Abstract
The Staphylococcus aureus cidA and lrgA genes have been shown to affect cell lysis under a variety of conditions during planktonic growth. It is hypothesized that these genes encode holins and antiholins, respectively, and may serve as molecular control elements of bacterial cell lysis. To examine the biological role of cell death and lysis, we studied the impact of the cidA mutation on biofilm development. Interestingly, this mutation had a dramatic impact on biofilm morphology and adherence. The cidA mutant (KB1050) biofilm exhibited a rougher appearance compared with the parental strain (UAMS-1) and was less adherent. Propidium iodide staining revealed that KB1050 accumulated more dead cells within the biofilm population relative to UAMS-1, indicative of reduced cell lysis. In agreement with this finding, quantitative real-time PCR experiments demonstrated the presence of 5-fold less genomic DNA in the KB1050 biofilm relative to UAMS-1. Furthermore, treatment of the UAMS-1 biofilm with DNase I caused extensive cell detachment, whereas similar treatment of the KB1050 biofilm had only a modest effect. These results demonstrate that cidA-controlled cell lysis plays a significant role during biofilm development and that released genomic DNA is an important structural component of S. aureus biofilm.
Keywords: autolysis, extracellular DNA, programmed cell death, holin
One of the primary causative agents of nosocomial infections is Staphylococcus aureus, and increasing accounts of community-acquired methicillin-resistant S. aureus have recently been described. Many S. aureus infections are thought to involve the formation of biofilm, surface-associated communities of microbes encompassed by an extracellular matrix (1, 2). Several S. aureus genes have been reported to contribute to its biofilm-forming ability, including agr (3–5), sarA (5–7), sigB (8), ica (9), rbf (10), tcaR (11), arlRS (12, 13), and alsSD (14). Microarray analyses of S. aureus (15, 16), Pseudomonas aeruginosa (17, 18), Bacillus subtilis (19, 20), and Escherichia coli (21) have also illustrated that there are major differences in gene expression between biofilm and planktonic cultures of these organisms. These results underscore the need to study biofilm physiology as a way to better understand how the bacteria exist in vivo.
The S. aureus cidABC and lrgAB operons regulate cell lysis and antibiotic tolerance in an opposing manner: lrgAB has an inhibitory effect on murein hydrolase activity and promotes antibiotic tolerance (22), whereas cidA has a positive effect on murein hydrolase activity and decreases tolerance to various antibiotics (23, 24). Expression of cid and lrg is responsive to physiological signals (24, 25) and to genetic regulation by SigB (26), LytSR (27, 28), and CidR (29, 30). Although their specific functions are unknown, CidA and LrgA share structural similarities to bacteriophage holins, a family of small membrane proteins that control the timing and onset of phage-induced cell lysis (22, 23, 27, 31). Furthermore, the cid and lrg operons have been proposed to encode components of a bacterial programmed cell death and lysis mechanism (32, 33). Unfortunately, the biological role of the Cid/Lrg system, as well as programmed cell death and lysis, remains elusive.
In the study presented here, we demonstrate that disruption of cidA decreases the ability of S. aureus to form an adherent biofilm in both in vitro and in vivo models of biofilm growth. Furthermore, the cidA mutant biofilm displayed decreased cell lysis and release of extracellular genomic DNA (eDNA), a component of the biofilm matrix that has been recently reported to be important for biofilm formation in P. aeruginosa (34, 35). This study provides evidence that eDNA is an important structural component of the S. aureus biofilm matrix and suggests a biological role for bacterial programmed cell death and cidA-mediated lysis.
Results
The S. aureus cidA Gene Affects in Vitro Biofilm Formation.
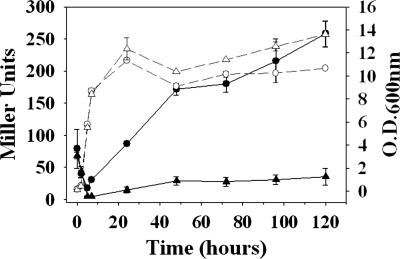
Previous work in our laboratory has shown that planktonic cultures of the cidA mutant KB1050 (24) display a near-complete loss of extracellular murein hydrolase activity (24) and decreased stationary-phase lysis (36) relative to its parental strain, the clinical isolate UAMS-1 (37). This latter result was also demonstrated here by measuring cell lysis as a function of β-galactosidase release into planktonic culture supernatants of UAMS-1 and KB1050 (each harboring the constitutively expressing lacZ reporter plasmid pAJ22) (Fig. 1). As expected, low levels of β-galactosidase activity were observed in the cidA mutant supernatant throughout stationary phase, whereas β-galactosidase activity in the UAMS-1 supernatant steadily increased over time, reaching maximum activity at 120-h incubation.
Fig. 1.
β-galactosidase release assay. Cell lysis of planktonic cultures of UAMS-1 (circles) and KB1050 (cidA mutant; triangles), each harboring pAJ22, was monitored by measuring the release of β-galactosidase into the culture supernatant (solid black lines), and its activity was quantified by incubation with o-nitrophenyl-β-d-galactopyranoside and reported in Miller units (70). The growth curves of each strain are also indicated (dashed gray lines). These results represent the average of two independent experiments.
Although these results emphasize the importance of CidA in mediating cell lysis, the biological significance of this regulation remains unknown. Given that cell death and lysis are important during biofilm development in other organisms (38–40), we compared the in vitro biofilm-forming ability of UAMS-1 and KB1050 when grown for 24 h in a static biofilm assay (Fig. 2). Despite the fact that UAMS-1 and KB1050 display similar levels of biofilm growth (OD600 = 0.49 ± 0.021 vs. 0.50 ± 0.0088, respectively), KB1050 retained ≈50% less biofilm relative to UAMS-1 after the wells were washed and stained [Fig. 2 and supporting information (SI) Fig. 7]. This finding was not caused by a defect in the initial attachment of KB1050, as similar levels of attached bacteria were observed between UAMS-1 and KB1050 at 30 and 60 min postinoculation (SI Fig. 8). Previously, we were unable to complement the antibiotic tolerance and murein hydrolase phenotypes of KB1050 (24), which was also the case in the present study, as the biofilm-defective phenotype was not complemented by supplying cidA on a plasmid (data not shown). However, this mutant phenotype is unlikely caused by a secondary site mutation because similar cidA mutations in different S. aureus genetic backgrounds also decreased their ability to form adherent biofilm (SI Fig. 7). This phenotype was also not caused by a polar effect on the downstream cid genes, because isogenic cidBC and cidC mutants grown under the same conditions produced biofilm comparable to that of UAMS-1 (data not shown).
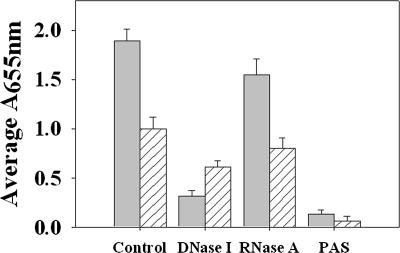
Fig. 2.
The role of cell lysis and eDNA in S. aureus biofilm adherence. UAMS-1 (filled bars) and KB1050 (cidA mutant; striped bars) were grown for 24 hr in a static biofilm assay as described in Materials and Methods. Where indicated, DNase I, RNase A, or PAS was added at the time of inoculation. Biofilm formation was quantified after washing with PBS, fixation with ethanol, and staining with crystal violet, by measuring the A655nm of each well. Mean values from four (RNase A, DNase I, and PAS treatment) or six (untreated control) independent experiments, each performed in triplicate, are shown. Error bars represent the SEM.
Cell Lysis and DNA Release Affect S. aureus Biofilm Adherence.
Given that KB1050 has been previously shown to undergo decreased cell lysis in stationary phase but undergoes cell death at levels similar to UAMS-1 (36), we hypothesized that a cidA mutant biofilm contains a larger proportion of dead (but not lysed) cells relative to a UAMS-1 biofilm. To test this idea, unwashed 24-hr static biofilm of each strain was disrupted, stained with propidium iodide (PI), and the percentage of dead cells in each biofilm was calculated by dividing the number of dead (PI-stained) cells by the total number of cells (see SI Text). This analysis demonstrated that a cidA mutant biofilm contains ≈4-fold more dead cells relative to biofilm of the parental strain (1.06% versus 0.24%, respectively; see SI Fig. 9) and is consistent with the observation that the cidA mutant exhibited less cell lysis (Fig. 1).
Studies performed in other biofilm-forming organisms (34, 35, 41) have demonstrated that eDNA is an important component of the biofilm matrix, and a potential source for this eDNA comes from cell lysis (35). Therefore, the cidA mutation may affect S. aureus biofilm formation by reducing the amount of eDNA present in the biofilm. To evaluate the importance of eDNA in S. aureus biofilm, UAMS-1 and KB1050 biofilm were treated with DNase I at the time of inoculation and grown for 24 h (Fig. 2). DNase treatment significantly (P < 0.05, Dunn's test) decreased UAMS-1 biofilm by 6-fold relative to untreated biofilm, whereas the same treatment resulted in a 1.6-fold decrease in KB1050 biofilm that was not statistically significant (P > 0.05, Dunn's test). In contrast, treatment of UAMS-1 and KB1050 biofilm with RNase A did not significantly affect the amount of biofilm formed by either strain (P > 0.05, Dunn's test), suggesting that RNA is not a significant component of the biofilm matrix. To ensure that the effects of DNase were not caused by an effect on cell viability, the bacteria from DNase-treated and untreated wells were harvested before washing the wells to determine their cfu/ml. As expected, the results of this analysis (data not shown) indicated that DNase treatment did not affect cell viability.
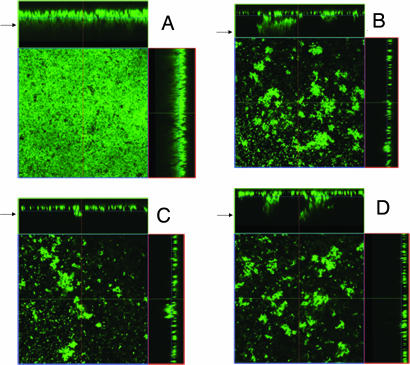
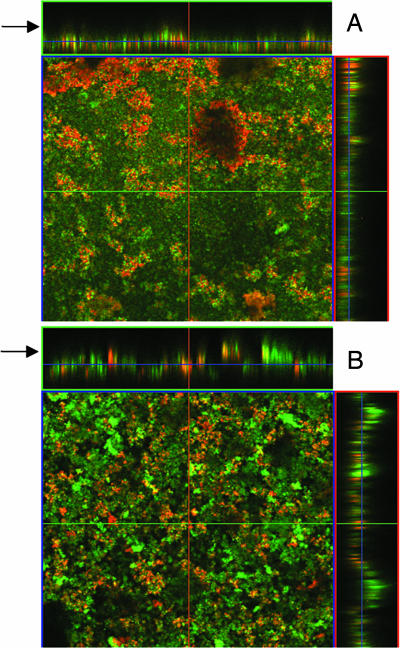
UAMS-1 and cidA mutant biofilm were also assessed by confocal laser scanning microscopy (CLSM). As shown in Fig. 3, several differences were observed between the UAMS-1 and cidA mutant biofilm after they were washed and stained with the fluorescent dye SYTO 9. The UAMS-1 biofilm had a much smoother appearance relative to the KB1050 biofilm (Fig. 3 A and B, respectively), which was reflected by the KB1050 biofilm having a larger roughness coefficient compared with the UAMS-1 biofilm (SI Table 1). The average thickness of the UAMS-1 biofilm after washing (30.52 ± 1.96 μm) was ≈2.5-fold more than that of KB1050 (12.41 ± 1.14 μm). Given that there are nearly equal amounts of biomass present before washing (OD600 = 0.49 ± 0.021 vs. 0.50 ± 0.0088 for UAMS-1 and KB1050, respectively), these results indicate that much of the KB1050 biofilm had detached during the wash step. Interestingly, DNase treatment of the UAMS-1 biofilm also caused detachment during the wash step, resulting in a thinner (9.59 ± 1.23 μm) biofilm with a rough appearance nearly identical to the control KB1050 biofilm (Fig. 3 B and C). Also, DNase treatment of the KB1050 biofilm had only a minor effect on thickness (9.64 ± 1.14 μm) and appearance after washing (Fig. 3, compare B and D, and see SI Table 1). These results suggest that eDNA is an important structural component of the staphylococcal biofilm.
Fig. 3.
CLSM of static biofilm. UAMS-1 and KB1050 (cidA mutant) were grown as static biofilm as described in Materials and Methods. Where indicated, 28 units of DNase I was added to each well at the time of inoculation. After 24-h growth, the biofilms were washed and stained with SYTO 9, and z-stacks of each were acquired by CLSM with a Plan-Neofluar ×40/1.3 oil objective lens. Orthogonal views of z-stacks of control UAMS-1 and KB1050 biofilm (A and B, respectively) and DNase-treated UAMS-1 and KB1050 biofilm (C and D, respectively) are shown at ×400 magnification and represent 12 fields of view acquired in two independent experiments. Arrows indicate the top of the biofilm.
Quantification of eDNA Release in S. aureus Biofilm.
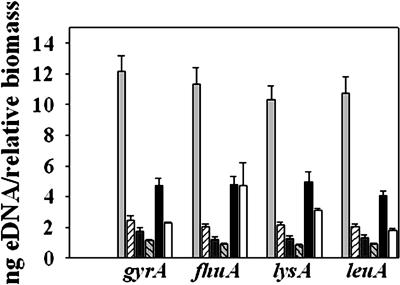
The amount of eDNA present in unwashed 24-h static UAMS-1 and KB1050 biofilms was quantified by real-time PCR using four different primer pairs specific for four randomly selected chromosomally encoded genes and reported as eDNA per relative biomass to account for the amount of bacteria present in the unwashed biofilm of each sample (Fig. 4). The average amount of eDNA present in the untreated UAMS-1 biofilm (11.15 ± 0.49 ng) was 5-fold greater than that present in the cidA mutant biofilm (2.18 ± 0.11 ng), a statistically significant difference (P < 0.05, Dunn's test). Furthermore, the amount of eDNA in the DNase-treated UAMS-1 biofilm was reduced 8-fold compared with the control biofilm, whereas DNase treatment of the KB1050 biofilm resulted in only a 2.3-fold reduction in eDNA (Fig. 4). UAMS-1 and KB1050 biofilm were also grown in the presence of sodium polyanethole sulfonate (PAS), a compound that has previously been shown to inhibit S. aureus autolysis without affecting viability (42, 43). PAS treatment severely reduced biofilm adherence by UAMS-1 (Fig. 2) and likewise decreased the amount of eDNA by 2.4-fold (Fig. 4). Although PAS treatment clearly impacted the release of eDNA, we cannot exclude the possibility that PAS also interferes with other factors that may contribute to biofilm formation. In support of this idea is the observation that growth of KB1050 in the presence of PAS also reduced the amount of adherent biofilm relative to the untreated KB1050 control (Fig. 2) even though the effect on eDNA was not significant (Fig. 4; P > 0.05, Dunn's test).
Fig. 4.
Quantification of eDNA present in unwashed UAMS-1 and cidA mutant static biofilm. eDNA was isolated from untreated UAMS-1 (light gray bars) and KB1050 (cidA mutant; striped bars), DNase-treated UAMS-1 (dark gray bars) and KB1050 (gray striped bars), and PAS-treated UAMS-1 (black bars) and KB1050 (white bars) 24-hr unwashed biofilms as described in Materials and Methods. The total eDNA present in each biofilm was quantified by real-time PCR, using primer pairs specific for gyr (gyrase A), fhu (ferrichrome transport ATP-binding protein A), lys (diaminopimelate decarboxylase A), and leu (2-isopropylmalate synthase). The values are expressed as nanogram of eDNA per relative biofilm biomass, as described in Materials and Methods. Mean values from a minimum of three independent experiments, each performed in triplicate, are shown. Error bars represent the SEM.
The cidA Mutation Affects Biofilm Formation in Other Models of Biofilm Growth.
To better reflect the conditions under which biofilm growth occurs in vivo, UAMS-1 and KB1050 biofilms were grown on polycarbonate coupons in flow cell chambers for 24 h, stained with SYTO 9 and PI, and analyzed by CLSM (Fig. 5). Under these conditions, the cidA mutant biofilm had a rough appearance and did not display confluent attachment to the coupon (Fig. 5B), whereas the UAMS-1 biofilm appeared as a relatively flat mat of cells interspersed with occasional 3D structures (Fig. 5A).
Fig. 5.
CLSM of flow cell biofilm. UAMS-1 (A) and KB1050 (B) biofilm were grown on polycarbonate coupons housed in flow cell chambers for 24 h and stained with SYTO 9 and PI as described in Materials and Methods, and z-stacks of each were acquired by CLSM with a Plan-Apochromat ×10/0.45 objective lens. Representative orthogonal views of UAMS-1 and KB1050 biofilm are shown at ×100 magnification and are representative of two independent experiments. Arrows indicate the top of the biofilm.
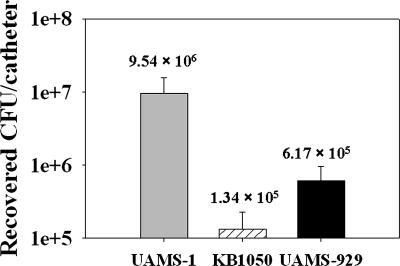
The effect of the cidA mutation on biofilm development was also assessed in vivo in a previously described murine model of catheter-associated infection (15). After 10 days, the infected catheters of each group of mice were removed and the bacteria recovered from each was quantified by dilution plating and reported as the average cfu per catheter (Fig. 6). Even though the cidA mutant KB1050 was still able to colonize the implanted catheters, the recovered CFU per catheter was decreased by 2 log units relative to UAMS-1. This difference was statistically significant (P < 0.05, Dunn's test), suggesting that KB1050 forms less biofilm in vivo. As reported (15), an isogenic sarA mutant (UAMS-929) was also impaired in its ability to form a biofilm in this model of infection, displaying a 1 log decrease in cfu per catheter relative to UAMS-1 (Fig. 6). Collectively, these results suggest that CidA-mediated cell lysis contributes to S. aureus biofilm formation in vitro and in vivo.
Fig. 6.
Effect of the cidA mutation on S. aureus in vivo biofilm formation. The ability of UAMS-1 (gray bar), KB1050 (cidA mutant; striped bar), and UAMS-929 (sarA mutant; black bar) to form biofilm in vivo was assessed in a previously published murine model (15) of catheter-associated infection. Biofilm formation in vivo is expressed as the mean number of bacteria recovered from the implanted catheters of each group of mice (recovered cfu per catheter) after 10 days of infection. Error bars represent the SEM. The numbers located above each bar represent the mean recovered cfu per catheter.
Discussion
Previous studies of the S. aureus cid and lrg genes have illustrated their importance in mediating cell death and/or lysis in stationary phase (36) and in response to antibiotic treatment (23, 24). In this present study, analysis of the clinical S. aureus isolate UAMS-1 and its isogenic cidA mutant (KB1050) in the context of biofilm growth led to the discovery that CidA contributes to biofilm adherence both in vitro and in vivo by affecting cell lysis and the release of genomic DNA. This study examines and demonstrates the role of eDNA in S. aureus biofilm formation and also provides evidence for the biological role of the Cid/Lrg system in this organism.
The contribution of eDNA to the stability and development of biofilm has been documented in other organisms (recently reviewed in ref. 44). Several studies have shown that eDNA is an abundant component of the extracellular matrix of P. aeruginosa biofilm (34, 35, 45). DNase treatment has been shown to disrupt P. aeruginosa biofilm grown in vitro (35, 46) and is used in combination with antibiotics to treat P. aeruginosa infections of cystic fibrosis patients (47). Treatment of streptococcal biofilm with DNase has also been shown to have a negative effect on biofilm adherence (41, 48, 49). These results correlate with our findings that DNase treatment had a greater ability to interfere with biofilm adherence in the wild-type S. aureus strain relative to the cidA mutant, presumably because the mutant biofilm contains less eDNA (Figs. 2–4). A recent study by Allesen-Holm et al. (35) using CLSM to analyze P. aeruginosa biofilm also showed that eDNA contributes to biofilm architecture: In young biofilm the eDNA appeared as a grid-like structure on the substratum as well as surface of the microcolonies, whereas in mature biofilm, the eDNA appeared as discrete layers located in mushroom-shaped structures. Because our study focused on an early time point in S. aureus biofilm development (24 h), the cidA mutant phenotype and its effect on eDNA release likely reflects the ability to interfere with the early stages of attachment and/or microcolony formation during biofilm development.
What is the actual mechanism of eDNA release in bacterial biofilm? In P. aeruginosa, eDNA release depends on quorum sensing (35), and there is evidence to suggest that cell lysis itself may be achieved by prophage induction within a biofilm (38, 50), or alternatively, as a consequence of the release of membrane vesicles that contain bacteriolytic activity (51, 52) as well as DNA (53). In Streptococcus mutans and Streptococcus pneumoniae, DNA is released from a lysing subfraction of the bacterial population in response to competence development, a physiological process that also depends on quorum sensing (54–59). Both the actions of bacteriocins (59–61) and autolysins (56, 58) have been implicated in the lysis of DNA-releasing cells during this process. Intriguingly, a higher frequency of natural transformation was observed in S. mutans biofilm relative to planktonic cultures (62), and biofilm formation depends on the com system in both S. mutans (63) and S. pneumoniae (64). Collectively, these observations support an emerging paradigm of eDNA release via cell lysis during biofilm formation, and our findings have implicated CidA as an additional regulator of this process in S. aureus.
In Staphylococcus epidermidis and S. aureus, disruption of the atl gene, encoding the primary murein hydrolase of these bacteria, causes a dramatic decrease in their abilities to form biofilm (65, 66). Inactivation of a gene encoding an Atl homolog in S. mutans also reduced its biofilm-forming ability (67, 68). In S. epidermidis, the reduction in biofilm formation was attributed to loss of adhesive functions of Atl important in the initial stages of attachment, because this protein was found to exhibit vitronectin-binding activity (65). In light of the findings described here, the possibility should also be considered that the impact of the atl mutations on biofilm formation is a result of decreased lysis and eDNA release.
One of the primary findings of this study is that cell death and lysis is a necessary and, apparently, controlled process during the development of S. aureus biofilm. Given the presence of the cid/lrg genes in a wide variety of bacterial species (23), including both Gram-positive and Gram-negative bacteria, it is possible that the role of these genes in biofilm development is widely conserved. At a fundamental level, this process is similar to the role of apoptosis in the development of more complex multicellular organisms. Indeed, functional similarities between the cid/lrg system and the Bax/Bcl proteins involved in the control of apoptosis in eukaryotic organisms have been noted (32). Further investigations into the cid/lrg system should provide greater insight into the role and significance of bacterial cell death and lysis, as well as their functional relationship to more complex eukaryotic systems.
Materials and Methods
Strains and Growth Conditions.
The bacterial strains and plasmids used in this study are presented in SI Table 2. S. aureus strains were grown in either tryptic soy broth (TSB) containing 0.25% (wt/vol) glucose or in TSB containing 3.5% (wt/vol) NaCl and 0.75% (wt/vol) glucose (TSB-NaCl/Glc), as indicated. Broth cultures were grown as described (36).
Measurement of β-Galactosidase Activity in Culture Supernatants.
Overnight cultures of UAMS-1 and KB1050, each harboring plasmid pAJ22 (69), were diluted to an OD600 of 0.1 in TSB (no antibiotic) and grown for 120 h. At various time points, supernatants from each culture were harvested by centrifugation, and the corresponding OD600 was measured. β-galactosidase activity in the supernatants was determined as described (54) using o-nitrophenyl-β-d-galactopyranoside as the substrate and reported in Miller units (70).
Growth and Quantification of Static Biofilm.
Overnight cultures of S. aureus grown in TSB-NaCl/Glc were diluted to an OD600 of 0.05 in fresh TSB-NaCl/Glc, and 200 μl of each culture, supplemented with 500 μg·ml−1 of PAS, 28 units of DNase I (Qiagen, Valencia, CA), or 200 μg of DNase-free RNase A (Sigma, St. Louis, MO) where indicated, was transferred to wells in Costar 3596 plates (Corning Life Sciences, Acton, MA) and incubated for 24 h at 37°C. Unless otherwise indicated, the wells were coated overnight at 4°C with 20% human plasma (Sigma) in bicarbonate buffer before inoculation.
Biofilm quantification was performed as described (6, 71) except that biofilm formation was quantified by measuring the A655 nm of each well (containing crystal violet-stained biofilm) with a model 680 microplate reader (Bio-Rad, Hercules, CA).
CLSM of Static Biofilm.
UAMS-1 and KB1050 were grown in Costar 3614 plates (Corning Life Sciences) exactly as described above. Where indicated, 28 units of DNase I was added to the wells at the time of inoculation. The next day, the wells were gently washed three times with 0.85% (wt/vol) NaCl, followed by staining with 1.25 μM SYTO 9 (Invitrogen, Carlsbad, CA) for 18 min. After removing the stain, the wells were gently washed once with 0.85% (wt/vol) NaCl. Biofilm images were collected by CLSM using a LSM 510 META confocal scanning system (Zeiss, Thornwood, NY) and AxioPlan 2IE MOT motorized upright microscope (Zeiss). SYTO 9 fluorescence was detected by excitation at 488 nm, and emission was collected with a 500- to 530-nm bandpass filter. All z-sections were collected at 1-μm intervals by using a Plan-Neofluar ×40/1.3 oil objective lens. Image acquisition and processing was performed by using a LSM Image Browser (Zeiss). Quantification of the z-stacks was done with the computer program COMSTAT (72).
Purification and Quantification of eDNA.
UAMS-1 and KB1050 were grown in Costar 3596 plates exactly as described above. After 24 h, the plates were chilled at 4°C for 1 h, and 1 μl of 0.5M EDTA was added to each well. The supernatants were discarded, and unwashed biofilm were harvested by resuspension in 50 mM Tris·HCl/10 mM ETDA/500 mM NaCl, pH 8.0 and transferred into chilled tubes. After centrifugation for 5 min at 4°C and 18,000 × g, 100 μl of each supernatant was transferred to a tube containing 300 μl of TE buffer (10 mM Tris·HCl/1 mM EDTA, pH 8.0), and extracted once with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) and once with chloroform/isoamyl alcohol (24:1). The aqueous phase of each sample was then mixed with 3 vol of ice-cold 100% (vol/vol) ethanol and 1/10 volume of 3 M Na-acetate (pH 5.2) and stored at −20°C. The next day, the ethanol-precipitated DNA was collected by centrifugation for 20 min at 4°C and 18,000 × g, washed with ice-cold 70% (vol/vol) ethanol, air-dried, and dissolved in 20 μl of TE buffer.
eDNA was quantified by real-time PCR using the four primer pairs listed in SI Table 3. PCRs were performed on 10−1 dilutions of each sample with the LightCycler DNA Master SYBR Green I kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer's recommendations, using 5 μM of each primer in the reaction. Purified UAMS-1 genomic DNA at known concentrations was also subjected to quantitative RT-PCR with each primer pair to generate a standard curve used to calculate the concentration of eDNA in the unknown samples. PCR was performed in a Lightcycler 1.0 by using the following parameters: one cycle of 95°C for 30 s, 45 cycles of 95°C for 1 s, 55°C for 5 s, and 72°C for 20 s. To account for potential differences in biomass, the average OD600 of each unwashed biofilm was determined and used to calculate the relative OD600 of each biofilm with respect to the OD600 of the untreated UAMS-1 biofilm (whereby the relative OD600 of the UAMS-1 biofilm = 1). The nanogram of eDNA per relative biomass of each biofilm was then calculated by dividing its total eDNA (ng) by its relative OD600.
Cultivation and CLSM of Flow Cell Biofilm.
Eighteen-hour TSB cultures of UAMS-1 and KB1050 were diluted in TSB to an OD600 = 0.05 and used to inoculate polycarbonate coupons housed in a BST FC 270 Flow Cell apparatus (BioSurface Technologies, Bozeman, MT), whose setup was performed according to the manufacturer's recommendations. Each flow cell was coated with 20% (vol/vol) human plasma overnight at 4°C before use. Culture media (0.1 × TSB, 0.25% glucose, and 4 μM PI) were pumped through the flow cells at a rate of 0.35 ml·min−1 for 1 h before inoculation. After inoculation, the flow cell reactors were incubated statically for 60 min at 37°C, and media flow was reinitiated at the same rate. After growth at 37°C for 24 h, each biofilm was stained for 25 min with 1.3 μM SYTO 9. Images were collected by CLSM with a LSM 510 META system and AxioPlan 2IE MOT motorized upright microscope (Zeiss). SYTO 9 fluorescence was detected as described above. PI fluorescence was detected by excitation at 488 nm, and emission was collected with a 565- to 615-nm bandpass filter. Z-sections were collected at 1.0-μm intervals with a Plan-Apochromat ×10/0.45 lens. Image acquisition and processing was performed as described above.
In Vivo Biofilm Assay.
UAMS-1 and KB1050 biofilm formation in vivo was assessed in a murine model of catheter-associated infection as described (15).
Acknowledgments
We thank Dr. Alex O'Neill (University of Leeds, Leeds, United Kingdom) for S. aureus AJ22; Arne Heydorn (Technical University of Denmark, Kongens Lyngby, Denmark) for COMSTAT software; Janice Taylor of the CLSM Core Facility at the University of Nebraska Medical Center (supported by the Nebraska Research Initiative) for assistance with CLSM; and Dr. Heather Jensen-Smith for assistance with CLSM performed at the Integrative Biological Imaging Facility (constructed with support from C06 Grant RR17417-01 from the National Center for Research Resources, National Institutes of Health) at Creighton University, Omaha, NE. This work was funded by National Institutes of Health Grants R01AI038901 (to K.W.B.) and R01AI43356 (to M.S.S.), Department of Defense Grant DAAD 19-03-1-0191 (to K.W.B.), and an American Health Association Predoctoral Fellowship (to J.E.C.).
Abbreviations
- eDNA
extracellular genomic DNA
- CLSM
confocal laser scanning microscopy
- PI
propidium iodide
- PAS
polyanethole sulfonate
- TSB
tryptic soy broth.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610226104/DC1.
References
- 1.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 3.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vuong C, Saenz HL, Gotz F, Otto M. J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 5.Pratten J, Foster SJ, Chan PF, Wilson M, Nair SP. Microbes Infect. 2001;3:633–637. doi: 10.1016/s1286-4579(01)01418-6. [DOI] [PubMed] [Google Scholar]
- 6.Beenken KE, Blevins JS, Smeltzer MS. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 8.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. Infect Immun. 2001;69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim Y, Jana M, Luong TT, Lee CY. J Bacteriol. 2004;186:722–729. doi: 10.1128/JB.186.3.722-729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jefferson KK, Pier DB, Goldmann DA, Pier GB. J Bacteriol. 2004;186:2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I. J Bacteriol. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fournier B, Hooper DC. J Bacteriol. 2000;182:3955–3964. doi: 10.1128/jb.182.14.3955-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang SJ, Rice KC, Bayles KW, Smeltzer MS. Microbiology. 2006;152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 15.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Resch A, Rosenstein R, Nerz C, Gotz F. Appl Environ Microbiol. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 18.Waite RD, Papakonstantinopoulou A, Littler E, Curtis MA. J Bacteriol. 2005;187:6571–6576. doi: 10.1128/JB.187.18.6571-6576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanley NR, Britton RA, Grossman AD, Lazazzera BA. J Bacteriol. 2003;185:1951–1957. doi: 10.1128/JB.185.6.1951-1957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren D, Bedzyk LA, Setlow P, Thomas SM, Ye RW, Wood TK. Biotechnol Bioeng. 2004;86:344–364. doi: 10.1002/bit.20053. [DOI] [PubMed] [Google Scholar]
- 21.Schembri MA, Kjaergaard K, Klemm P. Mol Microbiol. 2003;48:253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 22.Groicher KH, Firek BA, Fujimoto DF, Bayles KW. J Bacteriol. 2000;182:1794–1801. doi: 10.1128/jb.182.7.1794-1801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice KC, Firek BA, Nelson JB, Yang S-J, Patton TG, Bayles KW. J Bacteriol. 2003;185:2635–2643. doi: 10.1128/JB.185.8.2635-2643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice KC, Nelson JB, Patton TG, Yang SJ, Bayles KW. J Bacteriol. 2005;187:813–821. doi: 10.1128/JB.187.3.813-821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton TG, Yang SJ, Bayles KW. Mol Microbiol. 2006;59:1395–1404. doi: 10.1111/j.1365-2958.2006.05034.x. [DOI] [PubMed] [Google Scholar]
- 26.Rice KC, Patton TG, Yang S-J, Dumoulin A, Bischoff M, Bayles KW. J Bacteriol. 2004;186:3029–3037. doi: 10.1128/JB.186.10.3029-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunskill EW, Bayles KW. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunskill EW, Bayles KW. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang SJ, Rice KC, Brown RJ, Patton TG, Liou LE, Park YH, Bayles KW. J Bacteriol. 2005;187:5893–5900. doi: 10.1128/JB.187.17.5893-5900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SJ, Dunman PM, Projan SJ, Bayles KW. Mol Microbiol. 2006;60:458–468. doi: 10.1111/j.1365-2958.2006.05105.x. [DOI] [PubMed] [Google Scholar]
- 31.Bayles KW. Trends Microbiol. 2000;8:274–278. doi: 10.1016/s0966-842x(00)01762-5. [DOI] [PubMed] [Google Scholar]
- 32.Bayles KW. Trends Microbiol. 2003;11:306–311. doi: 10.1016/s0966-842x(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 33.Rice KC, Bayles KW. Mol Microbiol. 2003;50:729–738. doi: 10.1046/j.1365-2958.2003.t01-1-03720.x. [DOI] [PubMed] [Google Scholar]
- 34.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 35.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 36.Patton TG, Rice KC, Foster MK, Bayles KW. Mol Microbiol. 2005;56:1664–1674. doi: 10.1111/j.1365-2958.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 37.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mai-Prochnow A, Evans F, Dalisay-Saludes D, Stelzer S, Egan S, James S, Webb JS, Kjelleberg S. Appl Environ Microbiol. 2004;70:3232–3238. doi: 10.1128/AEM.70.6.3232-3238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mai-Prochnow A, Webb JS, Ferrari BC, Kjelleberg S. Appl Environ Microbiol. 2006;72:5414–5420. doi: 10.1128/AEM.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen FC, Tao L, Scheie AA. J Bacteriol. 2005;187:4392–4400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wecke J, Lahav M, Ginsburg I, Kwa E, Giesbrecht P. Arch Microbiol. 1986;144:110–115. doi: 10.1007/BF00414719. [DOI] [PubMed] [Google Scholar]
- 43.Yabu K, Kaneda S. Curr Microbiol. 1995;30:299–303. doi: 10.1007/BF00295505. [DOI] [PubMed] [Google Scholar]
- 44.Spoering AL, Gilmore MS. Curr Opin Microbiol. 2006;9:133–137. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Steinberger RE, Holden PA. Appl Environ Microbiol. 2005;71:5404–5410. doi: 10.1128/AEM.71.9.5404-5410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemoto K, Hirota K, Murakami K, Taniguti K, Murata H, Viducic D, Miyake Y. Chemotherapy. 2003;49:121–125. doi: 10.1159/000070617. [DOI] [PubMed] [Google Scholar]
- 47.Gibson RL, Burns JL, Ramsey BW. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 48.Petersen FC, Pecharki D, Scheie AA. J Bacteriol. 2004;186:6327–6331. doi: 10.1128/JB.186.18.6327-6331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moscoso M, Garcia E, Lopez R. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webb JS, Lau M, Kjelleberg S. J Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadurugamuwa JL, Beveridge TJ. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z, Clarke AJ, Beveridge TJ. J Bacteriol. 1996;178:2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Renelli M, Matias V, Lo RY, Beveridge TJ. Microbiology. 2004;150:2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 54.Steinmoen H, Knutsen E, Havarstein LS. Proc Natl Acad Sci USA. 2002;99:7681–7686. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinmoen H, Teigen A, Havarstein LS. J Bacteriol. 2003;185:7176–7183. doi: 10.1128/JB.185.24.7176-7183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moscoso M, Claverys JP. Mol Microbiol. 2004;54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- 57.Guiral S, Henard V, Granadel C, Martin B, Claverys JP. Microbiology. 2006;152:323–331. doi: 10.1099/mic.0.28425-0. [DOI] [PubMed] [Google Scholar]
- 58.Guiral S, Mitchell TJ, Martin B, Claverys JP. Proc Natl Acad Sci USA. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreth J, Merritt J, Shi W, Qi F. Mol Microbiol. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Ploeg JR. J Bacteriol. 2005;187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kreth J, Merritt J, Zhu L, Shi W, Qi F. FEMS Microbiol Lett. 2006;265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 62.Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida A, Kuramitsu HK. Appl Environ Microbiol. 2002;68:6283–6291. doi: 10.1128/AEM.68.12.6283-6291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G. Mol Microbiol. 2006;61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heilmann C, Hussain M, Peters G, Gotz F. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 66.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. FEMS Microbiol Lett. 2006;259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 67.Shibata Y, Kawada M, Nakano Y, Toyoshima K, Yamashita Y. Infect Immun. 2005;73:3512–3520. doi: 10.1128/IAI.73.6.3512-3520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahn SJ, Burne RA. J Bacteriol. 2006;188:6877–6888. doi: 10.1128/JB.00536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Neill AJ, Miller K, Oliva B, Chopra I. J Antimicrob Chemother. 2004;54:1127–1129. doi: 10.1093/jac/dkh476. [DOI] [PubMed] [Google Scholar]
- 70.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 71.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]