Abstract
The metabolically versatile Gram-negative bacterium Pseudomonas aeruginosa inhabits terrestrial, aquatic, animal-, human-, and plant-host-associated environments and is an important causative agent of nosocomial infections, particularly in intensive-care units. The population genetics of P. aeruginosa was investigated by an approach that is generally applicable to the rapid, robust, and informative genotyping of bacteria. DNA, amplified from the bacterial colony by circles of multiplex primer extension, is hybridized onto a microarray to yield an electronically portable binary multimarker genotype that represents the core genome by single nucleotide polymorphisms and the accessory genome by markers of genomic islets and islands. The 240 typed P. aeruginosa strains of diverse habitats and geographic origin segregated into two large nonoverlapping clusters and 45 isolated clonal complexes with few or no partners. The majority of strains belonged to few dominant clones widespread in disease and environmental habitats. The most frequent genotype was represented by the sequenced strain PA14. Core and accessory genome were found to be nonrandomly assembled in P. aeruginosa. Individual clones preferred a specific repertoire of accessory segments. Even the most promiscuous genomic island, pKLC102, had integrated preferentially into a subset of clones. Moreover, some physically distant loci of the core genome, including oriC, showed nonrandom associations of genotypes, whereas other segments in between were freely recombining. Thus, the P. aeruginosa genome is made up of clone-typical segments in core and accessory genome and of blocks in the core with unrestricted gene flow in the population.
Keywords: bacterial evolution, chip technology, population genetics
Pseudomonas aeruginosa is a metabolically versatile Gram-negative bacterium, which inhabits terrestrial, aquatic, animal-, human-, and plant-host-associated environments (1). This opportunistic pathogen is the most dominant bacterium causing chronic infections in the cystic fibrosis (CF) lung (2) and has emerged as an important causative agent of nosocomial infections, particularly in intensive-care units (3).
The P. aeruginosa genome is a mosaic of a conserved core and variable accessory segments (4). The core genome is characterized by a conserved synteny of genes, a low average nucleotide divergence of 0.5%, and multiple alleles at a few loci that are subject to diversifying selection (4–6). The accessory genome consists of a variable set of genomic islets and genomic islands, most of which belong to an ancient tRNA-integrated island type (4, 7,9–11). Genome size ranges from 5.2 to 7 Mbp in the P. aeruginosa population (4).
Typing informative traits allows identification of bacterial isolates to the strain level and provides basic information about the evolutionary biology, population biology, taxonomy, ecology, and genetics of bacteria. Typically, strains of bacteria, including P. aeruginosa, have been differentiated on the basis of specific phenotypic traits or anonymous genome fingerprinting techniques such as (macro)restriction fragment-pattern analysis or PCR-based generation of multiple differently sized DNA amplicons (12). These typing schemes, however, are poorly portable because they index variation that is difficult to compare among laboratories. In contrast, multilocus sequence typing (MLST), which uses nucleotide sequence data of internal fragments of, typically, seven housekeeping genes, is a portable, universal, and definitive method for characterizing bacteria (13, 14). MLST schemes and strain databases have been published for more than 30 species (14), including two for P. aeruginosa (15, 16). One should note, however, that MLST only scans the genetic diversity of the core genome and relies on the comparably expensive and slow DNA-sequencing technology of purified PCR products, which still cannot be performed under point-of-care or rural field conditions.
Here, we report on an approach for the rapid, inexpensive, robust, and informative genotyping of bacteria that was developed for the typing of P. aeruginosa strains in both the conserved core and flexible accessory genome. Labeled DNA is directly generated from the bacterial colony by cycles of multiplex primer extension reactions and then hybridized onto a microarray to yield an electronically portable 58-binary marker genotype. We typed a representative strain collection of diverse habitats and geographic origin to describe the global population structure of P. aeruginosa. The data indicate that the majority of P. aeruginosa strains belong to a few dominant clones widespread in disease and environmental habitats. Most studied loci of the genome are freely recombining with each other, but some physically distant loci exist in fixed combinations of genotypes, suggesting that the free flow of genes in the P. aeruginosa population is tolerated for most, but not all, loci of the genome.
Results and Discussion
A Portable Method for the Genotyping of Bacteria: Design and Optimization of Device and Protocol for the Typing of P. aeruginosa Strains.
Selection of markers.
The genotype of the core genome of a P. aeruginosa strain was represented by 13 single-nucleotide polymorphisms (SNPs) at seven conserved loci (17) and two multiallelic loci [flagellin fliC (18) and pyoverdine receptor fpvA (6)]. The selected SNPs are evenly distributed on the P. aeruginosa PAO1 chromosome (19) and are informative with a frequency of ≥15% for the rarer allele in the P. aeruginosa population (17). Assuming linkage equilibrium between the loci (15, 20), the multilocus SNP genotype was predicted (17) to differentiate between unrelated clones with a false-positive rate of <0.01%. The 16 binary SNP genotypes were represented by a four-digit hexadecimal code. The 16 SNPs were divided into four groups of four SNPs each, and the 16 possible combinations in each group were differentiated by 16 characters: 0–9, A–F. The conversion of the SNP pattern to the hexadecimal code is explained in detail in the supporting information (SI) Appendix.
The composition of the accessory genome was tested with a set of 38 genetic markers that identify 10 genomic islets (8, 21) and six types of genomic islands (4, 7, 9–11) whereby subtypes of the latter were differentiated by markers that are variably present in the respective genomic island. The localization of the markers on the microarray is shown in SI Fig. 7.
Outline of the protocol.
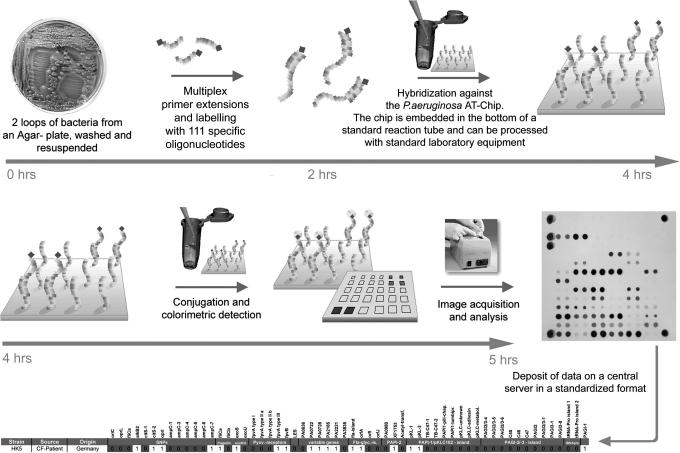
For the outline of the protocol, see Fig. 1.
Fig. 1.
Flow chart of P. aeruginosa genotyping with the low-resolution microarray.
Preparation of targets.
All 58 targets for hybridization are amplified from P. aeruginosa colony DNA by cycles of multiplex primer extension reactions with Therminator-DNA-polymerase (22) whereby the nascent strands are randomly labeled by incorporation of biotin-16-dUTP. Primer design was guided by in silico predictions of matching hybridization kinetics (23). All primers (see SI Table 1) had CC, GC, CG, or GG at the 3′ end, comparable length (20- to 22-mers), and identical free enthalpy of duplex formation (theoretical value for 1 M: melting temperature Tm = 385.75 ± 1.21 K). Multiplex primer extension reactions were performed with two nested primers in each cycle. Two-versus-one primer for each target increased and synchronized the yield of biotinylated product for all markers, probably because the temperature-sensitive strand displacement activity of the thermophilic DNA polymerase is low at the annealing temperature but high at the elongation temperature (24). The binding sites of the two nested primers terminated at a median genomic distance of 30 bp (range 1–87 bp) and 61 bp (range 11–209 bp) upstream of the 5′ end of the binding site of the hybridization probe. Further critical issues that ensured the successful multiplex amplification of all markers in equal yields are a molar ratio of dTTP:biotin-16-dUTP of 3:1 for target labeling and the choice of a DNA polymerase that tolerates the bulky biotin-16-dUTP as substrate (22).
Hybridization and detection.
The multiplex amplificate is hybridized under high stringency with the oligonucleotide microarray of target sequences (see SI Figs. 7–9 and Table 1) that is inserted into the tip of a standard Eppendorf-like microtube. This design allows all hybridization and analysis procedures to be performed with standard laboratory equipment. The hybridization signals are detected by colorimetry with streptavidin-horseradish peroxidase conjugate and tetramethylbenzidine. The hybridization signals are automatically converted to the multilocus genotype either on site or by a Web-based server that compares the genotype with all other entries in the database.
A step-by-step instruction manual for array-tube genotyping of P. aeruginosa is provided in SI Materials and Methods.
Pilot Studies of Microarray Analysis.
During the initial phase of array development, the time course of the colorimetric signal was monitored to optimize the probe and primer sequences for the absence of false-negative signals, high signal-to-noise ratios, and comparable kinetics of signal intensity for all targets (see SI Table 2). Of the primarily selected targets, nine markers were not incorporated into the final fourth-generation array.
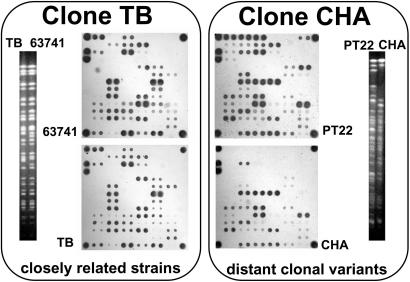
When the final protocol for microarray genotyping had been established (see SI Materials and Methods), we tested its performance, reliability, and robustness at five partner institutions in one-day practical workshops. Test strains were consistently typed at all sites. At our home laboratory, we first typed a collection of 71 P. aeruginosa isolates of diverse habitats and geographical origins that had been analyzed before in their genomic SpeI macrorestriction pattern and in the same 13 SNPs that had been selected for the chip (17). Genotyping with the microarray or the restriction digestions of individual PCR products produced matching SNP patterns for all 71 strains, indicating that the multiplex amplification from bacterial colonies and the subsequent hybridization of the amplificate on the microarray yielded unequivocal signals for all SNP alleles. Ten SNP genotypes were found in two or more strains having different SpeI genotypes. In all cases, the chip genotyping of these strain pairs or trios uncovered an identical marker genotype of the core genome but a different composition of the accessory genome. Fig. 2 illustrates the extremes of minor and major intraclonal differences detected in isolates of two abundant clones. Fig. 2 Left shows two highly related clone-TB strains that differ in just one marker for one phage-related genomic islet. The two distantly related clone-CHA strains (Right) are divergent in genome size and SpeI fragment pattern because several genomic islands are present in one strain but absent from the other.
Fig. 2.
Pulsed-field gel electrophoresis-separated SpeI fragment and microarray hybridization patterns of two closely related strains of the TB clone [Left; isolates from burn wound and CF lungs (29)] and of two distantly related strains of the CHA clone [Right; isolates from river and CF lungs (30)]. Clonal variants share identical SNP genotypes in the core genome (lower part of the array), but they differ in either few (Left) or many (Right) markers of the accessory genome.
In the second pilot study, we typed P. aeruginosa isolates that had been collected from 55 CF twin and sibling pairs at 35 CF centers in 11 European countries (25). CF siblings are known to share highly related strains in the family setting (26). Hence, this panel of strains seemed to be appropriate to compare the performance of microarray genotyping with that of SpeI macrorestriction analysis, the current gold standard of the genotyping of P. aeruginosa (4), to recognize highly related strains. All strains were consistently classified in their genetic relatedness by SpeI and microarray genotypes. Twenty-two of the 28 cocolonized patient pairs shared clones, verifying the single CF center data (26) that CF siblings in a family are prone to cross-infection. Six of the total 52 clones were found in two or more CF clinics in Europe, leading to the tentative conclusion that several major clones may be abundant in the CF patient population.
Dominant Clones in the P. aeruginosa Population.
This finding led to the hypothesis that dominant clones are a general feature of the P. aeruginosa population. To address this issue, a collection of 240 strains from diverse habitats and geographical origins was genotyped with the microarray. These strains had been isolated during the last 60 years. The collection consisted of 20 environmental isolates from Japan, North America, and Europe; 136 CF isolates from 40 CF clinics in Europe; 58 isolates from 13 intensive-care units in six countries; 23 isolates from other human infections sampled at seven hospitals; and three ATCC strains of unknown origin (see SI Table 3). Genotyping identified 103 clones, 38 of which were found more than once. The most frequent genotype was determined in 13 strains, including the completely sequenced reference strain, PA14 (27). Further common clones were C (7), K (11), M (28), TB (29), CHA (30), and the Midlands and Liverpool epidemic strains (31) (see SI Table 3). Primary microarray hybridization data of the most frequent clones are shown in SI Fig. 9.
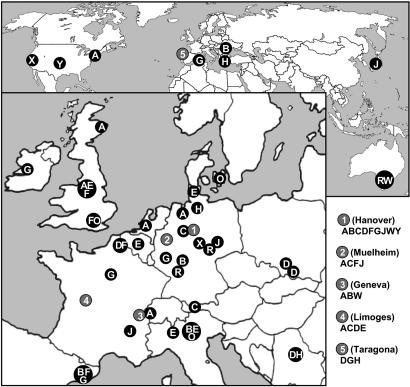
The 16 most common clones made up half of the strain panel and were found to be widespread in diverse habitats and locations. The PA14 clone, first described in the United States (27), was isolated at numerous locations in Europe from patients and aquatic habitats (Fig. 3). Further examples for transcontinental abundance are: clone CHA, isolated from soil in Japan, rivers in Germany, and several CF patients in Central Europe; the sequenced reference strain, PAO (19), which had originally been isolated >50 years ago in Melbourne and was reisolated recently in Swiss and German hospitals from tracheal secretions; and two clones designated X and Y in Fig. 3, which were retrieved from clinical and polluted environmental habitats in the United States and in hospitals and patients in Germany, respectively. Hence, the data shown in Fig. 3 and SI Table 3 indicate that major clones are just as versatile in their habitat and geographic origin as the whole species. This conclusion is in accordance with phenotypic data that isolates from the inanimate environment and clinical habitats share the same chemotaxonomic profile (32) and repertoire of metabolic and virulence traits (33). Irrespective of their origins, isolates from diseases and the environment were similarly proficient in the degradation of environmental pollutants and the secretion of virulence factors (33).
Fig. 3.
Widespread geographic distribution of major P. aeruginosa clones. Clones are depicted by uppercase letters and are arranged by decreasing frequency in alphabetical order. Source and origin of strains are listed in SI Table 3. The most abundant clone, A, is represented by the sequenced strain PA14 (27).
If the major clones are widespread in clinical and environmental habitats, they should be detected in representative samples from a single habitat. A recent field study (34) has indeed demonstrated that the local P. aeruginosa community in a small river was almost as diverse as the global population and harbored clones that were also represented among 73 clinical and environmental isolates collected across the world (35).
CF and Non-CF Isolates Differ in Their Allele Distributions at the Pyoverdine Locus and in pKLC102-Type Islands.
If the whole P. aeruginosa population behaves like the major abundant clones, there should be no habitat- or disease-associated clones. We tested this hypothesis by comparing the marker allele frequencies in non-CF and CF isolates. CF lungs were the only habitat represented by a sufficiently large number of strains in our panel to allow a meaningful statistical analysis. The allele distribution was statistically indistinguishable for 50 of 58 markers, including all housekeeping genes. All eight markers with a CF-specific frequency map to hypervariable regions of the P. aeruginosa genome (4), i.e., to the pyoverdine locus (5, 6) and the regions 3′ of the two identical tRNALys genes adjacent to PAO1 homologs, PA0976 and PA4541 (10, 11, 36).
Pyoverdines are iron(III) chelators and signal molecules for the production of virulence factors (37). Pyoverdine outer membrane type I FpvA receptors dominated in the global population, but isolates from CF lungs carried all three FpvA receptor types with similar frequency (P < 0.01). Pyoverdine-negative mutants accumulate in CF lungs with colonization time but retain the capacity to take up the siderophore pyoverdine (38), and type II and type III strains can also use type I pyoverdine (39). The pyoverdine region is the most divergent region of the core genome, and FpvA is the only known case of positive Darwinian selection in P. aeruginosa with substantial intratype variation (4). The pyoverdine region is subject to speciation and coevolution (6) hence, habitat-specific microevolution of this locus in CF lungs appears plausible.
Four markers that are diagnostic for subtypes of tRNALys-integrated genomic islands were significantly (P < 0.01) underrepresented in CF isolates. The marker genes were selected from pKLC102 (11) and from genomic islands adjacent to PA0976 in the sequenced strains PAO1 (19) and PA14 (10). The two latter islands probably have originated from a single ancestral mobile pKLC102-related genetic element but then underwent numerous alterations, including deletions, inversions, and acquisition of additional insertion sequences and transposons, leading to pronounced genomic diversity of this region (36). Given that other pKLC102-related markers were detected with similar frequency in CF and non-CF isolates (SI Table 3), pKLC102-type islands (11, 40) apparently undergo purifying selection in the CF respiratory tract.
P. aeruginosa Population Structure.
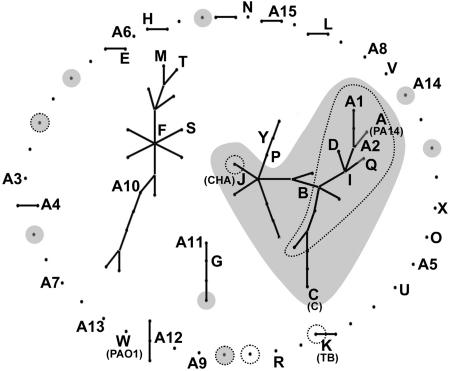
Bacteria can have population structures ranging from the fully sexual to the highly clonal. Two independent MLST studies on unrelated strain collections had consistently calculated a low index of association of 0.29 (15) and 0.31 (20), indicating that P. aeruginosa has a nonclonal population structure. The eBurst analysis (14, 41) of our significantly larger dataset of markers and clonal complexes uncovered a more differentiated structure of the P. aeruginosa population (Fig. 4). The 240 typed strains segregated into two large nonoverlapping clusters and 45 isolated clonal complexes with few or no partners. Seven of the 16 most common clones are members of the largest cluster, including the four most abundant clones (see SI Table 3).
Fig. 4.
Clonal complex structures of 240 P. aeruginosa strains collected from diverse habitats and geographic origins (see SI Table 3). Clonal complexes were calculated from the 15-marker genotype of the core genome by the eBurst algorithm (41). Clones represented by two or more isolates in the strain panel are depicted by uppercase letters and are sorted by decreasing frequency in alphabetical order (see SI Table 3). Clones shaded in gray harbor oriC allele 1, and clones encircled by dots are exoU-positive.
The exoS and exoU genes encoding type III secretion virulence effector proteins occur in a mutually exclusive manner in the P. aeruginosa genome (21), and interestingly, most exoU-positive clones are members of the major cluster (Fig. 4). The basis for the incompatibility of exoU and exoS within the same P. aeruginosa genome is not known, but comparative sequence analysis has suggested that exoU was incorporated by horizontal gene transfer followed by a targeted deletion of exoS (36). Consistent with this proposal, exoU is found in tRNALys-integrated genomic islands. Most exoU-positive strains (encircled by dots in Fig. 4) belong to the major clade of P. aeruginosa with a clonal population structure, and all but two exoU strains were harboring the oriC allele 1 (shaded in gray in Fig. 4). In contrast, the exoS strains carried either one of the two oriC alleles.
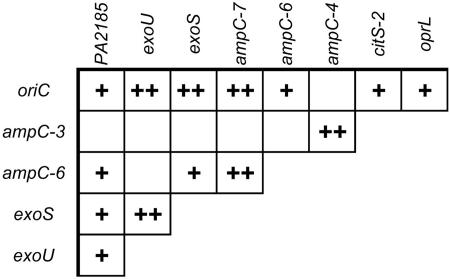
This almost absolute linkage between oriC and the >1-Mbp distant exoU urged us to search for further nonrandom associations between the 23 markers of the PAO1 genome represented on the chip. Besides the linkage disequilibrium between the adjacent SNPs 3 and 4, and 6 and 7 in the ampC gene, 12 nonrandom allelic associations between physically distant loci were detected (P < 0.05 after Bonferroni correction for multiple testing; Fig. 5). Absolute linkage was seen between oriC and ampC-7. In the other 11 cases, the distribution of two-marker haplotypes was significantly different from a random combination of marker alleles expected for linkage equilibrium. In particular, the oriC allele 1 was typically found in genomes with allele 0 in ampC-7 (PA4110), allele 0 in citS2 (PA0795), allele 1 in oprL (PA0973), and the absence of PA2185. The latter gene is part of a genomic islet upstream of exoY (PA2191) encoding the type III secretion-effector protein exoenzyme Y (21). PA2185 is typically present in exoS strains and absent from exoU strains, i.e., the genomic environment of exoY is different in exoU and exoS strains.
Fig. 5.
Nonrandom association of genetic variants in the P. aeruginosa genome. Fourteen two-marker combinations showed significant over- or underrepresentation of marker allele haplotypes (+, P < 0.05; ++, P < 0.01 after Bonferroni correction for multiple testing). The SNPs amp-3, amp-4, amp-6, and amp-7 are located within the ampC gene, but the other six genes are evenly distributed on the chromosome (see Fig. 6).
This nonrandom association of genetic variants over large genomic distances suggests that within the chromosome, a backbone of blocks is conserved and only infrequently disrupted by recombination. These conserved blocks are in linkage disequilibrium and define lineages. In contrast, genomic segments in between these blocks are freely recombining and are in linkage equilibrium with each other.
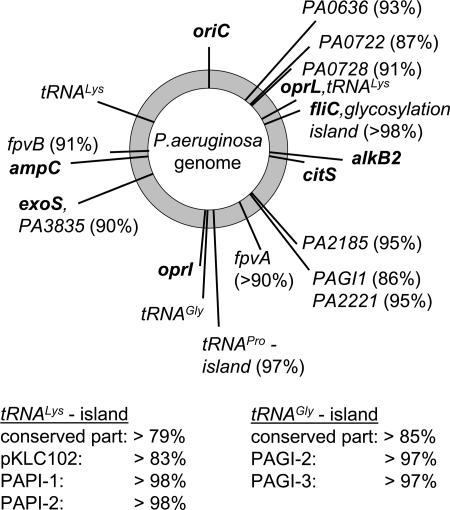
Consistent with the nonrandom association of alleles in parts of the core genome, the components of the accessory genome were also found to be nonrandomly distributed among the clonal complexes of P. aeruginosa. Clones were scored to either carry or lack the marker for a specific genomic islet or island. Extreme values of 100% and 50% would indicate absolute and random associations between the SNP genotype of the core genome and the respective element of the accessory genome. Fig. 6 shows that the markers of all analyzed islets and islands are nonrandomly associated with the genotype of the core genome. Each clone in P. aeruginosa is characterized by a specific repertoire of its accessory genome. The association is strong for all genomic islets with values ≥90%. Exact copies of large genomic islands such as PAPI-1 (10) or PAGI-2 (7) are even clone specific. The mobile genomic island pKLC102 (11) was the only exception. pKLC102 integrated into a broad range of chromosomal hosts but still showed significant preference for a subset of clones. Genomic islands of the pKLC102/PAGI-2-type carry strain-specific ORFs and a conserved syntenic set of ORFs, the majority of which are conserved hypotheticals of unknown function or related to DNA replication or mobility genes (40). Genes of this conserved part are diagnostic for the presence of large genomic islands in the region of largest plasticity of the P. aeruginosa chromosome (4, 7, 11, 40). Interestingly, the members of the abundant family of pKLC102/PAGI-2-type genomic islands (40) were nonrandomly distributed in the analyzed strain panel. This family of large tRNA-integrated genomic islands is present only in a subset of clones.
Fig. 6.
Nonrandom association of the P. aeruginosa SNP genotype of the core genome with elements of the accessory genome. The chromosomal map position of genes of the core and accessory genome is given in boldface and lightface font, respectively. The numbers in parentheses behind the loci of the accessory genome indicate the percentage of the 240 typed strains with a fixed association between the presence or absence of this locus and the clonal frame of the core genome defined by the hexadecimal SNP genotype. The two columns indicate the association between SNP genotype and the presence or absence of members of the abundant PAGI-2/pKLC102 family of genomic islands (7, 10, 11, 40). The extreme values of 50% and 100% indicate no or absolute linkage between the clonal frame of the core genome and the respective element of the accessory genome.
In summary, core and accessory genome are nonrandomly assembled in P. aeruginosa. Individual clones prefer a specific repertoire of accessory segments. Moreover, some parts of the core genome tolerate only a subset of the possible combinations of sequence variants, whereas other segments are freely recombining. It is the latter part where P. aeruginosa exhibits a nonclonal population structure. Thus, the P. aeruginosa genome is made up of clone-typical segments in core and accessory genome and of blocks in the core with unrestricted gene flow in the population.
Materials and Methods
The P. aeruginosa strain collection (see SI Table 3) consisted of strains from public collections, the 71-strain panel of unrelated genotypes (17), the isolates of the European CF Twin and Sibling Study (26), and isolates from the intensive-care units of the study by Anbics Management-Service (Zürich, Switzerland; Anb 006-2001: Proof-of-concept study to investigate the impact of azithromycin iv. vs. placebo on the prevention of pneumonia in ventilated patients colonized with P. aeruginosa). Strains were typed in their pulsed-field gel electrophoresis-separated SpeI fragment pattern, as described in ref. 17. For the manufacturing of the arrays, 3′ C7 amino-modified oligonucleotides (Metabion, Martinsried, Germany) at a final concentration of 10 μM in spot buffer (SCHOTT Nexterion, Jena, Germany) were spotted at room temperature in 1-nl aliquots onto a 3 × 3 mm surface-coated Borofloat 33 glass (SCHOTT, Jena, Germany) with a Microgrid II spotting machine (Zinsser Analytic, Frankfurt, Germany) (42). After production, arrays were inserted into standard microreaction tubes. A step-by-step protocol for ArrayTube (CLONDIAG, Jena, Germany) genotyping of P. aeruginosa is provided in SI Materials and Methods. The hexadecimal code of array genotype is explained in SI Appendix. The relatedness of strains by multilocus genotype was calculated by the eBurst algorithm (http://eburst.mlst.net) (41). Nonrandom association of marker haplotypes was tested by using Monte Carlo simulations (43).
Acknowledgments
We thank Eugen Ermantraut for continuous support of the project; Fernando Rojo and Pierre Cornelis for valuable suggestions for the processing of samples and the selection of targets for the microarray; Engin Gürlevik and Aneta Meyer for providing experimental help during the initial stages of the project; the core teams of the European CF Twin and Sibling Study for taking on-site throat swabs and sputum samples from CF twins and siblings; Frauke Stanke for the documentation and evaluation of clinical and genetic data; Gisela Rademacher for the processing of the samples; all the patients, their families, physicians, and staff at the clinics and intensive-care units for their time, cooperation and assistance; and Craig Winstanley (University of Liverpool, Liverpool, U.K.) for the supply of strains. The isolates from the intensive-care units were obtained without restriction from Anbics Management-Service. This work has been supported in part by grants from the German Federal Ministry of Education and Research (Infektiologie der cystischen Fibrose, Förderkennzeichen: 01 KI 0216) and the Vth Framework Program of the European Union (QLK2-CT-2001-01339). N.C. received a stipend from the Deutsche Forschungsgemeinschaft-supported International Research Training Group, Pseudomonas: Biotechnology and Pathogenicity. C.v.D. was supported by the Swiss National Science Foundation Grants 320000-108106/1 and 3200-067262.
Abbreviations
- CF
cystic fibrosis
- MLST
multilocus sequence typing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609213104/DC1.
References
- 1.Ramos JL, editor. Pseudomonas. Vol 1–3. New York: Kluwer; 2004. [Google Scholar]
- 2.Lyczak JB, Cannon CL, Pier GB. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trautmann M, Lepper PM, Haller M. Am J Infect Control. 2005;33(Suppl 1):S41–S49. doi: 10.1016/j.ajic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Tümmler B. In: Pseudomonas. Ramos JL, Levesque RC, editors. Vol 4. Heidelberg: Springer; 2006. pp. 35–68. [Google Scholar]
- 5.Spencer DH, Kas A, Smith EE, Raymond CK, Sims EH, Hastings M, Burns JL, Kaul R, Olson MV. J Bacteriol. 2003;185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. J Bacteriol. 2005;187:2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larbig KD, Christmann A, Johann A, Klockgether J, Hartsch T, Merkl R, Wiehlmann L, Fritz HJ, Tümmler B. J Bacteriol. 2002;184:6665–6680. doi: 10.1128/JB.184.23.6665-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. Proc Natl Acad Sci USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora SK, Wolfgang MC, Lory S, Ramphal R. J Bacteriol. 2004;186:2115–2122. doi: 10.1128/JB.186.7.2115-2122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J, Baldini RL, Deziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG. Proc Natl Acad Sci USA. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klockgether J, Reva O, Larbig K, Tümmler B. J Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Bruijn FJ, Lupski JR, Weinstock GM, editors. Bacterial Genomes: Physical Structure and Analysis. New York: Chapman & Hall; 1998. [Google Scholar]
- 13.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, et al. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiden MC. Annu Rev Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 15.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. J Clin Microbiol. 2004;42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernez I, Hauser P, Bernasconi MV, Blanc DS. FEMS Immunol Med Microbiol. 2005;43:29–35. doi: 10.1016/j.femsim.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Morales G, Wiehlmann L, Gudowius P, van Delden C, Tümmler B, Martinez JL, Rojo F. J Bacteriol. 2004;186:4228–4237. doi: 10.1128/JB.186.13.4228-4237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spangenberg C, Heuer T, Bürger C, Tümmler B. FEBS Lett. 1996;396:213–217. doi: 10.1016/0014-5793(96)01099-x. [DOI] [PubMed] [Google Scholar]
- 19.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 20.Kiewitz C, Tümmler B. J Bacteriol. 2000;182:3125–3135. doi: 10.1128/jb.182.11.3125-3135.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Microbiology. 2001;147:2659–2669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]
- 22.Gardner AF, Jack WE. Nucleic Acids Res. 2002;30:605–613. doi: 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SantaLucia J. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southworth MW, Kong H, Kucera RB, Ware J, Jannasch HW, Perler FB. Proc Natl Acad Sci USA. 1996;93:5281–5285. doi: 10.1073/pnas.93.11.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bronsveld I, Mekus F, Bijman J, Ballmann M, de Jonge HR, Laabs U, Halley DJ, Ellemunter H, Mastella G, Thomas S, et al. J Clin Invest. 2001;108:1705–1715. doi: 10.1172/JCI12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grothues D, Koopmann U, von der Hardt H, Tümmler B. J Clin Microbiol. 1988;26:1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, et al. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Römling U, Wingender J, Müller H, Tümmler B. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tümmler B, Koopmann U, Grothues D, Weissbrodt H, Steinkamp G, von der Hardt H. J Clin Microbiol. 1991;29:1265–1267. doi: 10.1128/jcm.29.6.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dacheux D, Attree I, Schneider C, Toussaint B. Infect Immun. 1999;67:6164–6167. doi: 10.1128/iai.67.11.6164-6167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott FW, Pitt TL. J Med Microbiol. 2004;53:609–615. doi: 10.1099/jmm.0.45620-0. [DOI] [PubMed] [Google Scholar]
- 32.Foght JM, Westlake DWS, Johnson WM, Ridgway HF. Microbiology. 1996;142:2333–2340. doi: 10.1099/00221287-142-9-2333. [DOI] [PubMed] [Google Scholar]
- 33.Alonso A, Rojo F, Martínez JL. Environ Microbiol. 1999;1:421–443. doi: 10.1046/j.1462-2920.1999.00052.x. [DOI] [PubMed] [Google Scholar]
- 34.Pirnay JP, Matthijs S, Colak H, Chablain P, Bilocq F, Van Eldere J, De Vos D, Zizi M, Triest L, Cornelis P. Environ Microbiol. 2005;7:969–980. doi: 10.1111/j.1462-2920.2005.00776.x. [DOI] [PubMed] [Google Scholar]
- 35.Pirnay JP, De Vos D, Cochez C, Bilocq F, Vanderkelen A, Zizi M, Ghysels B, Cornelis P. Environ Microbiol. 2002;4:898–911. doi: 10.1046/j.1462-2920.2002.00321.x. [DOI] [PubMed] [Google Scholar]
- 36.Kulasekara BR, Kulasekara HD, Wolfgang MC, Stevens L, Frank DW, Lory S. J Bacteriol. 2006;188:4037–4050. doi: 10.1128/JB.02000-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Proc Natl Acad Sci USA. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Vos D, de Chial M, Cochez C, Jansen S, Tümmler B, Meyer JM, Cornelis P. Arch Microbiol. 2001;175:384–388. doi: 10.1007/s002030100278. [DOI] [PubMed] [Google Scholar]
- 39.Ghysels B, Dieu BT, Beatson SA, Pirnay JP, Ochsner UA, Vasil ML, Cornelis P. Microbiology. 2004;150:1671–1680. doi: 10.1099/mic.0.27035-0. [DOI] [PubMed] [Google Scholar]
- 40.Klockgether J, Würdemann D, Reva O, Wiehlmann L, Tümmler B. J Bacteriol. 2007;189:2443–2459. doi: 10.1128/JB.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mönecke S, Leube I, Ehricht R. Genome Lett. 2003;2:106–118. [Google Scholar]
- 43.Sham PC, Curtis D. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]