suggests that an acquired mutation occurred in the nontargeted gene-trapped allele. (C) B8 GEM astrocytomas at the age of 1 month (LGA, Upper) and 3 months (HGA, Lower) (H&E). Gata6 is expressed in 1-month LGA (Upper Right) but absent in 3-month HGA (Lower Right), suggesting a role in tumor progression. β-Gal staining (Center) confirms expression of the GFAP:V12HaRas-IRES-LacZ transgene in the transformed astrocytes. GATA6 is abundantly expressed in wild-type age-matched brains (13). (Magnifications: ×400 for B and C and ×40 for Insets in B Left. Scale bars: 25 μm for B and C and 250 μm for Insets in B Left.) (D) Western blot analysis demonstrating that p19ARF and p53 expression is relatively unchanged in the B8 gene-trapped clones (GT1, GT2, and GT3) when compared with the B8-P0 parental cells. p19ARF expression is absent in the B8–3mth astrocytoma cells. Approximately 40 μg of protein lysates was loaded in each well. GAPDH was used to assess the amount of protein loaded.
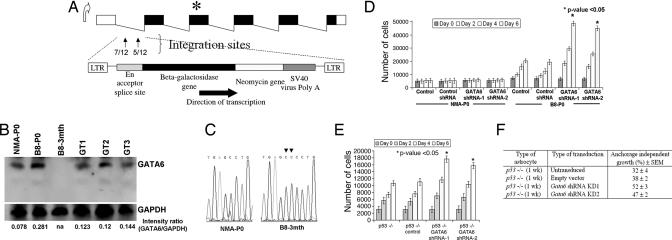
The retroviral gene trap cassette (Fig. 2A), consisting of a splice acceptor immediately upstream of β-gal–neomycin resistance fusion gene (βgeo) and the control (empty) vector, was used to transduce 1.2 × 106 NMA-P0 and B8-P0 astrocytes (passage 4) with an ≈1.5 multiplicity of infection. Growth in soft agarose was not detected with control retrovirus, whereas ≈0.00375% of gene-trapped B8-P0, but not NMA-P0 astrocytes, grew in soft agar (SI Table 1). Inverse PCR on 45 anchorage-independent gene-trapped B8-P0 clones yielded 15 clones with trapped genes. In 3/15 clones the murine Elys gene (GenBank accession no. NM_026375.1) was trapped. Elys is speculated to have a major role in development of extraembryonic membranes and hematopoietic cell lineages (10–12). We ruled out the human ELYS gene as a relevant astrocytoma tumor suppressor gene by demonstrating expression by RT-PCR in a panel of established human GBM specimens (data not shown). In 12/15 clones the retroviral gene trap cassette integrated within two sites of the first intron of the murine Gata6 gene (GenBank accession no. NM_010258.2), in the same orientation as the Gata6 promoter (Fig. 2A). Quantitative real-time PCR estimated two to three copies of the gene trap vector per cell (SI Table 2) in three of these Gata6-trapped B8-P0 clones (GT1, GT2, and GT3).
Fig. 2.
Characteristics of gene-trapped astrocytes. (A) Schematic of the murine Gata6 transcript isoform expressed in the CNS. Integrations of the retroviral gene-trap vector were identified within the first intron of Gata6 and in the same orientation as the endogenous promoter. The number of integrations per site is shown in the first intron. ∗ represents the map position of a homozygous frameshift mutation identified in exon 3 of B8–3mth astrocytes. (B) Western blot analyses on protein lysates isolated from primary astrocyte cultures, with 40 μg of total protein lysate loaded per lane. Absent Gata6 expression is noted in B8–3mth astrocytoma cells (consistent with findings in Fig. 1 B and C). GAPDH was used as a positive control for loading and normalization of the densitometric analyses with Fluor Chem software. Gata6 expression was reduced between ≈50% and 58% among the three B8-P0 gene-trapped clones analyzed in detail (GT1, GT2, and GT3). (C) Chromatograms demonstrating a 1641_1642InsCC mutation in exon 3 of Gata6 in the B8–3mth astrocytes encoding the DNA binding domain. (D) Gata6 expression was constitutively knocked-down in NMA and B8-P0 astrocytes by ≈90% compared with controls by Western blot analysis (data not shown), with two shRNAs (mapping to murine exons 2 and 4) expressed from a stably integrated pSIREN-RetroQ vector. Parental or NMA and B8-P0 astrocytes engineered with a negative shRNA vector (control) were used as negative controls for the MTT proliferation assays. Gata6 knockdown induced a significant (∗, P < 0.05) proliferation advantage after day 2, only in the B8-P0 astrocytes and not in NMA. (E) Gata6 knockdown in homozygous null murine astrocytes induced a proliferative advantage (P < 0.05) and increased anchorage-independent growth in soft agar (F) compared with parental p53−/− astrocytes or those transduced with a negative shRNA vector (control).
Characterization of the Gata6 Gene-Trapped B8-P0 Clones.
From the 12 Gata6-trapped B8-P0 clones we characterized three (GT1, GT2, and GT3) clones in detail. Gata6 expression was reduced by ≈50–58% (Fig. 2B), with an accompanying decrease in RNA expression by quantitative real-time RT-PCR (SI Table 3) compared with parental B8-P0 astrocytes. In NMA-P0 astrocytes Gata6 expression was low but present, whereas it was completely absent in B8–3mth astrocytoma cells (Fig. 2B). Ras-GTP levels in the Gata6-trapped clones were comparable to B8-P0 astrocytes (SI Fig. 8); however, the trapped clones had increased proliferation (P < 0.05) compared with parental or control vector transduced NMA-P0 and B8-P0 astrocytes, approaching those of B8–3mth cells (Fig. 1A). In contrast to NMA-P0 or B8-P0 astrocytes, ≈7% (n = 3/45) of mice injected with these Gata6-trapped clones grew intracranial invasive malignant astrocytomas, pathologically similar to mice injected with B8–3mth cells (SI Table 1 and Fig. 1B). Gata6 expression was absent from the astrocytomas that developed from both the B8–3mth astrocytes and the B8-P0 gene-trapped clones (Fig. 1B), suggesting that in the gene-trapped clones an acquired loss-of-function mutation in the nontargeted Gata6 allele had occurred. Of additional interest, in contrast to loss of p19ARF and mutations in p53 described previously in B8 HGA and derived B8–3mth cells (9), these were not found in B8-P0 or the Gata6 gene-trapped clones (Fig. 1D).
Gata6 Alterations in Transgenic Murine Astrocytes.
Mutational screen of all six exons of the mouse Gata6 gene did not reveal any deletions or insertions in NMA-P0, B8-P0, or the three gene-trapped clones, whereas the B8–3mth astrocytoma cells harbored a 1641_1642InsCC mutation in the third exon of Gata6 encoding the DNA binding domain (Fig. 2C and SI Table 6). This mutation was not a naturally occurring polymorphism, because it was not identified in 50 normal chromosomes analyzed from 25 different mice (data not shown). We also examined Gata6 expression in the B8 GEM astrocytomas at different times in astrocytoma development. Gata6 was abundantly expressed in LGA (B8–1mth) but absent in the HGA RasB8 tumors (Fig. 1C), suggesting a role for loss of Gata6 expression as a late progression factor.
Effects of Decreased Gata6 Expression in Transgenic Murine Astrocytes.
Gata6 expression was decreased by constitutive and stable expression of two shRNAs in parental NMA-P0 and B8-P0 astrocytes, targeting the second and fourth exons of the Gata6 gene, resulting in >90% reduced expression (data not shown). This resulted in a significant proliferative advantage in the B8-P0 but not in NMA-P0 astrocytes (P < 0.05) (Fig. 2D), which was paralleled by the ability to grow in soft agar (SI Table 1). Loss of Gata6 expression promoting transformation was not restricted to murine astrocytes with oncogenic p21-Ras, because a proliferative advantage and ability to grow in soft agar also resulted in p53−/− astrocytes (passage 3), but not in empty vector transduced control cells (Fig. 2 E and F).
GATA6 Alterations in Human Astrocytomas.
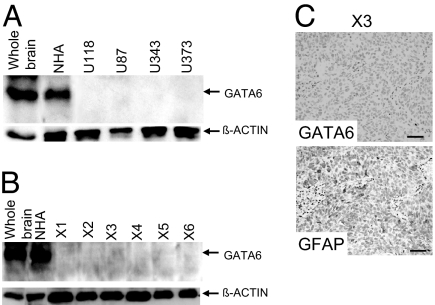
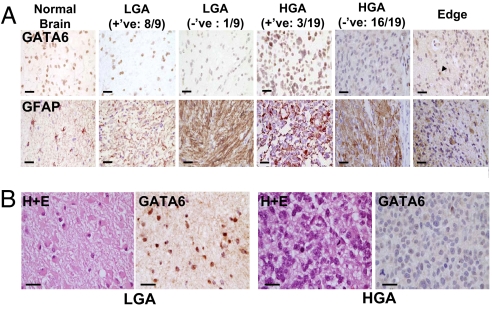
Western blot analyses, with an antibody that recognizes all murine and human GATA6 isoforms, failed to detect any expression in a panel of well characterized established human GBM lines (Fig. 3A). GATA6 expression was also absent in a panel of early- and late-passage human GBM s.c. xenograft explants (Fig. 3 B and C). An additional panel of 22 human GBM flash-frozen operative specimens was screened with RT-PCR, with GATA6 expression not detectable in ≈90% (20/22) (Fig. 4A). Insufficient amounts of frozen tissue were available to undertake Western blot analysis on each specimen; however, we undertook immunohistochemistry (IHC) analysis on a panel of nine LGA and 19 GBM (Fig. 5A), the latter a subset from the 22 GBM analyzed by RT-PCR (Fig. 4A). GATA6 expression was lost in only ≈10% (1/9) of LGA while absent in ≈85% of GBM (16/19). We also examined a patient who presented with a secondary GBM and found GATA6 expression in nontransformed astrocytes and the LGA at initial presentation but absent in the subsequent GBM (Fig. 5B). This preliminary evaluation in adult human LGA and HGA suggests that loss of GATA6 expression is associated with astrocytoma progression rather than initiation.
Fig. 3.
Alterations in GATA6 expression among human GBM lines and explant xenografts. (A) Western blot analysis demonstrating loss of GATA6 expression in all four established human GBM lines. Each lane has 40 μg of total protein lysate, with β-actin used as a positive loading control. Whole-brain sample was obtained from a head injury patient requiring surgery, whereas NHA represents human hTERT immortalized but nontransformed astrocytes. (B) GATA6 expression was absent in six human GBM explant xenografts. Each lane has 40 μg of total protein lysate, with β-actin used as a positive loading control. (C) GATA6 expression in human GBM explant xenograft (X3) determined by IHC. A rabbit GATA6 polyclonal primary antibody and a protein G-HRP secondary antibody were used. None of the xenografts expressed GATA6, with 50% immunopositive for the differentiated astrocyte marker GFAP (rabbit GFAP polyclonal antibody from DAKO). No immunoreactivity was detected on the GBM explant xenograft specimens using only a secondary antibody. (Magnification: ×200.)
Fig. 4.
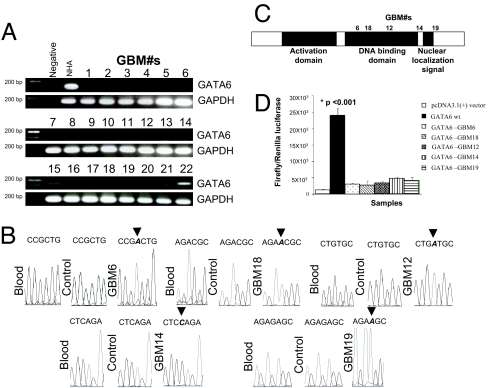
Screening of human GBM operative specimens for alterations in expression and mutations. (A) RT-PCR screening of a panel of 22 human GBM operative specimens. GATA6 expression was absent in 20/22 samples tested. GAPDH was used as a positive control marker. (B) Chromatograms of homozygous frameshift mutations identified in the GATA6 DNA binding domain and C terminus of five GBM specimens. These specimens also had loss of GATA6 RNA expression by RT-PCR (A), with loss of GATA6 protein expression in 19 specimens available for IHC evaluation (Fig. 5A). (C) Schemata of the human GATA6 protein with locations of homozygous frameshift mutations identified in the DNA binding domain (3/5) and C terminus (2/5) of GBM specimens and associated with LOH (SI Fig. 10). (D) DNA binding assays of GATA6 patient GBM mutations using the p450-Cytochrome:C17 promoter:pGL3 firefly luciferase reporter in U87 cells. Negative controls included nontransfected U87 plus transfection with the pcDNA3.1+ vector lacking only the GATA6 gene. The data (expressed as mean ± SEM) demonstrate that wild-type GATA6 induced significant expression (P < 0.001) of the firefly luciferase reporter from the p450-Cytochrome:C17 promoter. However, the GATA6 frameshift mutations identified from the GBM specimens resulted in minimal nonsignificant induction of the firefly luciferase reporter from the p450-Cytochrome:C17 promoter.
Fig. 5.
Screening of human astrocytomas operative specimens for alterations in expression using IHC. (A) GATA6 expression was tested by IHC in normal human brain (from head injury patient), nine LGA, and 19 human GBM paraffin-embedded specimens. One of nine (≈10%) LGA and 16/19 (≈85%) GBM had loss of GATA6 expression, suggesting that loss of GATA6 is involved in astrocytoma progression. Pictures show GATA6 (Upper) and GFAP (Lower) immunostaining. The rightmost panels demonstrate the invading edge of a GBM with loss of GATA6 expression, while nontransformed astrocytes and other cells retain GATA6 expression (arrowhead). (Magnification: ×400. Scale bars: 25 μm.) (B) GATA6 expression is analyzed in a patient with a pathologically documented secondary GBM. GATA6 expression is abundant in the initial resected LGA (Left) but absent when the patient recurred with a GBM (Right). Shown are H&E immunostaining and GATA6 immunostaining. (Magnification: ×400. Scale bars: 25 μm.)
Mutation analyses across the entire seven exons plus 5′ flanking region of the human GATA6 gene were undertaken in five GBM samples demonstrated to have loss of GATA6 transcript (Fig. 4A and SI Tables 7 and 8). Mutations were identified in 3/5 GBM within the DNA binding domain (exon3) and 2/5 harbored mutations in the C terminus of the protein (SI Table 4, Fig. 4 B and C, and SI Fig. 9A). Presence of mutation was also confirmed by using a second set of primers and PCR-RFLP analysis (SI Fig. 9). The mutations were not naturally occurring polymorphisms, as they were not identified in 50 normal chromosomes analyzed from 25 normal volunteers. Second, these mutations were not germ-line because they were absent in normal peripheral blood leukocytes from corresponding GBM patients (Fig. 4B). To discern whether LOH was evident, we selected a panel of inter- and intragenic SNP genetic markers mapping within and flanking the GATA6 gene to genotype the patient's blood and GBM (SI Fig. 10 and SI Table 9). LOH of SNP markers (both inter- and intragenic) were noted to be associated with the GATA6 gene that had mutations within the protein-coding exons.
Because all five GBM lacked GATA6 transcripts by RT-PCR (Fig. 4A), we postulated that these identified human mutations (associated with LOH) in the DNA binding domain and C terminus result in nonsense frameshift mutation-mediated mRNA decay pathway/and or a GATA6 protein with loss of function. To test for the functional consequence of the GATA6 mutations found in the five GBM sequenced, we used a 227-bp fragment of the p450-Cytochrome:C17 promoter containing GATA6 transcriptional regulatory elements (13) using a dual luciferase assay. Wild-type GATA6 induced the p450-Cytochrome:C17 promoter (Fig. 4D) after transient transfection in U87 cells lacking GATA6 expression (Fig. 3A). However, induction by the five identified human GBM GATA6 mutations was reduced by ≈5- to 8-fold (Fig. 4D) (P < 0.001) and was only ≈2- to 4-fold greater than background induction from transient transfection with the pcDNA3.1+ vector only.
GATA6 Functional Studies in Human Astrocytes.
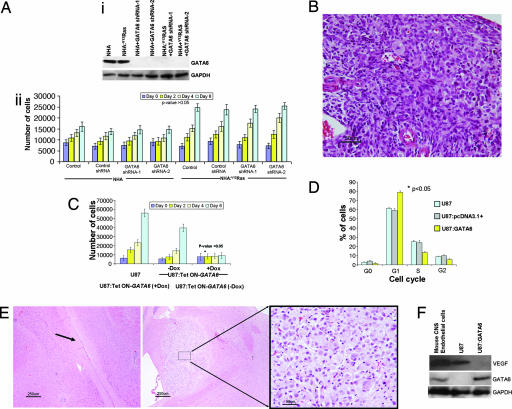
Knockdown of GATA6 expression was undertaken by constitutive and stable expression of shRNAs in hTERT immortalized normal human astrocytes (NHA) (14) and NHA stably transfected to express V12Ha-Ras (NHA:V12Ha-Ras), both of which are unable to form soft agar colonies and grow intracranial tumors (data not shown). The shRNAs targeted the fourth and fifth exons of the human GATA6 gene, resulting in reduced RNA expression and subsequent absent protein expression (Fig. 6Ai and SI Table 5). Parental NHA:V12Ha-Ras had an overall increased proliferation rate compared with NHA astrocytes (P < 0.05), but GATA6 knockdown did not increase the in vitro proliferation of either line (P > 0.05) (Fig. 6Aii). The difference in proliferation properties compared with the murine astrocytes (Fig. 2D) is most likely due to the B8-P0 astrocytes harboring many genetic alterations other than V12Ha-Ras (8), many of which are uncharacterized. These genetic alterations are sufficient to create a greater susceptibility for enhanced proliferation compared with only hTERT and V12Ha-Ras expression in NHA. However, intracranial injection of GATA6 knockdown NHA:V12Ha-Ras astrocytes (1 × 106) (n = 3), but not of NHA:V12Ha-Ras transduced with vector only or GATA6 knockdown NHA astrocytes (n = 3 of each), grew invasive astrocytomas in Nod-Scid mice at ≈4 months (Fig. 6B).
Fig. 6.
Functional studies of human GATA6 shRNA knockdown and reexpression. (Ai) GATA6 expression was knocked down by two shRNAs (human exons 4 and 5) from a stably integrated pSIREN-RetroQ vector. Parental astrocytes and those expressing control shRNA were used as negative controls. (Aii) MTT proliferation assay on hTERT immortalized nontransformed NHA and NHA:V12Ha-Ras human astrocytes. GATA6 knockdown did not induce a significant in vitro proliferation advantage in the NHA or NHA:V12Ha-Ras cells (P > 0.05). (B) Intracranial injection into Nod-Scid mice with 106 NHA:V12Ha-Ras + GATA6 shRNA knocked-down astrocytes, but not controls, resulted in in vivo malignant astrocytomas (H&E; Magnification: ×200), demonstrating tumor cells with nuclear atypia and mitosis. (C) GATA6 reexpression induced antiproliferative effects on U87:Tet ON-GATA6 lines treated with Dox (P < 0.05). Leaky expression of GATA6 in the pREV-TRE vector can account for slight inhibition of proliferation even in untreated cells compared with parental U87 cells over the 6-day analysis period. (D) Cell cycle flow cytometry analysis of U87 cells transiently expressing GATA6 under control of the cytomegalovirus promoter (pcDNA3.1+). Nontransfected and empty vector (pcDNA3.1+) transient transfected U87 cells served as negative controls. Transient reexpression of GATA6 induced a significant (P < 0.05) increase in G1 arrested cells compared with the negative controls. (E) Intracranial xenografts with 106 U87:Tet ON-GATA6 cells with or without Dox in the drinking water. Tumor growth was not seen after ≈4 months in mice treated with Dox (Left; arrow shows injection tract). In contrast, mice without Dox had to be killed at 4 weeks, with development of large tumor nodules at the site of injection (Center; Right shows a magnified view of the tumor). (Magnification: ×40 in Left and Center and ×400 in Right. Scale bars: 250 μm in Left and Center and 25 μm in Right.) (F) GATA6 regulation of VEGF expression. U87 cells express VEGF (similar to control mouse brain endothelial cells) but not GATA6. Transient reexpression of GATA6 in the U87 cells inhibits VEGF expression.
Overexpression of GATA6 was undertaken by tetracycline [doxycycline (Dox)]-On-induced expression in U87 and U373 human GBM cells, which do not express GATA6 (Fig. 3A). Expression of GATA6 in U87 cells by administration of Dox resulted in inhibition of in vitro proliferation, measured by MTT assay, over 6 days (Fig. 6C) and also growth in soft agar (data not shown). Transient overexpression of GATA6 in U87 cells resulted in a significant increase in the number of cells arrested in G1 when compared with U87 cells or U87 transiently transfected with an empty vector (P < 0.05) (Fig. 6D), but no significant changes were noted in the percentage of cells in the subG1 phase, indicative of apoptotic cells. Intracranial injections of 1 × 106 U87:Tet On-GATA6 cells into the frontal lobe of Nod-Scid mice reproducibly produced gliomas within ≈4 weeks without Dox, (n = 3) (Fig. 6E Right). With Dox administered in the drinking water to express GATA6, gliomas did not develop in the mice by 4 months (n = 3) (Fig. 6E Left). Inhibition of tumorigenesis was also noted with Dox-induced GATA6 expression in U373 GBM lines (data not shown). We have begun to study GATA6-regulated effectors that may play a functional role in glioma progression, such as those linked to astrocytoma angiogenesis, a recognized key differentiating factor between low- and high-grade gliomas (2). Transient constitutive reexpression of GATA6 in U87 cells inhibited VEGF expression (Fig. 6F), recognized as a critical mediator of angiogenesis in gliomas.
Discussion
GEM models of cancers, based on known genetic alterations of human tumors, have been used to enhance our knowledge of tumor biology, genetic interactions, progression, and preclinical therapeutic evaluation. This study demonstrates how these models may also be used as gene-discovery reagents for finding novel genetic alterations in human cancers. Using retroviral gene-trapping on newborn (P0) transgenic astrocytes, established from a GFAP:V12HaRas GEM astrocytoma model, we identified Gata6 as a transforming modifier with V12HaRas in astrocytes. Expression, mutational, and functional analyses all demonstrate that GATA6 is a novel tumor suppressor gene not only in the GEM model, but also more importantly in human GBM.
GATA6 is one of six members of the mammalian GATA family of transcription factors, all containing two highly conserved zinc-finger DNA binding domains that interact with a canonical DNA motif (G/A)GATA(A/T) (15). Expression of Gata6 in the developing murine brain at embryonic days 11 and 15 and postnatal day 7 has been undertaken as part of the Brain Expression Map Project (www.stjudebgem.org/web/mainPage/mainPage.php). We and others have recently demonstrated GATA6 expression in a variety of adult murine and human cells of the nervous system, such as the choroid plexus epithelium, neurons, astrocytes, and endothelial cells (16, 17).
Our interest in Gata6 stemmed not only from its prevalence in the gene-trapped RasB8-P0 clones (12/15) (Fig. 2A), but also from prior evidence of its role as a potential tumor suppressor gene. Loss of Gata6 expression was reported in a murine model of adrenocortical tumors (18), and ≈82% of human ovarian cancers had complete absence or mislocalization of the GATA6 protein (19). Although Gata6 was obtained from a gene-trap screen of a GEM astrocytoma model, this report is the first where all major criteria for GATA6 to be a valid tumor suppressor gene in a human cancer have been obtained. (i) GATA6 protein expression was absent in several established human GBM lines and explant xenografts (Fig. 3). (ii) GATA6 RNA expression was absent in 20/22 (≈90%) human GBM operative specimens (Fig. 4A). (iii) GATA6 protein expression by IHC was absent in the majority (≈85%) of human GBM specimens and a much lower number (≈10%) of LGA (Fig. 5A). In a patient demonstrating a secondary GBM, GATA6 expression was absent in the GBM lesion but present in the LGA lesion (Fig. 5B), similar to what we found with the B8 GEM (Fig. 1C). These findings collectively suggest loss of GATA6 to play a role in astrocytoma progression rather than initiation. (iv) Nonpolymorphic GATA6 mutations were found in all five GBMs sequenced, with resultant loss of GATA6 transcriptional activity (Fig. 4 B–D). These mutations were associated with LOH (SI Fig. 10). (v) Stable reduction of GATA6 expression by two shRNA in nontransformed V12Ha-Ras immortalized mouse and human astrocytes resulted in in vitro and in vivo transformation (Figs. 2D and 6 A and B). (vi) Replacement of wild-type GATA6 into human GBM cell lines lacking GATA6 expression reverted in vitro and in vivo growth potentiation, as a consequence of increasing the number of cells arresting in G1 and decreasing VEGF expression (Fig. 6 C–F).
Loss of GATA6 and resultant tumor progression is not restricted to astrocytes with only p21-Ras mutations or elevated levels of Ras-GTP activity. Mutations and loss of GATA6 expression were present in human GBM specimens, which, though harboring elevated levels of Ras-GTP activity, do not harbor p21-Ras mutations (5). Furthermore, there was no significant difference in Ras-GTP levels in the B8-P0, corresponding B8-P0 gene-trapped clones (GT1, GT2, and GT3), or B8–3mth astrocytes (SI Fig. 8), although only the latter two cell lines grew in soft agar and made in vivo tumors. Last, Gata6 shRNA knockdown induced proliferation and transformation in both V12HaRas and p53−/− (without p21-Ras mutations) murine astrocytes (Fig. 2 E and F).
Reexpressing GATA6 in human GBM cells (U87), with resultant decreased proliferation, demonstrates accumulation in the G1 phase of the cell cycle (Fig. 6 C and D). This is similar to reports in rat glomerular mesangial cells and murine vascular smooth muscle cells plus embryonic fibroblasts (20, 21). In rat glomerular mesangial cells, GATA6 overexpression inhibited CDK2 kinase activity, with an increase in the steady-state levels of p21Cip and attenuation of cyclin A, leading to G1 arrest (20). Consistent with the above report, loss of GATA6 in ≈85–90% of human GBM (Figs. 4A and 5A) is also associated with a decrease or absence of p21Cip immunoreactivity in ≈50% of GBM (22). In addition, we found that knockdown of GATA6 in the gene-trapped B8-P0 astrocytes did not alter expression of p19ARF or p53 (Fig. 1D). However, in the tumors that arise from these intracranial injected gene-trapped B8-P0 cells (data not shown), B8 HGA, and derivative B8–3mth astrocytoma cells, there is loss of p19ARF expression and p53 mutations (8, 9). This suggests that additional GATA6 independent secondary genetic alterations resulting in altered p19ARF and p53 expression are collectively involved in promoting in vivo tumorigenesis.
GATA6-regulated transcripts involved in promoting astrocytoma progression are of biological and potential therapeutic interest. Restoration of GATA6 expression in U87 cells inhibited their growth in intracranial xenograft models, which may result from decreased angiogenic capacity because of inhibition of VEGF expression (Fig. 6 E and F). Increased angiogenesis and VEGF expression are closely linked to malignant astrocytoma progression, especially GBM (2).
GATA6 maps to mouse chromosome 18A1 and human chromosome 18q11.1-q11.2 (www.ensembl.org). LOH in the proximal region of human chromosome 18q has been reported in human squamous cell, ovarian, and prostate cancers (23–25). Moreover, LOH of human chromosome 18q has been reported in astrocytomas specimens (26, 27). cDNA macroarray and microarray experiments have not yet reported a significant loss of GATA6 expression in human GBM due to a lack GATA6 on the arrays or to the method of analysis (28–33). However, using a nonbiased gene-discovery approach with a GEM astrocytoma model, we have identified, and verified with expression, mutational, and functional analyses, GATA6 to be a novel human astrocytoma tumor suppressor gene involved in tumor progression. In a broader perspective, this work also illustrates how random insertional mutagenesis strategies most commonly used to generate mouse germ-line mutations, such as gene trapping, RAGE, and Sleeping Beauty (34, 35), can be used in relevant GEM models of human diseases to decipher novel loss- or gain-of-function alterations in the corresponding human disease.
Methods
Transformation Assays.
MTT viability assay with triplicate plating of 3,000–10,000 cells into each well of a 96-well microtiter plate with 100 μl of astrocyte medium was done to evaluate anchorage dependent growth (9). The assay was performed at 2-day intervals for a period of 6–10 days. Statistical analysis was undertaken by using the unpaired two-tailed Student t test, with P values <0.05 considered significant. Ability to form colonies in soft agar after 4 weeks was used to evaluate anchorage-independent growth by plating ≈106 cells in soft agar as described (9). Dox (1 μg/ml; Sigma, St. Louis, MO) was used to induce expression of GATA6 in U87 and U373 lines in vitro (pREV-TRE:GATA6), with controls grown in the absence of Dox. To evaluate in vivo transformation, stereotactic intracranial injection of ≈106 cells into the frontal lobe of Nod-Scid mice was done. At specified time points or as per animal care guidelines, the mice were killed, and brains were harvested and fixed in 10% formamide. The brains were embedded in paraffin, cut into 7-μm sections, and evaluated by using IHC. For these in vivo experiments, 10 mg/ml Dox was given in the drinking water of mice injected with U87 and U373 (pREV-TRE:GATA6) astrocytoma cells to induce GATA6 expression.
IHC.
Sections (7 μm) of paraffin-embedded specimens [six human GBM explant xenografts, 19 human GBM, nine LGA, secondary GBM specimens (low- and high-grade astrocytoma from a single patient), B8 gene-trapped induced intracranial tumors, B8–3mth induced intracranial tumors, and B8 GEM brains (LGA-1mth and HGA-3mth)] were subjected to antigen retrieval by pressure cooking for 15 min in antigen unmasking solution (Vector Laboratories, Burlingame, CA) used with standard IHC protocols (9). Specific dilutions used included 1:100 of rabbit GATA6 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), 1:600 of rabbit GFAP polyclonal antibody (Dako), 1:300 of β-gal mouse ascites (Sigma), and 1:300 of protein G-HRP secondary antibody (Zymed) as a negative control. Detection was with a protein G-HRP secondary antibody (Zymed) and a peroxidase kit (Vector Laboratories); protein G-avidin secondary antibody and a biotin detection kit (Vector Laboratories); or the Vector Elite avidin–biotin complex method detection system (Vector Laboratories). Sections were counterstained briefly in hematoxylin for 5 sec, dehydrated in 70%, 80%, and 100% ethanol series, followed by a brief wash in xylene, and mounted in Permount (Fisher). Sections were also stained with H&E (Fisher) by using standard protocols. Images were captured with Nikon ACT-1 software and a Nikon E-600 microscope mounted with a digital camera.
Additional Details.
Additional methods are described in SI Text.
Acknowledgments
We thank Ms. V. Lee and Mrs. J. Ma for technical assistance, Dr. P. Soriano (Fred Hutchinson Cancer Institute, Seattle, WA) for providing the pGEN− retroviral vector cassette, Dr. W. C. Skarnes (Sanger Institute, Cambridge, U.K.) for providing the pGT1.8βgeo plasmid-based gene trap vector, Dr. T. Mak (University of Toronto) for providing p53−/− mice, and Dr. R. Pieper (University of California, San Francisco, CA) for providing hTERT immortalized NHA. D.K. is a recipient of funds/awards from Restracomp, the National Sciences and Engineering Research Council of Canada, and the Canadian Institutes of Health Research. W.L.S. is the Canada Research Chair in Stem Cell Biology and Functional Genomics. A.G. is the Hudson Chair in Neurooncology at the University of Toronto. This project was funded by grants to A.G. from the National Cancer Institute of Canada, the Cleveland Foundation, and the National Institutes of Health.
Abbreviations
- GBM
glioblastoma multiforme
- GEM
genetically engineered mouse
- IHC
immunohistochemistry
- LOH
loss of heterozygosity
- NHA
normal human astrocyte
- NMA
normal murine astrocyte
- P0
newborn
- shRNA
short hairpin RNA
- Dox
doxycycline
- LGA
low-grade astrocytoma
- HGA
high-grade astrocytoma
- GFAP
glial fibrillary acidic protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7737.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611669104/DC1.
References
- 1.Mahaley MS, Jr, Mettlin C, Natarajan N, Laws ER, Jr, Peace BB. J Neurosurg. 1989;71:826–836. doi: 10.3171/jns.1989.71.6.0826. [DOI] [PubMed] [Google Scholar]
- 2.Kleihues P, Cavenee WK, editors. WHO Classification of Tumors: Pathology and Genetics of Tumours of the Nervous System. Lyon, France: Int Agency for Res on Cancer; 2000. [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Gusella JF. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- 5.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Oncogene. 1997;15:2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 6.Feldkamp M, Lau N, Guha A. Oncogene. 1999;18:7514–7526. doi: 10.1038/sj.onc.1203105. [DOI] [PubMed] [Google Scholar]
- 7.Feldkamp M, Lau N, Roncari L, Guha A. Cancer Res. 2001;61:4425–4431. [PubMed] [Google Scholar]
- 8.Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, Gutmann DH, Squire JA, Nagy A, Guha A. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- 9.Shannon P, Sabha N, Lau N, Kamnasaran D, Gutmann DH, Guha A. Am J Pathol. 2005;167:859–867. doi: 10.1016/S0002-9440(10)62057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura N, Takizawa M, Okita K, Natori O, Igarashi K, Ueno M, Nakashima K, Nobuhisa I, Taga T. Genes Cells. 2002;7:435–446. doi: 10.1046/j.1365-2443.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- 11.Okita K, Nobuhisa I, Takizawa M, Ueno M, Kimura N, Taga T. Biochem Biophys Res Commun. 2003;305:327–332. doi: 10.1016/s0006-291x(03)00772-1. [DOI] [PubMed] [Google Scholar]
- 12.Okita K, Kiyonari H, Nobuhisa I, Kimura N, Aizawa S, Taga T. Genes Cells. 2004;9:1083–1091. doi: 10.1111/j.1365-2443.2004.00791.x. [DOI] [PubMed] [Google Scholar]
- 13.Fluck CE, Miller WL. Mol Endocrinol. 2004;18:1144–1157. doi: 10.1210/me.2003-0342. [DOI] [PubMed] [Google Scholar]
- 14.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, Pieper RO. Cancer Res. 2001;61:4956–4960. [PubMed] [Google Scholar]
- 15.Lowry JA, Atchley WR. J Mol Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- 16.Kamnasaran D, Guha A. Dev Brain Res. 2005;160:90–95. doi: 10.1016/j.devbrainres.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Brewer C, Nemer G, Gove C, Rawlins F, Nemer M, Patient R, Pizzey J. Gene Expr Patterns. 2002;2:123–131. doi: 10.1016/s0925-4773(02)00302-7. [DOI] [PubMed] [Google Scholar]
- 18.Rahman NA, Kiiveri S, Siltanen S, Levallet J, Kero J, Lensu T, Wilson DB, Heikinheimo MT, Huhtaniemi IT. Reprod Biol. 2001;1:5–9. [PubMed] [Google Scholar]
- 19.Capo-chichi CD, Roland IH, Vanderveer L, Bao R, Yamagata T, Hirai H, Cohen C, Hamilton TC, Godwin AK, Xu XX. Cancer Res. 2003;63:4967–4977. [PubMed] [Google Scholar]
- 20.Nagata D, Suzuki E, Nishimatsu H, Yoshizumi M, Mano T, Walsh K, Sata M, Kakoki M, Goto A, Omata M, Hirata Y. Circ Res. 2000;87:699–704. doi: 10.1161/01.res.87.8.699. [DOI] [PubMed] [Google Scholar]
- 21.Perlman H, Suzuki E, Simonson M, Smith RC, Walsh K. J Biol Chem. 1998;273:13713–13718. doi: 10.1074/jbc.273.22.13713. [DOI] [PubMed] [Google Scholar]
- 22.Korkolopoulou P, Kouzelis K, Christodoulou P, Papanikolaou A, Thomas-Tsagli E. Acta Neuropathol (Berlin) 1998;95:617–624. doi: 10.1007/s004010050848. [DOI] [PubMed] [Google Scholar]
- 23.Bodey B, Siegel SE, Kaiser HE. In Vivo. 2006;20:511–518. [PubMed] [Google Scholar]
- 24.Latil A, Baron JC, Cussenot O, Fournier G, Soussi T, Boccon-Gibod L, Le Duc A, Rouesse J, Lidereau R. Genes Chromosomes Cancer. 1994;11:119–125. doi: 10.1002/gcc.2870110208. [DOI] [PubMed] [Google Scholar]
- 25.Chenevix-Trench G, Leary J, Kerr J, Michel J, Kefford R, Hurst T, Parsons PG, Friedlander M, Khoo SK. Oncogene. 1992;7:1059–1065. [PubMed] [Google Scholar]
- 26.Jones JW, Raval JR, Beals TF, Worsham MJ, Van Dyke DL, Esclamado RM, Wolf GT, Bradford CR, Miller T, Carey TE. Arch Otolaryngol Head Neck Surg. 1997;123:610–614. doi: 10.1001/archotol.1997.01900060052009. [DOI] [PubMed] [Google Scholar]
- 27.Bayani J, Pandita A, Squire JA. Neurosurg Focus. 2005;19:1–36. doi: 10.3171/foc.2005.19.5.2. [DOI] [PubMed] [Google Scholar]
- 28.Kotliarov Y, Steed ME, Christopher N, Walling J, Su Q, Center A, Heiss J, Rosenblum M, Mikkelsen T, Zenklusen JC, Fine HA. Cancer Res. 2006;66:9428–9436. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Colella S, Kurrer M, Yonekawa Y, Kleihues P, Ohgaki H. Cancer Res. 2000;60:6868–6874. [PubMed] [Google Scholar]
- 30.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 31.Otomo T, Hishii M, Arai H, Sato K, Sasai K. J Radiat Res (Tokyo) 2004;45:53–60. doi: 10.1269/jrr.45.53. [DOI] [PubMed] [Google Scholar]
- 32.Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 33.Qi ZY, Li Y, Ying K, Wu CQ, Tang R, Zhou ZX, Chen ZP, Hui GZ, Xie Y. J Neurooncol. 2002;56:197–208. doi: 10.1023/a:1015079705841. [DOI] [PubMed] [Google Scholar]
- 34.Stanford WL, Cohn JB, Cordes SP. Nat Rev Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 35.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Nature. 2005;14:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]