Abstract
Fruit body formation in filamentous fungi is a complex and yet hardly understood process. We show here that protein turnover control is crucial for Aspergillus nidulans development. Deletion of genes encoding COP9 signalosome (CSN) subunits 1, 2, 4, or 5 resulted in identical blocks in fruit body formation. The CSN multiprotein complex controls ubiquitin-dependent protein degradation in eukaryotes. Six CSN subunits interacted in a yeast two-hybrid analysis, and the complete eight-subunit CSN was recruited by a functional tandem affinity purification tag fusion of subunit 5 (CsnE). The tagged CsnE was unable to recruit any CSN subunit in a strain deleted for subunit 1 or subunit 4. Mutations in the JAMM metalloprotease core of CsnE resulted in mutant phenotypes identical to those of csn deletion strains. We propose that a correctly assembled CSN including a functional JAMM links protein turnover to fungal sexual development.
Keywords: development, filamentous fungi
Fungal fruit bodies are sexual reproduction structures that generate meiotic spores. The model mold Aspergillus nidulans develops a closed spherical fruit body (cleistothecium) including different tissue types: Hülle cells surround and nurse the growing cleistothecium, pericarp cells develop the protecting wall, and inner ascogenous cells mature into sexual spores (1, 2). Massive reconstruction of vegetative hyphae is required to build the complex three-dimensional fruit body. The regulation of this development is hardly understood in any fungus (3). A genetic screen recently identified csnD and csnE resembling genes for subunits of the COP9 signalosome (CSN) of animals and plants to be essential for fruit body formation of A. nidulans (4).
CSN is a multiprotein complex composed of proteins containing PCI and MPN interaction domains (5, 6). Csn5/Jab1 is the only subunit conserved in all eukaryotes, and it carries an MPN+ domain containing the JAMM motif conferring metalloprotease (deneddylation) activity (6, 7). CSN controls by its MPN+ domain the activity of cullin-RING E3 ligases by cleaving the ubiquitin-like protein Nedd8/Rub1 from the cullin (8, 9). Neddylated E3 ubiquitin ligases are key mediators of posttranslational labeling of pro-teins for the proteasome (10). The CSN thus controls eukaryotic ubiquitin-dependent protein degradation.
The complete eight-subunit CSN, composed of six PCI and two MPN domain proteins, was described for eukaryotes as humans (11), mice (12), plants (13), flies (14), and Dictyostelium (15). In fungi, definitive evidence for an eight-subunit CSN is lacking so far. CSN complex purification from Neurospora crassa revealed subunits 1–7, but subunit 8 was identified neither in the purification experiment nor in the genome sequence by bioinformatics means (16). In fission yeast subunits 6 and 8 have not been identified yet (17), and in the CSN-related complex of Saccharomyces cerevisiae only subunit Csn5 (yeast Rri1p) is well conserved (18).
The fungal CSN complexes known to date are not essential for viability but are involved in cellular processes like circadian clock regulation, cell cycle progression, and the pheromone response (16, 18, 19). In contrast, CSN dysfunction leads to severe defects in cell proliferation of Dictyostelium discoideum (15) and embryonic lethality of mice, plants, and flies (20–22), indicating a function of CSN in regulation of basal developmental processes. Here we demonstrate the existence of the first complete fungal eight-subunit CSN. In A. nidulans, CSN complex formation and a functional JAMM deneddylase motif are critical for development of fruit bodies.
Results
The A. nidulans Genome Encodes Eight Proposed CSN Subunits.
In the genomes of three aspergilli (23–25) we identified genes for 18 PCI, five MPN, and three MPN+ domain proteins. Surprisingly, we found eight subunits for CSN, which were designated csnA–H (Table 1), and which characterize Aspergillus as the first fungus where all eight subunits are clearly recognizable by bioinformatic means on the genome level. The other genes encode putative subunits of the LID of the proteasome, of translation factor eIF3, (26, 27), the AMSH-like ubiquitin isopeptidase (28), and the Prp8-like splicing factor (29) [see supporting information (SI) Table 3)]. Only A. nidulans csnD, csnE, and csnG/acoB had been previously described (4, 30). We amplified all csn cDNAs from a vegetative cDNA library (31) to verify that all eight genes are transcribed (data not shown). Intron positions and lengths were determined by comparison with the corresponding genomic sequences (Table 1). In silico analyses revealed that the composition and sequence similarity of the A. nidulans CSN more closely resembles that of humans and plants than that of yeasts.
Table 1.
The A. nidulans CSN subunits
| CSN subunit | A. nidulans protein no. | A. nidulans protein name | Domain | e value | % amino acid identities to: |
Positions of introns and PCI/MPN domain | Coding regions, bp | Protein, kDa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| hs | at | sp | sc | ||||||||
| CSN1 | AN1491 | CsnA | PCI | 2.1 e−07 | 33.3 | 38.6 | 27.0 | — |  |
1,497 | 55.7 |
| CSN2 | AN4783 | CsnB | PCI | 1.5 e−16 | 51.1 | 48.1 | 46.1 | — | 1,521 | 58.1 | |
| CSN3 | AN5798 | CsnC | PCI | 6.0 e+01 | 26.0 | 23.4 | 20.1 | — | 1,491 | 55.2 | |
| CSN4 | AN1539 | CsnD | PCI | 1.1 e−03 | 38.5 | 41.2 | 19.4 | — | 1,227 | 44.9 | |
| CSN5 | AN2129 | CsnE | MPN+ | 6.6 e−37 | 55.2 | 54.4* | 38.8 | 27.8 | 1,008 | 37.8 | |
| CSN6 | AN2233 | CsnF | MPN | 3.9 e−01 | 31.1 | 29.9* | — | — | 1,161 | 42.1 | |
| CSN7 | AN3623 | CsnG | PCI | 7.6 e−15 | 28.8* | 29.5 | 27.0 | — | 984 | 35.3 | |
| CSN8 | AN10208 | CsnH | PCI | 1.4 e+02 | 22.4 | 18.9 | — | — | 639 | 23.9 | |
CsnA–CsnH contain PCI and MPN domains (SMART-ID: SM00088/SM00232); poor E values are indicated by italics. Percentages of amino acid identities to the sequences of Homo sapiens (hs), Arabidopsis thaliana (at), Schizosaccharomyces pombe (sp), and S. cerevisiae (sc) (26) are given (sequence IDs are in SI Table 3). Positions of PCI/MPN (blue) and introns (red) within coding regions are indicated.
*Isoforms: hsCSN7B (27.8%), atCSN5B (53.2%), and atCSN6B (31.1%).
Different Δcsn Strains Share the Same Phenotypes.
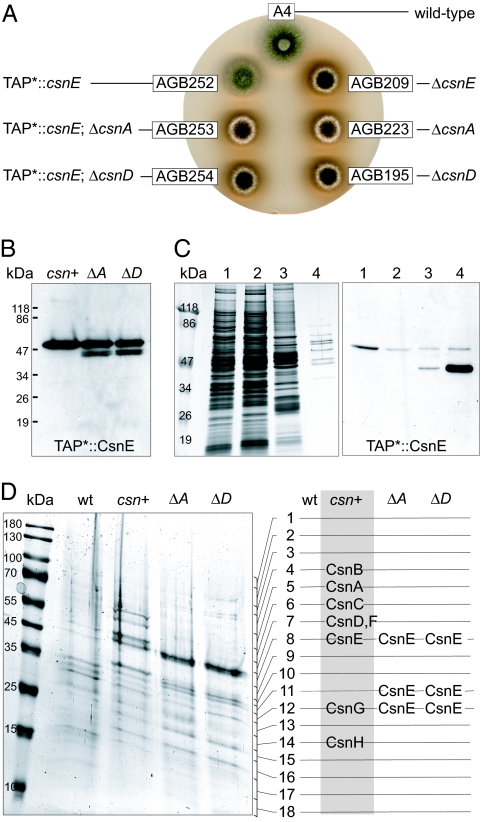
We found previously that A. nidulans strains deleted for csnD (AGB195) or csnE (AGB209) stop fruit body formation at the level of primordia and produce an aberrant red dye (4) (Fig. 1A). To survey whether the CSN mutant phenotypes are restricted to these subunits, we used the ΔcsnA strain AGB223 and deleted the complete coding sequences of csnB (strain AGB238). Additionally, we constructed a double deletion strain lacking subunits csnA and csnB (strain AGB250). The five different csn deletion strains were able to initiate fruit body formation by production of Hülle cells and primordia like the wild-type (Fig. 1B). However, further maturation of primordia was aborted, and mature cleistothecia were never observed. Therefore, like in higher eukaryotes, deletion of any csn gene resulted in early developmental defects. Additionally, the Δcsn strains produced an aberrant red color within distinct hyphae after ≈48 h of growth on an air–medium interface (data not shown). The mutant phenotypes were complemented in the Δcsn strains (Table 2) by ectopic integration of the corresponding csn wild-type alleles, respectively (data not shown).
Fig. 1.
A. nidulans Δcsn mutant phenotypes. Strains AGB223 (ΔcsnA), AGB238 (ΔcsnB), AGB195 (ΔcsnD), AGB209 (ΔcsnE), and AGB250 (ΔcsnAB) were compared with wild type (A4). (A) Maturation of primordia (p), accompanied by Hülle cells (h), includes development of the pericarp (pc) into the cleistothecium wall (cw) and of ascogenous hyphae (ah) into ascospores (as). (B) csn deletion strains developed sexual cell types on an air-limited liquid medium surface (96 h at 37°C), but not mature fruit bodies and ascospores. (Scale bars: 40 μm.)
Table 2.
A. nidulans strains
| Strain | Relevant genotype | Source/construction |
|---|---|---|
| A. nidulans wild-type and control strains | ||
| A4 | Glasgow wild type | Fungal Genetics Stock Center |
| AGB152 | pyrG98; pyroA4 | 4 |
| A. nidulans strains with csn deletions (A. fumigatus pyrG, N. crassa pyr-4) | ||
| AGB223 | ΔcsnA::zeo-pyrG-zeo; pyrG98; pyroA4 | pME2505 in AGB152 |
| AGB238 | ΔcsnB::pyrG; pyrG98; pyroA4 | pME2814 in AGB152 |
| AGB195 | ΔcsnD::pyr-4; pyrG98; pyroA4 | 4 |
| AGB209 | ΔcsnE::pyr-4; pyrG98; pyroA4 | 4 |
| AGB234 | ΔcsnA; pyrG98; pyroA4 | 5-FOA counterselection in AGB223 |
| AGB250 | ΔcsnA; ΔcsnB::pyrG; pyrG98; pyroA4 | pME2969 in AGB234 |
| A. nidulans strains with Δcsn complementation | ||
| AGB237 | ΔcsnA; PalcA::csnA::This2B::ble; pyrG98; pyroA4 | pME2937 in AGB234 |
| AGB239 | ΔcsnB::pyrG; csnB::ble; pyrG98; pyroA4 | pME2815 in AGB238 |
| AGB203 | ΔcsnD::pyr-4; csnD::ble; pyrG98; pyroA4 | 4 |
| AGB211 | ΔcsnE::pyr-4; csnE::ble; pyrG98; pyroA4 | 4 |
| A. nidulans strains for TAP tag analysis | ||
| AGB190 | PalcA::gfp::This2B::pyr-4; pyrG98; pyroA4 | pMCB32 in AGB152 |
| AGB255 | PalcA::gfp::cTAP*::This2B::pyr-4; pyrG98; pyroA4 | pME2985 in AGB152 |
| AGB256 | PalcA::nTAP*::gfp::This2B::pyr-4; pyrG98; pyroA4 | pME2986 in AGB152 |
| AGB252 | nTAP*::csnE; pyrG98; pyroA4 | pME2971 in AGB209, 5-FOA selection |
| AGB253 | nTAP*::csnE; ΔcsnA::zeo-pyrG-zeo; ΔcsnE; pyrG98; pyroA4 | pME2505 in AGB252 |
| AGB254 | nTAP*::csnE; ΔcsnD::pyr-4; ΔcsnE; pyrG98; pyroA4 | pME2342 in AGB252 |
| A. nidulans strains with mutant csnE alleles | ||
| AGB244 | csnE1::pyr-4; ΔcsnE; pyrG98; pyroA4 | pME2883 in AGB209 |
| AGB245 | csnE2::pyr-4; ΔcsnE; pyrG98; pyroA4 | pME2884 in AGB209 |
These data suggest that the integrity of the fungal CSN complex is more important for function than unique roles of individual subunits.
PCI Domain Proteins Form a Core Interaction Cluster in the Fungal CSN.
CSN complex integrity is presumably mediated by subunit interactions based on the characteristic PCI and MPN motifs (32). Therefore, we analyzed binary protein interactions of CSN subunits. A. nidulans csn cDNAs were fused reciprocally to the activation domains and DNA binding domains (DBD) of the yeast two-hybrid plasmids pEG202 and pJG4-5 (33, 34). Interactions were tested by two reporter systems based on leucine prototrophy and β-galactosidase activity (Fig. 2A). The CsnB::DBD construct conferred strong growth on its own; therefore, only the activity tests were taken into account for this construct (Fig. 2B). Six protein–protein interactions were shown by both complementary bait/prey pairs between CSN subunits A–B, A–D, B–D, D–G, E–F, and F–G, leading to a minimum binary interaction map (Fig. 2C). Subunits CsnC and CsnH, showing least overall similarities and lowest e values for PCI domain prediction (Table 1), did not interact in this experimental setup, indicating that they might need other subunits or specific posttranslational modifications for stable interactions. Our results suggest that the interaction cluster between the four PCI domain subunits A–B–D–G (1–2–4–7) forms a core that might be the basis for a stable CSN complex.
Fig. 2.
A. nidulans CSN yeast two-hybrid interactions. (A) Yeasts with all plasmid combinations of A. nidulans csn cDNAs (lanes A–H) and empty DNA binding domain (d) and activation domain (a) control vectors (0) were viable on SC plates with leu. Interactions were monitored as leu prototrophy and β-galactosidase activity. (B) Readouts of growth (red) and activity (blue) were evaluated as strong (bold +), weak (regular +), or absent (−), and combined readouts overall evaluated as positive were highlighted by gray boxes. (C) Arrows indicate a “minimum binary interaction map,” and solid lines indicate that both bait/prey combinations of a given protein pair were positive.
An Adapted Tandem Affinity Purification (TAP) Tag Enables Expression of a Functional CsnE Fusion Protein.
We wondered whether the catalytic subunit CsnE of A. nidulans is able to physically interact to all seven other subunits resulting in an eight-subunit CSN or whether we find only a reduced fungal CSN complex. The tags for the TAP method, which allows mild enrichment using IgG and calmodulin Sepharose (35), was codon-optimized for A. nidulans by site-directed mutagenesis. Expression and proper localization of the N- and C-terminal TAP tags (nTAP* and cTAP*) to csnE were validated as fusions to GFP in strains AGB255 and AGB256 by Western hybridization experiments with anti-GFP antibody and by fluorescence microscopy (data not shown).
The nTAP*-csnE fusion was integrated at its original locus as single copy into ΔcsnE strain AGB209 resulting in AGB252. Expression of the nTAP*::CsnE fusion protein was analyzed by Western experiments with the TAP*-recognizing calmodulin antibody. The expressed fusion gene fully complemented the red and acleistothecial mutant phenotypes of the parental ΔcsnE strain (Fig. 3 A and B).
Fig. 3.
CsnA and CsnD are essential for complex formation. (A) The nTAP*::csnE fusion of AGB252 (csn+) complements mutant phenotypes of AGB209. Deletion of csnA or csnD in AGB252 resulted in Δcsn phenotypes in AGB253 (ΔA) and AGB254 (ΔD). nTAP*::csnE was analyzed in 20-h-old vegetative mycelia. (B) Western experiments with anti-calmodulin and 20 μg of protein crude extract show expression of nTAP*::CsnE. (C) nTAP*::CsnE enrichment from 150 mg of crude protein of AGB252 was monitored by silver stain (50) and Western experiments using anti-calmodulin: crude extract (1), flow-through after binding to IgG (2) and calmodulin (3) beads, and final eluate (4). (D) Final eluates from 950 mg of crude proteins were separated on a gradient SDS gel and stained by EZBlue, and the complete lane was cut into 18 pieces for analysis. Only in the AGB252 eluate (gray box) were all eight CSN subunits detected by MS.
The Eight CSN Subunits of A. nidulans Form a Complex in Vivo.
Vegetative 20-h-old mycelia of A. nidulans AGB252 were used for TAP enrichment. The tag was cleaved from the full fusion protein (57.9 kDa) by TEV protease resulting in significant enrichment of the truncated (41.4 kDa) nTAP*::CsnE fusion protein (Fig. 3C).
A scaled-up enrichment procedure was applied to the nTAP*::CsnE strain (AGB252) and to the wild-type control (A4). Gel slices containing proteins of the complete final eluate were tryptically digested, and peptides were analyzed by nano-LC MS analysis. The wild-type lane contained typical “TAP contaminants” but no CSN subunits. However, all CSN subunits could be identified in the AGB252-derived eluate. The MS data were validated by using the Sequest peptide correlation coefficient (Xcorr), and the number of different peptides identified (pep) are given as best Xcorr/pep in parentheses: CsnA (4.56/11), CsnB (4.91/09), CsnC (5.61/11), CsnD (4.26/11), CsnE (5.02/04), CsnF (4.59/03), CsnG (2.39/01), and CsnH (4.09/04). The sizes of the subunits identified according to the gel slices (Fig. 3D) agreed with the calculated protein weights (Table 1). These results demonstrate the assembly of the complete eight-subunit CSN complex in A. nidulans.
Absence of CsnA or CsnD Prevents the Assembly of CsnE with Other CSN Subunits.
We questioned whether deletion of single CSN subunits affects the formation of the complex and whether we find CsnE-containing subcomplexes as described for mammals (36, 37). We therefore combined the nTAP*::CsnE fusion of AGB252 with the deletions of either csnA or csnD in strains AGB253 and AGB254, which showed the characteristic Δcsn phenotypes (Fig. 3A). They were analyzed as described above and resulted in multiple bands for the CsnE fusion protein (Fig. 3B). The expression level of CsnE did not seem to be significantly affected by the absence of CsnA or CsnD.
The TAP* enrichment procedure was applied to vegetative mycelia of strains AGB253 and AGB254. MS analysis of the final eluate identified solely the tagged CsnE (Fig. 3D), not only at the expected size (Xcorr/pep: 4.90/07 and 4.58/07) but additionally in two lower gel slices (4.97/5, 4.63/5 and 4.93/6, 4.89/5). The molecular weights of these additional CsnE forms matched the bands observed in Western experiments (Fig. 3B). The multiple CsnE bands hint at limited proteolysis of the subunit due to increased cellular exposure when the CSN complex is not formed. We did not identify any other proteins in the final eluate, and thus we observed no indications for CSN subunit assembly including CsnE, implying that deletion of single CSN subunits results in loss of the CSN complex. Nonetheless, we cannot exclude CsnE oligomers and proteins linked by weak interactions to CsnE that got lost during the purification procedure.
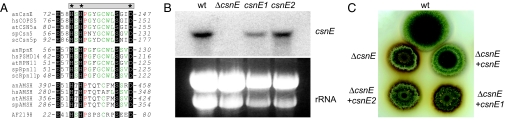
The JAMM Motif of CsnE Is Essential for Fungal Fruit Body Formation.
The MPN+ domain of CsnE spans amino acids 50–186 and includes the JAMM motif. The highly conserved JAMM active core (Fig. 4A) is the catalytic domain of the metalloprotease activity of CSN (7, 38, 39). To analyze whether csn deletion phenotypes can be attributed to the metalloprotease activity, we exchanged three of the highly conserved amino acids in the CsnE JAMM active core by site-directed mutagenesis: His-134, His-136, and Asp-147 (Fig. 4A). Two mutant csnE alleles, csnE1 (D147N) and csnE2 (H134A, H136A, D147N), were ectopically integrated into the ΔcsnE strain AGB209, resulting in strains AGB244 and AGB245, respectively. Both mutant alleles, driven by the endogenous csnE promoter, were transcribed as demonstrated in Northern experiments (Fig. 4B). In both cases, the JAMM defect caused the same block in fruit body formation as well as the accompanying phenotypes as in Δcsn strains. Thus, the CsnE-based metalloprotease is the key factor of CSN required for fungal development, and its function requires the entire eight-subunit CSN.
Fig. 4.
The JAMM motif of CsnE is essential for fruit body formation. (A) JAMM domain proteins CSN subunit 5, 26S proteasome LID subunit RPN11/PSMD14, and AMSH of A. nidulans (an), H. sapiens (hs), A. thaliana (at), S. pombe (sp), or S. cerevisiae (sc) were aligned by ClustalW (51) with the archaea JAMM protein AF2198 (38) (proteins are IDs in SI Table 3). JAMM is highlighted in black, and additional conserved residues are red (100%) or green (>40%). Residues mutated in JAMM (gray bar) are marked by asterisks. (B) csnE1 (D147N) and csnE2 (H134A, H136A, and D147N) were transcribed as shown by Northern hybridization (C) but did not rescue the ΔcsnE mutant phenotypes.
Discussion
A CSN consisting of eight subunits is typical for mammals, plants, and insects. The complex is essential for development, and defects in CSN result in embryonic death of these multicellular organisms (14, 37, 40). All reports on fungi, including N. crassa, Schizosaccharomyces pombe, and S. cerevisiae signalosomes, have so far described a signalosome of fewer than eight subunits that is involved in regulation of diverse pathways but has no significant impact on the fungal life cycle (16, 19, 41). This suggested that fungi that are highly valuable model systems for insights in numerous molecular processes in eukaryotes might not be suitable to understand CSN function in development of metazoa or higher plants. This view is challenged here, because we show that the mold A. nidulans also possesses a nonreduced eight-subunit CSN and that defects result in the abolishment of the potential to form fruit bodies. This phenotype is reminiscent of the embryonic lethality of higher eukaryotes. Therefore, the mold that is easily tractable in the laboratory and can be propagated by the asexual part of its life cycle is a promising model to learn more about CSN and its exact role in development. It might also be interesting to analyze CSN in other representatives of the fungal kingdom, when numerous fungal genomes that are currently annotated will be released.
The fifth subunit (CsnE/Jab1) of CSN includes the only known enzyme activity of the complex, a metalloprotease that binds zinc ions by its conserved JAMM motif. JAMM-derived metalloprotease activity is not restricted to multiprotein complexes, because an “orphan JAMM” like the endosome-associated AMSH also functions as a deubiquitinase (28). Small CSN subcomplexes including subunit 5 have been isolated (36, 37), but it seems that the entire complex is a prerequisite for deneddylase function of CsnE/Jab1 (40). In addition, large supercomplexes consisting of CSN, the LID of the proteasome, and cullin-RING E3 ligases have been described (42, 43). Rpn11 is the counterpart of CsnE/Jab1 in the LID and provides JAMM-mediated deubiquitylation activity. Similar to CsnE/Jab1, Rpn11 also requires complex formation to function as an enzyme (6, 44, 45). In the fungal system, CsnE/Jab1 is unable to recruit any other CSN subunit and is less stable when CSN subunits 1 or 4 are absent. This argues against stable CsnE subcomplexes in fungi, although it does not exclude the formation of CsnE oligomers or unstable CsnE-containing subcomplexes. The CsnE binary interaction with subunit 6 in the yeast two-hybrid analysis does not seem to be sufficient to allow copurification. The strong interactions between CSN subunits 1, 2, 4, and 7 in the yeast two-hybrid system hint at a role of these PCI domain proteins as a core for fungal CSN assembly. A. nidulans CsnE does not recruit non-CSN proteins at detectable amounts. Therefore, we have no evidence of CSN–LID–eIF3 supercomplexes or mixed complexes of CsnE with other associated proteins in fungi. In agreement with the same mutant phenotypes of different csn deletion strains and a JAMM motif mutant strain, these data suggest that primarily the deneddylase function of CSN is crucial for fungal sexual development. It remains to be seen whether there are subcomplexes of the fungal CSN that do not include the CsnE subunit or whether there are additional proteins that show weak interactions to the CSN complex.
The A. nidulans genome (23) reveals that all major genes for putative substrates of the CSN deneddylase activity are present. These include a proposed Nedd8 encoding sequence (AN6179) and the three putative cullins Cul1 (AN1019; CulA), Cul3 (AN3939; CulC), und Cul4 (AN10008; CulD). Cullins are part of cullin-RING E3 ligase cores and are known to assemble with various specific substrate-adaptors like F-box proteins (10). Interestingly, in a screen for A. nidulans strains defective in fruit body formation (4) we identified a mutant with insertion in the promoter region of CulA (unpublished data). We also showed that the A. nidulans F-box protein GrrA is specifically involved in sexual development (46). Therefore, the control of specific E3 ubiquitin ligases by CSN seems to be less critical for hyphal growth itself than for fungal developmental processes where hyphae have to be reconstructed to allow the maturation of fruit bodies. This suggests that the importance of a stringent control of ubiquitin-dependent protein degradation is less critical for the lifestyle of a modular multicellular form (hyphae) that grows by the repeated iteration of modules than for a unitary form with a determinate form (cleistothecium). This might be one reason why CSN is primarily important for organisms that exclusively live in a determinate form as unitary organisms.
We are confident that future analyses of the role of the fungal CSN in fruit body formation will provide further insights into specific functions of CSN and its link between development and degradation in multicellular organisms.
Materials and Methods
Strains are given in Table 2. Sequence IDs, plasmids, primer sequences, and recipes are listed in SI Tables 3 and 4.
A. nidulans Growth and Physiological Studies.
Strains were grown with 1 × 106 spores per milliliter in minimal medium at 37°C, and development was induced and monitored as described (4).
Computational and Molecular Methods.
BLAST analyses were performed at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov), aligned by ClustalW at Network Protein Sequence Analysis (http://npsa-pbil.ibcp.fr), and searched for PCI and MPN domains at the European Molecular Biology Laboratory (http://smart.embl-heidelberg.de). Sequences were analyzed at the Göttingen Genomics Laboratory. Standard molecular methods were applied as described by the Cold Spring Harbor Protocols. A. nidulans material was treated as described (4). For Western hybridization experiments, primary antibodies were mouse anti-GFP (Clontech–Takara Bio Europe, Saint-Germain-en-Laye, France), rabbit anti-calmodulin binding protein epitope tag (Upstate/Milliore, Schwalbach, Germany), and rabbit anti-actin C11 peptide (Sigma–Aldrich, Steinheim, Germany). Peroxidase-coupled secondary antibodies were anti-mouse and anti-rabbit IgG antibodies (Invitrogen/Molecular Probes, Karlsruhe, Germany).
Cloning of csn Genes.
Sequences were amplified by PCR from a vegetative A. nidulans cDNA library (31) and from genomic DNA and cloned into pBluescript II SK+ (Stratagene, La Jolla, CA): csnA (pME2987 and pME3204), csnB (pME2988 and pME3205), csnC (pME2989 and pME3206), csnD (pME2990 and pME3207), csnE (pME2991 and pME3208), csnF (pME2992 and pME3209), csnG (pME2993 and pME3210), and csnH (pME2994 and pME3211), respectively.
Generation of csn Deletion Strains.
Csn genes were deleted in AGB152 by homologous integration of cassettes that contained the csn 5′ and 3′ regions amplified by PCR with specific primer pairs from genomic DNA, flanking either the Aspergillus fumigatus pyrG of pME2409 (31) or the N. crassa pyr-4 of pRG3 (47). We constructed plasmid pME2814 (PcsnB::pyrG::TcsnB) and used pME2505 (PcsnA::pyrG::TcsnA), pME2342, and pME2369 (4). For complementation, csn wild-type loci were cloned into pME1510 or pME1565 (4) as plasmids pME2815 (PgpdA::ble::TtrpC, csnB), pME2937 (PalcA::csnA::This2B, pyr-4), pME2345, and pME2423 (4) and ectopically integrated into the csn deletion strains.
Two-Hybrid Analysis.
Csn cDNAs in pEG202 and pJG4–5 (33, 34) resulted in bait and prey constructs for csnA (pME2502 and pME2501), csnB (pME2972 and pME2978), csnC (pME2973 and pME2979), csnD (pME2355 and pME2357), csnE (pME2974 and pME2980), csnF (pME2975 and pME2981), csnG (pME2976 and pME2982), and csnH (pME2977 and pME2983). After transformation into S. cerevisiae EGY48-p1840, leucine prototrophy and β-galactosidase activity were analyzed (33) with 10-μl liquid cultures of OD546 = 0.01.
A. nidulans TAP* Tag.
Yeast TAP tags of pBS1479 and pBS1761 (35) in pGEM5 (Promega, Madison, WI) were optimized to A. nidulans codon usage (www.kazusa.or.jp/codon) by site-directed mutagenesis with the QuikChange Kit (Stratagene). Thirteen codons were changed in cTAP* (pME2967) and nTAP* (pME2968) (SI Table 4). As control, TAP* tags were fused to gfp of pMCB32 (48), resulting in pME2985 (PalcA::gfp::cTAP*::This2B, pyr-4) and pME2986 (PalcA::nTAP*::gfp::This2B, pyr-4), and ectopically integrated into AGB152.
The nTAP* of pME2968 was inserted into csnE of pME2237 (4), and the resulting pME2971 (PcsnE::nTAP*::csnE:TcsnE) was homologously integrated into AGB209 by counterselection against pyr-4 with 5-FOA. In the resulting strain AGB252, csnA and csnD were deleted with plasmids pME2505 (AGB253) and pME2342 (AGB254), respectively.
For nTAP*::csnE enrichment, 950 mg of total proteins was extracted from powdered 20-h-old vegetative mycelia of strains AGB252, AGB253, and AGB254 in fresh B* buffer and cleared twice (15 min at 5,000 × g and 10 min at 12,000 × g). The soluble protein fraction was incubated for 30 min on a rotator with 500 μl of Sepharose beads (Sepharose 6B; Sigma–Aldrich) in 50-ml Falcon tubes, spun down for 5 min at 1,104 × g, applied to fresh Falcon tubes, and incubated for 4 h on a rotator with 250 μl of IgG Sepharose 6 Fast Flow (Amersham Biosciences, Buckinghamshire, U.K.). Then, the National Center for Research Resources (University of Washington, Seattle, WA) protocol (step 14) was followed, except that no pump was used, cleaving was performed with 300 units of AcTEV, and the complete final eluate was suspended in SDS gel sample buffer.
In-Gel Digestion and Nano-ESI-MS.
Excised gel pieces were tryptically digested (49), and peptides were separated by water–acetonitrile gradients on Dionex-NAN75-15-03-C18-PM columns on an ultimate-nano-HPLC system (Dionex, Sunnyvale, CA). Online ESI-MS/MS2 spectra were generated on a LCQ-DecaXPplus mass spectrometer (ThermoFinnegan, San Jose, CA). Proteins were analyzed by MS2 spectra with SEQUEST/TURBOSEQUEST (Biowork Browser 3.1; ThermoFinnegan) by using the Broad A. nidulans genome database.
Mutant csnE Alleles.
Site-directed mutagenesis of csnE in pME2237 (4) was performed by using the QuikChange kit (Stratagene). The codons for His-134, His-136, and Asp-147 were substituted for Ala and Asn, respectively. The resulting mutant alleles csnE1 (H134A) and csnE2 (H134A, H136A, and D147N) were cloned BamHI/EcoRI into pME1510 (4), and resulting pME2883 and pME2884 were ectopically integrated as one copy into the A. nidulans csnE deletion strain AGB209.
Acknowledgments
We thank all members of the Department for Microbiology and Genetics, University of Göttingen, for discussions, and we value the initial work of L. Kozaczkiewicz. Funding was received from the Deutsche Forschungsgemeinschaft, the Research Center for Molecular Physiology of the Brain, the Volkswagen Stiftung, and the Fonds der Chemischen Industrie.
Abbreviation
- CSN
COP9 signalosome.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702108104/DC1.
References
- 1.Champe SP, Nagle DL, Yager LN. In: Aspergillus: 50 Years On, Progress in Industrial Microbiology. Martinelli SD, Kinghorn JR, editors. Vol 29. Amsterdam: Elsevier; 1994. pp. 429–454. [Google Scholar]
- 2.Sohn KT, Yoon KS. Mycobiology. 2002;30:117–127. [Google Scholar]
- 3.Braus GH, Krappmann S, Eckert SE. In: Molecular Biology of Fungal Development. Osiewacz HD, editor. New York: Dekker; 2002. pp. 215–244. [Google Scholar]
- 4.Busch S, Eckert SE, Krappmann S, Braus GH. Mol Microbiol. 2003;49:717–730. doi: 10.1046/j.1365-2958.2003.03612.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim T, Hofmann K, von Arnim AG, Chamovitz DA. Trends Plants Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- 6.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 8.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di X, Wang X, Rodriguez-Suarez RJ, Zhang H, Wei N. Curr Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 10.Petroski MD, Deshaies RJ. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 11.Hetfeld BK, Bech-Otschir D, Dubiel W. Methods Enzymol. 2005;398:481–491. doi: 10.1016/S0076-6879(05)98039-7. [DOI] [PubMed] [Google Scholar]
- 12.Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. Curr Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]
- 13.Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 14.Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, Chamovitz DA. Development (Cambridge, UK) 2002;129:4399–4409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- 15.Rosel D, Kimmel AR. Eur J Cell Biol. 2006;85:1023–1034. doi: 10.1016/j.ejcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 16.He Q, Cheng P, He Q, Liu Y. Genes Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mundt KE, Liu C, Carr AM. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maytal-Kivity V, Pick E, Piran R, Hofmann K, Glickman MH. Int J Biochem Cell Biol. 2003;35:706–715. doi: 10.1016/s1357-2725(02)00378-3. [DOI] [PubMed] [Google Scholar]
- 19.Mundt KE, Porte J, Murray JM, Brikos C, Christensen PU, Caspari T, Hagan IM, Millar JB, Simanis V, Hofmann K, Carr AM. Curr Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- 20.Castle LA, Meinke DW. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- 22.Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N. Mol Cell Biol. 2003;23:6790–6797. doi: 10.1128/MCB.23.19.6790-6797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 24.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 25.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 26.Scheel H, Hofmann K. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCullough J, Clague MJ, Urbe S. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis C, Champe SP. Microbiology. 1995;141:1821–1828. doi: 10.1099/13500872-141-8-1821. [DOI] [PubMed] [Google Scholar]
- 31.Krappmann S, Braus GH. Mol Genet Genomics. 2003;268:675–683. doi: 10.1007/s00438-002-0789-8. [DOI] [PubMed] [Google Scholar]
- 32.Kapelari B, Bech-Otschir D, Hegerl R, Schade R, Dumdey R, Dubiel W. J Mol Biol. 2000;300:1169–1178. doi: 10.1006/jmbi.2000.3912. [DOI] [PubMed] [Google Scholar]
- 33.Golemis E, Brent R. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmann JG, Smith AJ, Struhl K, editors. Vol 3. New York: Wiley; 1996. pp. 429–454. [Google Scholar]
- 34.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 35.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 36.Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N. FEBS Lett. 2005;579:1047–1054. doi: 10.1016/j.febslet.2004.12.076. [DOI] [PubMed] [Google Scholar]
- 37.Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. J Biol Chem. 2004;279:43013–43018. doi: 10.1074/jbc.M406559200. [DOI] [PubMed] [Google Scholar]
- 38.Ambroggio XI, Rees DC, Deshaies RJ. PLoS Biol. 2004;2:E2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gusmaroli G, Feng S, Deng XW. Plant Cell. 2004;16:2984–3001. doi: 10.1105/tpc.104.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dohmann EM, Kuhnle C, Schwechheimer C. Plant Cell. 2005;17:1967–1978. doi: 10.1105/tpc.105.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maytal-Kivity V, Piran R, Pick E, Hofmann K, Glickman MH. EMBO Rep. 2002;3:1215–1221. doi: 10.1093/embo-reports/kvf235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X, Hetfeld BK, Seifert U, Kahne T, Kloetzel PM, Naumann M, Bech-Otschir D, Dubiel W. FEBS J. 2005;272:3909–3917. doi: 10.1111/j.1742-4658.2005.04807.x. [DOI] [PubMed] [Google Scholar]
- 43.Peng Z, Shen Y, Feng S, Wang X, Chitteti BN, Vierstra RD, Deng XW. Curr Biol. 2003;13:R504–R505. doi: 10.1016/s0960-9822(03)00439-1. [DOI] [PubMed] [Google Scholar]
- 44.Lundgren J, Masson P, Realini CA, Young P. Mol Cell Biol. 2003;23:5320–5330. doi: 10.1128/MCB.23.15.5320-5330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, III, Koonin EV, Deshaies RJ. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 46.Krappmann S, Jung N, Medic B, Busch S, Prade RA, Braus GH. Mol Microbiol. 2006;61:76–88. doi: 10.1111/j.1365-2958.2006.05215.x. [DOI] [PubMed] [Google Scholar]
- 47.Waring RB, May GS, Morris NR. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 50.Blum H, Beier H, Gross HJ. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 51.Combet C, Blanchet C, Geourjon C, Deleage G. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]