Abstract
Proline-, glutamic acid-, and leucine-rich protein (PELP)1, also known as modulator of nongenomic actions of the estrogen receptor (MNAR), is a novel nuclear receptor coregulator with a multitude of functions. PELP1/MNAR serves as a scaffolding protein that couples various signaling complexes with nuclear receptors and participates in genomic and nongenomic functions. Recent data suggest that PELP1/MNAR expression is deregulated in several cancers, including breast, endometrial, prostate, and ovarian cancer, and that PELP1/MNAR interacts with several oncogenes. In this review, we summarize the emerging biological properties and functions of PELP1/MNAR.
Introduction
Researchers first identified proline-, glutamic acid-, and leucine-rich protein (PELP)1, as one of the several proteins that bind to the SH2 domain of Lck tyrosine protein kinase utilizing GST-SH2 pull down assays. In the SDS-Page gel this protein migrated as a 160-kDa protein, hence it was initially named p160 (not to be confused with the SRC/p160 family) (Figure 1 and [Joung et al., 1996]). Using peptide sequences derived from the purified 160-kDa protein as oligonucleotide probes, investigators cloned two cDNAs of different lengths: PELP1 and PELP2 [Vadlamudi et al., 2001]. Initial studies using the longer clone, PELP1 (3.8 kb), resulted in generation of a 160-kDa protein product and identification of coactivator activity for ERα transactivation [Vadlamudi et al., 2001]. Subsequent studies using ERα affinity chromatography identified a protein that is identical to PELP1 at the amino acid level, but differs slightly in cDNA length (3.4 kb). Therefore, researchers presumed that this was a new protein and named it modulator of nongenomic actions of estrogen receptor (MNAR) [Wong et al., 2002]. However, both the PELP1 and MNAR gene sequences map to the same chromosomal region, 17p13.2, and have identical sequences except for an additional 435-bp region in PELP1 [Balasenthil and Vadlamudi, 2003]. Further analysis of the PELP1 cDNA regions in 3.8 kb PELP1 suggested that the initially isolated PELP1 cDNA was an immature transcript and contained an extra 435-bp intron with consensus splice sites that artificially produced the disparity in length between the PELP1 and MNAR cDNAs. Later studies showed that PELP1 and MNAR code for the same proteins, and several studies reported that PELP1 and MNAR are identical proteins. HUGO Gene Nomenclature Committee (HGNC) approved PELP1 as the approved symbol for PELP1/MNAR. Functional studies revealed that in addition to Lck, PELP1/MNAR also interacts with other members of the Src kinase family, including c-Src [Wong et al., 2002]. Subsequent studies identified orthologs to PELP1/MNAR in mice and rats [Khan et al., 2005]. Homology data established that PELP1/MNAR is expressed in other mammals, including chimpanzees, dogs, and cats. A previous study also reported on a cDNA encoding Xenopus PELP1/MNAR [Haas et al., 2005].
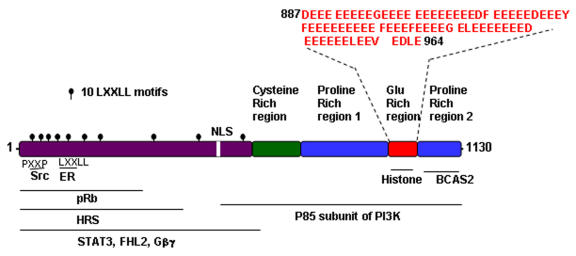
Figure 1. Schematic diagram of functional domains identified in PELP1/MNAR.
PXXP, SH3 binding domain; LXXLL, nuclear receptor interacting domain; glu-rich, histone binding domain. Putative regions of interaction with other proteins are shown.
Protein domain structure
PELP1/MNAR contains several motifs and domains that are commonly present in many transcriptional coactivators, including 10 nuclear receptor (NR)-interacting boxes (LXXLL motifs), a zinc finger, a glutamic acid-rich domain, and 2 proline-rich domains (Figure 1 and [Vadlamudi et al., 2001; Wong et al., 2002]). A unique feature of PELP1/MNAR is the presence of an unusual stretch of 70 acidic amino acids in the C-terminus that functions as a histone-binding region [Choi et al., 2004; Nair et al., 2004]. Interestingly, proline-rich regions contain several consensus PXXP motifs that may interact with signaling proteins containing SH3 domains. Analysis of the primary PELP1/MNAR sequence using the Eukaryotic Linear Motif server (http://elm.eu.org) revealed that PELP1 contained several conserved protein-protein interaction motifs that bind to FHA, SH2, SH3, PDZ, and WW domains. PELP1/MNAR encodes a protein of 1130 amino acids and has a predicted molecular weight of 120 kDa with an isoelectric point of 4.30, but because of its overall negative charge and excessive number of prolines, the protein migrates on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels as a 160-kDa protein.
Interactions
Interactions with NRs
PELP1/MNAR appears to function as a general coregulator, as it contains 10 NR-interacting boxes (LXXLL) and interacts with multiple NRs. Estrogen promotes PELP1 interaction with the AF2 domain of ERα [Vadlamudi et al., 2001]. LXXLL motifs 4 and 5 primarily mediate binding of PELP1/MNAR to ERα [Barletta et al., 2004]. PELP1/MNAR also interacts with ERβ [Wong et al., 2002]. Using receptor subtype-specific ligands, researchers showed that PELP1/MNAR acts as a coactivator for both ERα and ERβ [Vadlamudi et al., 2004b]. PELP1 also interacts with several other NRs, including androgen receptors, glucocorticoid receptors, and progesterone receptors, in a ligand-dependent manner [Vadlamudi et al., 2001; Wong et al., 2002]. PELP1/MNAR interacts with retinoid X receptor (RXR)α and enhances transactivation in response to 9-cis retinoic acid (RA) [Singh et al., 2006]. Furthermore, PELP1 functions as a corepressor of non-NR sequence-specific transcription factors, including activating protein 1, NFκB, and ternary complex factor/serum response factor [Choi et al., 2004]. PELP1 also interacts with the transcription factor signal transducer and activator of transcription (STAT)3 and enhances growth factor-mediated STAT3 transactivation functions [Manavathi et al., 2005].
PELP1/MNAR-interacting proteins
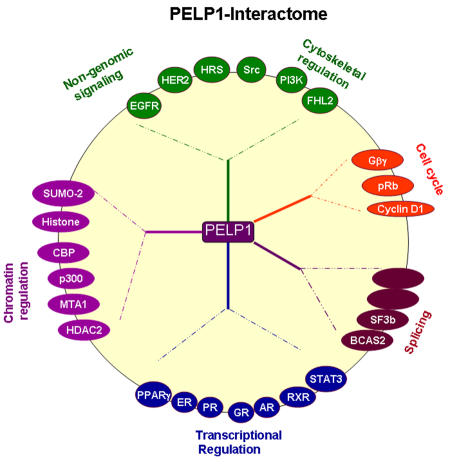
PELP1/MNAR interacts with several components of chromatin-modifying complexes, including CRE-binding protein-binding protein (CBP), p300, metastasis-associated protein (MTA)1, and histone deacetylase 2 [Choi et al., 2004; Vadlamudi et al., 2001], as well as with histones [Choi et al., 2004; Nair et al., 2004]. Recent studies showed that PELP1 interacts with Sumo-2 and forms a complex with the SET domain, bifurcated 1 and lysine-specific demethylase 1, which are a histone lysine methyltransferase and demethylase, respectively [Rosendorff et al., 2006]. PELP1/MNAR also interacts with the transcriptional activator four-and-a-half LIM-only protein 2 [Nair et al., 2007] and with several components of spliceosomes, including breast carcinoma amplified sequence 2 and splicing factor 3b, indicating that PELP1/MNAR may play a role in coupling transcriptional activation with splicing [Nair and Vadlamudi, 2007]. Also, PELP1/MNAR interacts with several key components that play a role in cell cycle progression, including CDK4, cyclin D1, and retinoblastoma protein (pRb) [Balasenthil and Vadlamudi, 2003; Haas et al., 2005; Manavathi et al., 2005; Vadlamudi et al., 2004b; Vadlamudi et al., 2001; Wong et al., 2002], and with the cytosolic kinases c-Src and phosphatidylinositol-3 kinase (PI3K) [Vadlamudi et al., 2005b; Wong et al., 2002]. Mitogenic signaling promotes the association of PELP1/MNAR with a number of signaling components, including epidermal growth factor (EGF) receptor (EGFR), HER2, STAT3, and hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) [Rajhans and Vadlamudi, 2006]. The diversity of the PELP1/MNAR-interacting proteins (Figure 2) and the ability of PELP1/MNAR to interact with histones and histone-modifying components and transcriptional regulators suggest that it plays a role in chromatin modifications and couples NRs with different signaling components.
Figure 2. PELP1/MNAR interactome.
Schematic representation of PELP1-interacting proteins, grouped on the basis of putative function.
Expression
PELP1/MNAR is expressed in a wide variety of tissues, with the highest levels of expression noted in the brain, testes, ovaries, and uterus [Greger et al., 2006; Khan et al., 2005; Pawlak and Beyer, 2005; Vadlamudi et al., 2001]. In the mouse brain, PELP1/MNAR expression is developmentally regulated, and researchers have observed widespread PELP1/MNAR expression in the cortex, hypothalamus, and midbrain, as well as in neurons and astroglia [Pawlak and Beyer, 2005]. In the rat brain, a study reported high-intensity PELP1/MNAR staining in the known targets of estrogen/steroid actions, including the hippocampus, cortex, hypothalamus, amygdala, and septum [Khan et al., 2005]. PELP1 expression is developmentally regulated in mammary glands, with the highest levels of expression found during pregnancy [Vadlamudi et al., 2001]. PELP1 expression is upregulated by estrogen signaling and differentially regulated by selective estrogen receptor modulators [Mishra et al., 2004a]. The PELP1/MNAR promoter region contains several consensus sites for retinoic acid receptor, and 9-cis RA transcriptionally upregulates the expression of the PELP1 gene and PELP1 protein [Singh et al., 2006].
Cellular mechanism and cellular biology
Subcellular distribution
PELP1/MNAR contains a central consensus nuclear localization site and exhibits both cytoplasmic and nuclear localization, depending on the tissue [Vadlamudi et al., 2001]. PELP1/MNAR predominantly resides in the nuclear compartment of hormonally responsive tissues [Nair et al., 2004; Vadlamudi et al., 2001]. Immunohistochemical studies using a variety of mouse tissues revealed PELP1 staining in all of the tissues examined, but that PELP1 was localized differently in each organ [Vadlamudi et al., 2001]. The protein was present in both the nuclei and cytoplasm to differing degrees, depending on the tissue. In ovaries and fallopian tubes in pregnant mice, PELP1/MNAR was localized predominantly in the nuclei of cells from the corpus luteum and the epithelium of fallopian tubes, but considerable cytoplasmic staining also occurred in these cells. In the lung, only parts of the epithelial cell nuclei were stained for PELP1/MNAR, whereas in the testis, PELP1/MNAR was prominently localized in the nuclei. Using silver-enhanced nanogold and immunofluorescence staining of rat brain tissue, researchers showed that PELP1/MNAR was localized primarily in the nuclei of cells in various brain regions, with scattered staining in the cytoplasm and plasma membrane compartments [Khan et al., 2005]. In endometrium, PELP1/MNAR exhibits distinct localization depending on the phase of the endometrium. In the proliferative and secretory phases, researchers showed that PELP1 was expressed in both the glandular and stromal compartments, and was localized in both the nucleus and cytoplasm of these cells [Vadlamudi et al., 2004b]. In contrast, in the postmenopausal phases, researchers observed PELP1 staining only in the glandular compartment, and that PELP1 expression was confined to the cytoplasm of these cells [Vadlamudi et al., 2004b]. Studies also reported PELP1/MNAR expression in all of the cell lines commonly used in laboratory research, including cancer cell lines; however, the level of expression varied among the cells [Greger et al., 2006; Vadlamudi et al., 2001]. In certain cancer cells, researchers reported the association of a small portion of PELP1/MNAR with the membrane [Greger et al., 2006; Vadlamudi et al., 2005b]. Within the nucleus, PELP1/MNAR appears to be present in several subcompartments, including the chromatin, nucleoplasm, and nuclear matrix [Nair et al., 2004].
Posttranslational modifications
PELP1/MNAR is a phosphoprotein and is phosphorylated on both serines/threonines and tyrosines. Bioinformatics analysis using the Motif Scan database (http://scansite.mit.edu/cgi-bin/motifscan_seq) indicated that PELP1/MNAR contains several potential sites for phosphorylation, including 8 tyrosine kinase/phosphatase sites (recognized by EGFR, platelet-derived growth factor receptor, insulin receptor, Src, Jak2, and SHP1) and 207 serine/threonine kinase/phosphatase motifs (recognized by AKT, glycogen synthase kinase, CDK, casein kinase 1, casein kinase 2, LKB1, mitogen-activated protein kinase [MAPK], protein kinase C, protein kinase A [PKA], and proline-directed kinases). A recent study showed that PELP1/MNAR is phosphorylated by PKA and that PKA phosphorylates PELP1 at Ser-350, Ser-415, and Ser-613 (Nagpal and Vadlamudi, unpublished results). Another study using phosphoproteomic analysis revealed that PELP1/MNAR is phosphorylated at Thr-745 in the developing brain [Ballif et al., 2004]. PELP1/MNAR phosphorylation is modulated by hormones and growth factor signaling. Accordingly, treatment with EGF promotes tyrosine as well as serine phosphorylation of PELP1 [Vadlamudi et al., 2005b]. Estrogen treatment is shown to promote tyrosine phosphorylation of PELP1 at Tyr920 [Greger et al., 2007].
Enzyme activity
No studies have shown an association between PELP1/MNAR and known enzymatic activity. However, emerging evidence suggests that PELP1/MNAR functions as a scaffolding protein by coupling various signaling complexes with NRs. PELP1 associates with chromatin and interacts with histones [Nair et al., 2004]. Although PELP1/MNAR itself has no histone acetyltransferase activity, it can recruit other coregulators with histone acetyltransferase activity, such as CBP and p300 [Vadlamudi et al., 2001]. In addition, PELP1/MNAR interacts with components of histone deacetylase complexes, including the nucleosome remodeling and histone deacetylation complex protein MTA1 [Mishra et al., 2003] and histone deacetylase 2 [Choi et al., 2004]. The ability of PELP1/MNAR to interact with histones, histone acetyltransferase enzymes, and histone deacetylase enzymes suggests that PELP1/MNAR promotes alteration in local chromatin structure in the vicinity of the NR target promoters by coupling NRs with chromatin-modifying enzymes and by displacing histone H1 [Nair et al., 2004]. A recent study also showed that PELP1 is associated with complexes that have a histone lysine methyltransferase and a histone lysine demethylase, suggesting that PELP1 has some function in these complexes [Rosendorff et al., 2006]. Furthermore, punctate subnuclear localization and nuclear matrix association of PELP1 suggest that PELP1 functions as a landing platform for several other chromatin remodeling complexes, thereby regulating gene expression. Emerging evidence indicates that PELP1/MNAR also plays a key role in nongenomic ER activity. PELP1/MNAR modulates the interaction of ER with Src, stimulating Src enzymatic activity and MAPK pathway activation [Wong et al., 2002]. Mechanistically, PELP1/MNAR interacts with the SH3 domain of c-Src via its N-terminal PXXP motif, and ER interacts with the SH2 domain of Src at phosphotyrosine 537; the MNAR-ER interaction further stabilizes this trimeric complex, leading to activation of the Ras/MAPK pathway [Barletta et al., 2004]. PELP1/MNAR also directly interacts with the p85 subunit of PI3K and enhances PI3K activity, leading to activation of the AKT pathway [Vadlamudi et al., 2005b].
Target genes
PELP1 does not appear to have a DNA-binding domain. However, its ability to interact with several NRs suggests that it enhances the transcription of target genes via its interactions with NRs [Vadlamudi et al., 2001; Wong et al., 2002]. Accordingly, chromatin immunoprecipitation analysis has shown that PELP1 is recruited to the promoters of ERα target genes, including pS2, PR, and IGF [Nair et al., 2004]. PELP1/MNAR regulates cyclin D1 expression at the transcriptional level; such regulation may involve functional interactions between PELP1 and pRb [Balasenthil and Vadlamudi, 2003]. Estrogen stimulation leads to enhanced recruitment of PELP1/MNAR to the MTA3 promoter chromatin, implying a role for PELP1/MNAR in the regulation of MTA3 [Mishra et al., 2004b]. Additionally, growth factor signals promote PELP1/MNAR interactions with STAT3; these interactions play an important role in growth factor-mediated activation of STAT3 target genes, including cyclin D1, fos, and jun [Manavathi et al., 2005]. In response to 9-cis RA stimulation, PELP1/MNAR enhances the expression of CRBP II via its interaction with RXR [Singh et al., 2006].
Chromatin remodeling and transcriptional activation
PELP1/MNAR exhibits a predominantly nuclear localization in many normal tissues. Within the nuclear compartment, PELP1 localizes to the nucleoplasm, chromatin, and nuclear matrix and is recruited to the promoters of several NR target genes [Nair et al., 2004]. PELP1/MNAR co-localizes with acetylated histones and interacts with the acetyltransferases, CBP and p300, and the PELP1/MNAR-associated histone acetyltransferase activity increases upon ligand-based treatment [Nair et al., 2004]. Researchers have shown reduced histone H1 residence in target gene promoters during the time that PELP1 occupies these promoters [Nair et al., 2004]. PELP1 is also associated with deacetylases, including components of nucleosome remodeling and the histone deacetylation complex [Mishra et al., 2003], and inhibition of deacetylase activity increases the PELP1 residency time at the target gene promoter [Nair et al., 2004]. Furthermore, a recent study reported that PELP1 exists as a complex with methyltransferases and methylases [Rosendorff et al., 2006]. PELP1 overexpression maintains NR-mediated nucleosomal alterations for an extended period [Nair et al., 2004]. Collectively, these data suggest that PELP1 contributes to alteration of the local chromatin structure required for the optimal transcriptional response by ligand-bound NRs via its interactions with histones and histone-modifying enzymes. In addition, PELP1/MNAR may play a role in NR-mediated splicing of mRNAs [Nair and Vadlamudi, 2007]. PELP1/MNAR interacts with several components of the spliceosome machinery, including breast carcinoma amplified sequence 2 and splicing factor 3b, and co-localizes with the splicing factor SC35 in nuclear speckles. PELP1/MNAR has an RNA-binding domain, interacts with RNA in in vitro assays, and enhances steroid hormone-mediated splicing in minigene assays [Nair and Vadlamudi, 2007]. These emerging data suggest that PELP1/MNAR mediates NR-mediated RNA splicing in addition to transcriptional activation functions.
Nongenomic signaling
Evolving evidence suggests that in addition to well-studied nuclear functions, the ERα also participates in nongenotropic (cytoplasmic and perhaps, membrane-mediated) signaling via formation of a multiprotein complex involving ERα, Src kinase, PELP1/MNAR, PI3K, SHC, and G-proteins, collectively called the “signalsome” [Song and Santen, 2006]. Substantial evidence suggests that PELP1/MNAR participates in cytoplasmic and membrane-mediated signaling events through stimulation of the Src kinase, MAPK, PI3K, and STAT3 [Manavathi and Kumar, 2006]. Emerging evidence suggests that PELP1/MNAR is one of the coregulators that facilitate NR interactions with cytoplasmic signaling components [Rajhans and Vadlamudi, 2006; Vadlamudi et al., 2004b]. Researchers initially identified PELP1/MNAR as a Src SH2-binding protein [Joung et al., 1996], and some portion of PELP1/MNAR exists in the cytoplasm and at the membranes [Nair et al., 2004; Vadlamudi et al., 2004b]. PELP1/MNAR modulates ER’s interaction with c-Src, stimulating c-Src enzymatic activity and leading to activation of the mitogen activated protein kinase (MAPK) pathway [Wong et al., 2002]. Mutational analysis of ERα and c-Src mutants revealed that MNAR interacts with the c-Src SH3 domain via its N-terminal PXXP motif. ERα interacts with Src’s SH2 domain at phosphotyrosine 537, and the MNAR-ER interaction further stabilizes this complex . In addition, PELP1/MNAR also directly interacts with the p85 subunit of PI3K and enhances PI3K activity [Vadlamudi et al., 2005b]. A recent study reported direct correlation between MNAR expression levels and E2-induced activation of PI3 and Akt kinases. In their model system, E2 treatment induced complex formation of endogenous MNAR, ERα, c-Src, and p85, the regulatory subunit of PI3 kinase. The interaction between p85 and MNAR required activation of c-Src and MNAR phosphorylation on Tyr920. These results suggest that PELP1/MNAR plays a key role in the E2-induced regulation of cell proliferation and apoptosis via its regulation of the PI3K-AKT pathway [Greger et al., 2007]. EGF promotes PELP1/MNAR association with the EGFR, resulting in the tyrosine phosphorylation of PELP1/MNAR [Vadlamudi et al., 2005b]. PELP1/MNAR can enhance EGF-mediated ERα transactivation, and mislocalization of PELP1/MNAR in the cytoplasm can increase ERα basal activity via the EGFR and PI3K pathways. Using a yeast two-hybrid screen, [Rayala et al., 2006a] demonstrated the physiological interaction between HRS and PELP1/MNAR. Interestingly, HRS sequesters PELP1/MNAR in the cytoplasm, leading to EGFR-dependent activation of MAPK. PELP1/MNAR can interact with several growth factor signaling components, including STAT3, and may have important functional implications in ERα/growth factor crosstalk [Vadlamudi et al., 2005b].
Cell cycle progression
Evidence suggests that PELP1/MNAR contributes to E2-mediated G1/S-phase progression [Balasenthil and Vadlamudi, 2003]. In this study, overexpression of PELP1/MNAR in breast cancer model cells was accompanied by persistent hyperphosphorylation of the cell cycle switch protein pRb in an E2-dependent manner, with increases in the rate of cell proliferation. PELP1/MNAR interacts with pRb via its C-terminal pocket domain, and PELP1/pRb interactions play a role in the maximal activation of E2 target genes such as cyclin D1. The results of this study suggested that PELP1/MNAR positively contributes to E2-mediated G1/S-phase progression and plays a permissive role in E2-mediated cell cycle progression, presumably via its regulatory interactions with the pRb pathway. Increased PELP1/MNAR expression in a mammary gland during pregnancy, when the rate of cell proliferation is high, supports a physiological role for PELP1/MNAR in E2-mediated cell cycle progression in mammary glands [Vadlamudi et al., 2001]. In addition to mitosis, emerging evidence indicates that PELP1/MNAR plays a role in meiosis. For example, in Xenopus oocytes, PELP1/MNAR interacts with androgen receptor (AR) and appears to mediate inhibition of meiosis via Gβγ signaling. PELP1/MNAR is widely expressed in oocytes, and reduction of its expression by RNA interference markedly enhances testosterone-triggered maturation and activation of MAPK. Furthermore, PELP1/MNAR appears to participate in maintaining meiotic arrest by directly enhancing G-mediated inhibition of meiosis and androgen binding to AR, releasing this inhibition, which allows maturation to occur [Haas et al., 2005].
Apoptosis and differentiation
PELP1/MNAR also appears to participate in cellular differentiation. In one study, PELP1/MNAR potentiated the effects of 9-cis RA and peroxisome proliferator-activated receptor (PPAR)-specific ligands by functioning as a coactivator of RXR homodimers and RXR-PPAR heterodimers [Singh et al., 2006]. Furthermore, the ligand 9-cis RA transcriptionally upregulated expression of the PELP1/MNAR gene. Functioning as a coactivator of the RXR-PPAR heterodimer, PELP1 was able to potentiate the differentiation-inducing effects of PPAR ligand, suggesting a novel facet of PELP1/MNAR function as a potentiator of antiproliferative and differentiation effects of RXR-specific ligands and PPAR-specific ligands. Because nongenomic functions of NRs are implicated in different cellular processes, including cell survival and apoptosis, PELP1/MNAR-mediated nongenomic actions may also play a role in PELP1-mediated apoptosis and differentiation.
Disease
Oncogenesis
Emerging evidence suggests that PELP1/MNAR has tumorigenic potential. PELP1/MNAR interacts with and modulates functions of several proto-oncogenes, including c-Src, STAT3, and EGFR [Rajhans and Vadlamudi, 2006]. In a recent study, PELP1 deregulation was shown to promote cell transformation in NIH3T3 fibroblast cell-based focus formation assays [Rajhans and Vadlamudi, 2006]. PELP1/MNAR also enhances the transformation potential of c-Src and other proto-oncogenes in focus formation assays. Similarly, PELP1-transfected (but not vector-transfected) rat kidney epithelial RK3E cells exhibited transformation potential. A PELP1 mutant that uniquely localizes in the cytoplasm also demonstrated focus formation, although to a lesser degree than that of PELP1-WT [Rajhans, In Press]. Vector-transfected RK3E cells are incapable of promoting anchorage-independent growth and the PELP1-WT-transfected RK3E cells showed a significant increase in colony formation. In addition, PELP1/MNAR promotes anchorage-independent growth of breast cancer cells in soft agar assays [Rajhans, In Press; Vadlamudi et al., 2005b]. In these studies, when injected subcutaneously into the mammary fat pad of mice in the absence of treatment with exogenous E2, MCF-7 human breast cancer cells stably expressing PELP1 showed tumor formation in 50% of the injected sites. These results suggest that deregulation of PELP1/MNAR expression is sufficient to promote hormone-independent growth of MCF-7 cells. Accordingly, PELP1/MNAR is widely expressed in breast cancer cells [Greger et al., 2006; Vadlamudi et al., 2001], and PELP1 expression and localization are deregulated in breast tumors [Vadlamudi et al., 2004a; Vadlamudi et al., 2001]. Also, PELP1/MNAR expression and localization are widely deregulated in endometrial cancers [Vadlamudi et al., 2004b]. In addition, PELP1/MNAR and ERα are localized predominantly in the cytoplasm in high-grade endometrial tumor cells, suggesting that PELP1/MNAR deregulation plays a role in endometrial cancer progression [Vadlamudi et al., 2004b]. Ongoing studies using human ovarian cancer tissue arrays have revealed that PELP1 is overexpressed twofold to threefold in 60% of ovarian tumors. PELP1 is deregulated in several ovarian tumor subtypes, including serous tumors, endometrioid tumors, clear cell carcinomas, and mucous tumors, and is predominantly localized in the cytoplasm in ovarian tumors (Chakravarty and Vadlamudi, unpublished results). Salivary duct carcinoma is a high-grade neoplasm with a morphology similar to that of mammary duct carcinoma. Interestingly, these tumors express PELP1/MNAR and ERα, and PELP1/MNAR signaling may therefore play a role in salivary tumorigenesis [Vadlamudi et al., 2005a]. Also, researchers reported elevated PELP1/MNAR expression in high-grade prostate tumors [Nair et al., 2007]. In that study, PELP1/MNAR staining was predominantly localized in the nuclear compartment, and increased PELP1 expression was correlated with increased prostate tumor grade.
Role of PELP1 in metastasis
PELP1/MNAR interacts with several proteins involved in cytoskeleton remodeling, including Src kinase, PI3K, and four-and-a-half LIM-only protein 2, and participates in E2-mediated nongenomic signaling pathways [Gururaj et al., 2006; Vadlamudi et al., 2005b; Wong et al., 2002]. Also, PELP1/MNAR overexpression uniquely enhances E2-mediated ruffles and filopodium-like structures. A recent study showed that in Boyden chamber assays, PELP1/MNAR-overexpressing MCF-7 cells displayed increased cell motility upon treatment with E2, whereas knockdown of PELP1/MNAR expression by small interfering RNA substantially reduced E2-mediated cell motility compared with that in control MCF-7 cells [Rajhans, In Press]. PELP1 modulates functions of MTA1, a protein implicated in metastasis. PELP1/MNAR also interacts with the MTA1-associated coactivator and promotes ERα-transactivation functions in a synergistic manner [Mishra et al., 2003]. Additionally, PELP1/MNAR modulates expression of MTA3, a gene implicated in the invasive growth of human breast cancers. The ability of PELP1/MNAR to recruit to the MTA3 promoter chromatin [Mishra et al., 2004b] and its interactions with other MTA family members, suggest that deregulation of PELP1/MNAR expression promotes metastasis. A recent study measured the expression levels of PELP1 by IHC examination and the results showed that when compared with node-negative specimens, node-positive and metastatic tumors exhibited increased PELP1 expression [Rajhans, In Press]. Statistical analysis revealed that PELP1 expression was positively correlated with cancer grade and node status. The number of samples with a high level (score 3) of PELP1 staining increased as tumors progressed from grade 1 to grade 2 or 3 (P = 0.005). Similarly, node-positive and metastatic tumors exhibited a greater expression of PELP1 than did node-negative tumors (P = 0.003). No significant correlation of PELP1 expression with ER, PR, patient age, or tumor stage was observed. These results suggest that PELP1 expression may be altered in higher grade node-positive and metastatic tumors [Rajhans, In Press]. The ability of PELP1/MNAR to interact with various enzymes that modulate the cytoskeleton and its deregulation in metastatic breast tumors suggest that PELP1/MNAR signaling plays a role in tumor cell migration and metastasis.
Role in hormonal independence
Although PELP1/MNAR is predominantly localized in the nuclei of hormonally responsive tissue cells, in one study it was shown to be localized in either the cytoplasm alone or the cytoplasm and nuclei in 58% of PELP1/MNAR-positive breast tumor cells [Vadlamudi et al., 2005b]. In that study, cells that mimicked PELP1/MNAR cytoplasmic localization in tumors were hypersensitive to E2, but resistant to tamoxifen. Altered localization of PELP1/MNAR to the cytoplasm was sufficient to trigger its interaction with the p85 subunit of PI3K, leading to activation of PI3K [Vadlamudi et al., 2005b]. Furthermore, PELP1/MNAR exhibits phase-dependent localization and expression in the endometrium and enhances tamoxifen-mediated partial agonist signaling in endometrial cancer cells [Vadlamudi et al., 2004b]. The normal, cytoplasmic localization of PELP1/MNAR in the endometrium of postmenopausal women suggests that PELP1/MNAR-mediated nongenomic functions may play a role in endometrial cell survival under low estrogen conditions. However, its exclusive cytoplasmic localization, as seen in a subset of breast cancer cells, may provide survival advantage to the cancer cells, and thus may contribute to tumorigenic potential. Furthermore, cytoplasmic PELP1/MNAR is also shown to interact with the trafficking molecule HRS to activate MAPK in the presence of EGFR [Rayala et al., 2006a]. In another study, clones of MCF-7 cells overexpressing cytoplasmic PELP1/MNAR were more sensitive to tumor necrosis factor-α-induced apoptosis than cells expressing wild-type nuclear PELP1/MNAR [Rayala et al., 2006b]. The results of this study suggest that altered localization of PELP1/MNAR modulates the hormonal sensitivity of breast cancer cells, thus paving the way for development of new treatment strategies for breast tumors with cytoplasmic PELP1 expression. Deregulation of PELP1/MNAR expression increases the expression of aromatase, an enzyme that produces estrogen locally [Rajhans and Vadlamudi, 2006]. Recent data suggest that AR, PELP1/MNAR, and Src form constitutive complexes in prostate cancer model cells that exhibit androgen independence, and constitutive activation of the Src/MAPK kinase 1/2/extracellular signal-related kinase 1/2/CRE-binding protein pathway is associated with the androgen-independence phenotype [Unni et al., 2004]. The mechanisms or modifications that promote PELP1 localization to the cytoplasm remain elusive. Since emerging evidence suggests that coregulator posttranslational modification may play a role in their localization, it is tempting to speculate that posttranslational modification of PELP1/MNAR may play a role in its exclusive cytoplasmic localization. Thus, deregulation of PELP1/MNAR expression has the potential to contribute to the hormonal therapy resistance seen in patients with hormone-dependent cancers by excessively activating nongenomic signaling pathways.
Summary
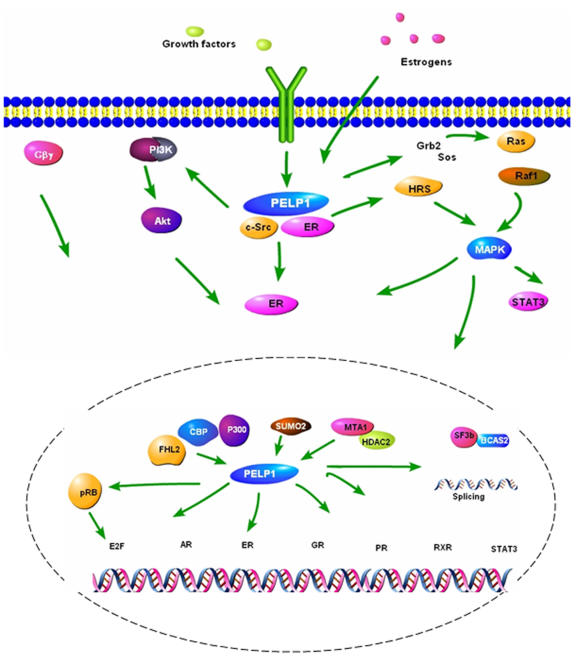
PELP1/MNAR appears to function as a scaffolding protein, coupling NRs with several proteins implicated in oncogenesis (Figure 3). With the enormous potential of PELP1/MNAR as a modulator of NRs and oncogenes, deregulation of PELP1/MNAR expression is likely to provide cancer cells with a survival, growth, metastasis, and hormonal independence advantage. However, the mechanism or mechanisms by which PELP1/MNAR promotes these functions are not clear. PELP1/MNAR functions appear to be regulated by a variety of binding proteins and by posttranslational modifications. Future studies elucidating the molecular mechanism or mechanisms of action of PELP1/MNAR in normal and tumor cells, characterizing the physiological function of PELP1/MNAR using Tg/KO mouse models, identifying PELP1/MNAR target genes, and profiling the expression of PELP1/MNAR in large numbers of tumor samples would allow use of this NR coregulator protein as a novel therapeutic target.
Figure 3. Schematic representation of the current understanding of the PELP1/MNAR signaling pathway.
PELP1 participation in multiple stages of nuclear receptor (genomic and nongenomic) signaling pathways suggests that PELP1 deregulation likely contributes to the development of hormonal independence, tumorigenesis and metastasis.
Acknowledgments
Work in the authors’ laboratories is supported by NIH grants CA095681 (RKV) and CA098823 (RK).
Abbreviations
- AR
Androgen receptor
- CBP
CRE-binding protein-binding protein
- CDK
cyclin-dependent kinase
- EGF
epidermal growth factor
- EGFR
EGF receptor
- ER
estrogen receptor
- HRS
hepatocyte growth factor-regulated tyrosine kinase substrate
- MAPK
mitogen-activated protein kinase
- MNAR
modulator of nongenomic actions of the ER
- MTA
metastasis-associated protein
- NR
nuclear receptor
- PELP
proline-, glutamic acid-, and leucine-rich protein
- PI3K
phosphatidylinositol-3 kinase
- PKA
protein kinase A
- PPAR
peroxisome proliferator-activated receptor
- pRb
retinoblastoma protein
- RA
retinoic acid
- RXR
retinoid X receptor
- SH
Src homology
- STAT
signal transducer and activator of transcription
References
- Balasenthil S., Vadlamudi R. K. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278:22119–27. doi: 10.1074/jbc.M212822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif B. A., Villen J., Beausoleil S. A., Schwartz D., Gygi S. P. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- Barletta F., Wong C. W., McNally C., Komm B. S., Katzenellenbogen B., Cheskis B. J. Characterization of the interactions of estrogen receptor and MNAR in the activation of cSrc. Mol Endocrinol. 2004;18:1096–108. doi: 10.1210/me.2003-0335. [DOI] [PubMed] [Google Scholar]
- Choi Y. B., Ko J. K., Shin J. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J Biol Chem. 2004;279:50930–41. doi: 10.1074/jbc.M406831200. [DOI] [PubMed] [Google Scholar]
- Greger J. G., Guo Y., Henderson R., Ross J. F., Cheskis B. J. Characterization of MNAR expression. Steroids. 2006;71:317–22. doi: 10.1016/j.steroids.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Greger J. G., Fursov N., Cooch N., McLarney S., Freedman L. P., Edwards D. P., Cheskis B. J. Phosphorylation of MNAR promotes estrogen activation of phosphatidylinositol 3-kinase. Mol Cell Biol. 2007;27:1904–13. doi: 10.1128/MCB.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gururaj A. E., Rayala S. K., Vadlamudi R. K., Kumar R. Novel mechanisms of resistance to endocrine therapy: genomic and nongenomic considerations. Clin Cancer Res. 2006;12:1001s–1007s. doi: 10.1158/1078-0432.CCR-05-2110. [DOI] [PubMed] [Google Scholar]
- Haas D., White S. N., Lutz L. B., Rasar M., Hammes S. R. The modulator of nongenomic actions of the estrogen receptor (MNAR) regulates transcription-independent androgen receptor-mediated signaling: evidence that MNAR participates in G protein-regulated meiosis in Xenopus laevis oocytes. Mol Endocrinol. 2005;19:2035–46. doi: 10.1210/me.2004-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung I., Strominger J. L., Shin J. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc Natl Acad Sci U S A. 1996;93:5991–5. doi: 10.1073/pnas.93.12.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. M., Hadman M., Wakade C., De Sevilla L. M., Dhandapani K. M., Mahesh V. B., Vadlamudi R. K., Brann D. W. Cloning, expression, and localization of MNAR/PELP1 in rodent brain: colocalization in estrogen receptor-α- but not in gonadotropin-releasing hormone-positive neurons. Endocrinology. 2005;146:5215–27. doi: 10.1210/en.2005-0276. [DOI] [PubMed] [Google Scholar]
- Manavathi B., Nair S. S., Wang R. A., Kumar R., Vadlamudi R. K. Proline-, glutamic acid-, and leucine-rich protein-1 is essential in growth factor regulation of signal transducers and activators of transcription 3 activation. Cancer Res. 2005;65:5571–7. doi: 10.1158/0008-5472.CAN-04-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B., Kumar R. Steering estrogen signals from the plasma membrane to the nucleus: two sides of the coin. J Cell Physiol. 2006;207:594–604. doi: 10.1002/jcp.20551. [DOI] [PubMed] [Google Scholar]
- Mishra S. K., Balasenthil S., Nguyen D., Vadlamudi R. K. Cloning and functional characterization of PELP1/MNAR promoter. Gene. 2004a;330:115–22. doi: 10.1016/j.gene.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Mishra S. K., Mazumdar A., Vadlamudi R. K., Li F., Wang R. A., Yu W., Jordan V. C., Santen R. J., Kumar R. MICoA, a novel metastasis-associated protein 1 (MTA1) interacting protein coactivator, regulates estrogen receptor-α transactivation functions. J Biol Chem. 2003;278:19209–19. doi: 10.1074/jbc.M301968200. [DOI] [PubMed] [Google Scholar]
- Mishra S. K., Talukder A. H., Gururaj A. E., Yang Z., Singh R. R., Mahoney M. G., Franci C., Vadlamudi R. K., Kumar R. Upstream determinants of estrogen receptor-α regulation of metastatic tumor antigen 3 pathway. J Biol Chem. 2004b;279:32709–15. doi: 10.1074/jbc.M402942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Vadlamudi R. K. Emerging significance of ER-coregulator PELP1/MNAR in cancer. Histol Histopathol. 2007b;22:91–6. doi: 10.14670/hh-22.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S. S., Mishra S. K., Yang Z., Balasenthil S., Kumar R., Vadlamudi R. K. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–23. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- Nair S. S., Guo Z., Mueller J. M., Koochekpour S., Qiu Y., Tekmal R. R., Schule R., Kung H. J., Kumar R., Vadlamudi R. K. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007a;21:613–24. doi: 10.1210/me.2006-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak J., Beyer C. Developmental expression of MNAR mRNA in the mouse brain. Cell Tissue Res. 2005;320:545–9. doi: 10.1007/s00441-005-1090-z. [DOI] [PubMed] [Google Scholar]
- Rajhans R., Vadlamudi R. K. Comprehensive analysis of recent biochemical and biologic findings regarding a newly discovered protein-PELP1/MNAR. Clin Exp Metastasis. 2006;23:1–7. doi: 10.1007/s10585-006-9019-9. [DOI] [PubMed] [Google Scholar]
- Rajhans R., Nair, S., Holden, A.H., Kumar, R., Tekmal, R., and Vadlamudi, R.K. Oncogenic potential of the nuclear receptor coregulator PELP1/MNAR. Cancer Res. In Press doi: 10.1158/0008-5472.CAN-06-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala S. K., Mascarenhas J., Vadlamudi R. K., Kumar R. Altered localization of a coactivator sensitizes breast cancer cells to tumor necrosis factor-induced apoptosis. Mol Cancer Ther. 2006b;5:230–7. doi: 10.1158/1535-7163.MCT-05-0276. [DOI] [PubMed] [Google Scholar]
- Rayala S. K., Hollander P., Balasenthil S., Molli P. R., Bean A. J., Vadlamudi R. K., Wang R. A., Kumar R. Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) interacts with PELP1 and activates MAPK. J Biol Chem. 2006a;281:4395–403. doi: 10.1074/jbc.M510368200. [DOI] [PubMed] [Google Scholar]
- Rosendorff A., Sakakibara S., Lu S., Kieff E., Xuan Y., DiBacco A., Shi Y., Shi Y., Gill G. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci U S A. 2006;103:5308–13. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. R., Gururaj A. E., Vadlamudi R. K., Kumar R. 9-cis-retinoic acid up-regulates expression of transcriptional coregulator PELP1, a novel coactivator of the retinoid X receptor α pathway. J Biol Chem. 2006;281:15394–404. doi: 10.1074/jbc.M601593200. [DOI] [PubMed] [Google Scholar]
- Song R. X., Santen R. J. Membrane initiated estrogen signaling in breast cancer. Biol Reprod. 2006;75:9–16. doi: 10.1095/biolreprod.105.050070. [DOI] [PubMed] [Google Scholar]
- Unni E., Sun S., Nan B., McPhaul M. J., Cheskis B., Mancini M. A., Marcelli M. Changes in androgen receptor nongenotropic signaling correlate with transition of LNCaP cells to androgen independence. Cancer Res. 2004;64:7156–68. doi: 10.1158/0008-5472.CAN-04-1121. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R. K., Balasenthil S., Broaddus R. R., Gustafsson J. A., Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004a;89:6130–8. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi R. K., Bagheri-Yarmand R., Yang Z., Balasenthil S., Nguyen D., Sahin A. A., den Hollander P., Kumar R. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004b;5:575–85. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R. K., Manavathi B., Balasenthil S., Nair S. S., Yang Z., Sahin A. A., Kumar R. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005a;65:7724–32. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi R. K., Wang R. A., Mazumdar A., Kim Y., Shin J., Sahin A., Kumar R. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor α. J Biol Chem. 2001;276:38272–9. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi R. K., Balasenthil S., Sahin A. A., Kies M., Weber R. S., Kumar R., El-Naggar A. K. Novel estrogen receptor coactivator PELP1/MNAR gene and ERbeta expression in salivary duct adenocarcinoma: potential therapeutic targets. Hum Pathol. 2005b;36:670–5. doi: 10.1016/j.humpath.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Wong C. W., McNally C., Nickbarg E., Komm B. S., Cheskis B. J. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc Natl Acad Sci U S A. 2002;99:14783–8. doi: 10.1073/pnas.192569699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]