Abstract
The use of the zebrafish as a model organism for the analysis of cardiac development is no longer proof-of-principle science. Over the last decade, the identification of a variety of zebrafish mutations and the subsequent cloning of mutated genes have revealed many critical regulators of cardiogenesis. More recently, increasingly sophisticated techniques for phenotypic characterization have facilitated analysis of the specific mechanisms by which key genes drive cardiac specification, morphogenesis, and function. Future enrichment of the arsenal of experimental strategies available for zebrafish should continue the yield of high returns from such a small source.
Keywords: heart, zebrafish, fate mapping, imaging, review
1. Benefits of the zebrafish as a model organism: making the most of a little heart
The road to creating a fully functional heart is complex. Heart formation begins with the specification of appropriate numbers and types of cardiac cells. Next, these cells coordinate their migration and interaction in order to assemble a simple, yet functional, heart tube. Further morphogenesis transforms the heart tube into morphologically and functionally discrete cardiac chambers. Even minor perturbations of cardiac specification or morphogenesis can have catastrophic consequences for cardiac function. To understand the etiology of congenital heart defects, it is essential to dissect the individual processes that combine to create the intricacy of the mature heart.
An ideal model organism for the study of heart formation would allow high-resolution inspection of cellular activities in conjunction with genetic analysis of regulatory influences. The zebrafish fulfills these criteria beautifully, and it has therefore been steadily gaining in popularity as a valuable model of cardiogenesis [1, 2]. A large part of its appeal derives from the transparency of the zebrafish embryo, which permits detailed examination of heart size, shape, and function. Additionally, the zebrafish presents excellent opportunities for conducting forward genetic screens, primarily due to its small size, fecundity, and brief generation time [3]. Screening for cardiac phenotypes is particularly convenient, since the zebrafish embryo does not require a functional cardiovascular system for survival during embryogenesis [4]. The first two large-scale zebrafish screens, completed in 1996, clearly illustrated the effectiveness of zebrafish genetics [5, 6]. Together, these screens found more than 1600 mutations affecting specific aspects of zebrafish embryogenesis; more than 100 of these mutations, representing 66 loci, disrupt cardiac form or function [7, 8].
The availability of a large collection of zebrafish mutations provided investigators with a plethora of opportunities to identify new genes with critical roles in cardiogenesis. Indeed, subsequent studies of these mutations, together with mutations found in independent screens (e.g. [9, 10]), have led to the cloning of more than 35 of the mutated genes, many of which are discussed below. This catalog of genes involved in cardiogenesis is impressive, yet it represents only the first step toward understanding how these genes function. Take, for example, a mutant exhibiting a small heart phenotype that is caused by a lesion in a gene encoding a transcription factor. An initial interpretation of these data would implicate this factor in the regulation of heart size, but deeper analysis would be required to reveal its precise role. Does this transcription factor promote the accumulation of cardiomyocytes by inducing cardiac specification or controlling proliferation? Does it influence heart size by regulating the size of individual cardiomyocytes? Does it control the three-dimensional organization of cells within the cardiac chambers? To distinguish between such hypothetical models of gene function, it is essential to have on hand the appropriate techniques for detailed characterization of mutant phenotypes.
In the past several years, a number of zebrafish laboratories have prevailed in moving beyond the level of gene discovery: in addition to using zebrafish genetics and genomics to rapidly clone mutated genes, they have taken advantage of the optical attributes of the embryo by developing experimental strategies and techniques suitable for high-resolution phenotypic analysis. In this review, we will highlight three categories of imaging-based techniques that have been particularly helpful in illuminating specific aspects of zebrafish cardiogenesis: fate mapping strategies for the analysis of cardiac specification, visualization of the dynamic cytoarchitecture that underlies cardiac morphogenesis, and high-speed imaging for the investigation of cardiac function. Far from being a comprehensive review, the studies recognized here serve a sample and a prelude of the potential for studying cardiac development in zebrafish.
2. Fate mapping: a retrospective approach to identifying progenitor potential
Though morphologically simpler than its mammalian counterpart, the physical components of the zebrafish heart are fundamentally similar [1]. The zebrafish heart has two major contractile chambers, a ventricle and an atrium. Each chamber is composed of an outer layer of myocardium and an inner layer of endocardium. During remodeling of the primitive heart tube, the ventricular myocardial wall thickens, supporting extensive trabeculation that is largely absent in the atrium [11]. In addition to displaying unique morphologies, the cardiac chambers are also molecularly distinct; for instance, they express chamber-specific myosin heavy chain genes, vmhc in the ventricle and amhc in the atrium [12, 13]. The formation of cardiac chambers has its roots in the specification and patterning of cardiac progenitors (CPs) [14]. Prior to their differentiation, CPs reside in bilateral heart fields where they receive inductive signals that promote their determination as well as their subdivision into ventricular and atrial populations. In addition to inductive signaling, the heart fields are also subject to inhibitory signaling that limits their effective size. Thus, cardiac specification requires a combination of signals and downstream effectors to insure the production of an appropriate quantity and variety of cardiomyocytes.
A number of zebrafish mutations have potent effects on cardiomyocyte production, and studies of these mutations have implicated several signaling pathways and downstream transcription factors in cardiac specification. In acerebellar (fgf8) and swirl (bmp2b) mutants, the number of cardiomyocytes is severely decreased; this deficiency is preceded by gene expression defects, including reduced expression of the transcription factor gene nkx2.5 [15, 16]. A similar phenotype is observed in one-eyed pinhead mutants, which lack an essential coreceptor in the Nodal signaling pathway [16]. Furthermore, in acerebellar and one-eyed pinhead mutants, the deficiency of ventricular tissue is more severe than the deficiency of atrial tissue, suggesting roles for Fgf8 and Nodals in chamber fate assignment [15, 16]. Mutations in the transcription factor gene faust (gata5) also result in reduced expression of nkx2.5 and severe deficiencies in cardiomyocyte production, especially for ventricular cardiomyocytes [17]. Epistasis experiments suggest that Gata5 functions downstream of Bmp2b and Nodal signaling [16]. Mutants lacking the transcription factor gene hands off (hand2) also produce too few cardiomyocytes but do not exhibit defects in nkx2.5 or gata5 expression, suggesting a downstream or parallel role for Hand2 in promoting cardiogenesis [18]. In contrast to mutations causing cardiomyocyte deficiencies, mutation of neckless (retinaldehyde dehydrogenase 2) causes a striking cardiomyocyte surplus [19]. Retinaldehyde dehydrogenase 2 controls the rate-limiting step of retinoic acid (RA) synthesis; thus, this mutant phenotype suggests that RA signaling plays an important role in restricting cardiomyocyte production.
The analysis of zebrafish mutations affecting cardiomyocyte formation suggests strong candidates for signaling pathways that induce or repress cardiac specification. However, there are other possible explanations for the abnormal numbers of cardiomyocytes observed in mutant embryos. For example, perhaps acerebellar mutants lack myocardium because Fgf8 is required for cardiomyocyte proliferation and viability, or perhaps the phenotype of neckless mutants could be explained by excess cardiomyocyte proliferation in the absence of RA signaling. To distinguish between various mechanisms influencing cardiomyocyte production, it is important to have a baseline understanding of where CPs originate in the early embryo and how each CP contributes to the total number of cardiomyocytes; that is, it is critical to define the fate map of CPs in the wild-type embryo.
Through refinement of an established fate mapping protocol [20], Keegan et al. have generated a high-resolution fate map of CPs within the late blastula, thereby providing a valuable reference for the analysis of pathways affecting cardiac specification [21]. This experimental strategy involves labeling individual blastomeres using a noninvasive optical method: laser-mediated activation of a caged fluorescein-dextran conjugate (Figure 1A,B). This lineage tracer can be distributed throughout the embryo via injection at the one-cell stage; cells are later selected for photoactivation simply by focusing the laser on any visible targets. Tracking the progeny of labeled cells indicates their paths through the embryo and their contributions to differentiated tissues (Figure 1C–E, H–J). By correlating cardiac contributions with initial blastomere positions, the wild-type fate map reveals the location of CPs within the lateral marginal zone (LMZ) of the late blastula, the density of CPs among the cells in this multipotential region, and the productivity of a typical CP in terms of the number of cardiomyocytes generated [21]. Additionally, the fate map demonstrates a distinct spatial organization of ventricular and atrial CPs prior to gastrulation: ventricular CPs tend to be located closer to the margin and more dorsal than are atrial CPs (Figure 1F). Although there is overlap between the areas containing ventricular CPs and atrial CPs (Figure 1F), individual blastomeres at this stage do not give rise to both ventricular and atrial cardiomyocytes [21], in accordance with an early separation of ventricular and atrial lineages [22]. Following gastrulation, CPs coalesce to form a portion of the anterior lateral plate mesoderm (ALPM) (Figure 1G,H). The relative organization of ventricular and atrial CPs persists within the ALPM, with ventricular CPs occupying a more medial portion of the ALPM than do ACPs, as demonstrated by a recent fate map of the ALPM during early somitogenesis stages (J.J.S. and D.Y., unpublished data) and by the regionalized expression patterns of vmhc and amhc once cardiac differentiation begins (Figure 1B,C,G,H) [12, 13].
Figure 1.
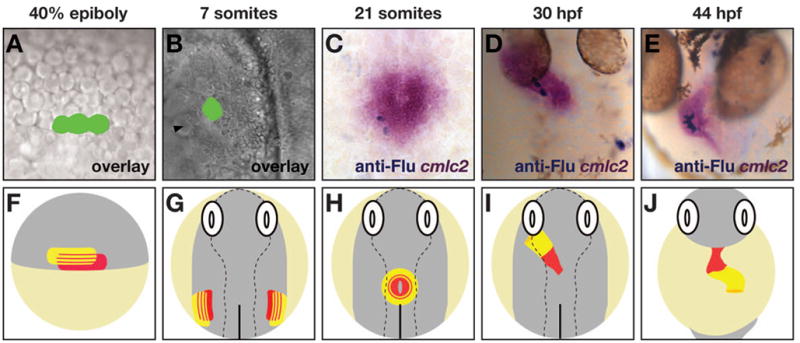
Fate mapping reveals that atrial and ventricular cardiomyocyte organization is preconfigured by atrial and ventricular CP organization preceding differentiation. (A–E) Images illustrating the generation and collection of fate map data. (F–J) Schematics illustrating atrial and ventricular CP organization. (A,B) Pseudo-colored overlay images of blastomeres (A) or ALPM cells (B) that were labeled by laser activation of caged fluorescein, as in [21]. (A) Lateral view, animal pole up, at 40% epiboly stage. In this example, three blastomeres in the first tier above the embryonic margin are labeled. (B) Dorsal view, anterior up, at the 7-somite stage. In this example, a cluster of 10–15 ALPM cells slightly interior to the ALPM edge (black arrowhead) were labeled. (C–E) Embryos processed to score cardiac progeny of labeled cells. Examples are at the 21-somite stage (C; dorsal view, anterior up), 30 hours post fertilization (hpf) (D; dorsal view), and 44 hpf (E; ventral view). In situ hybridization of cmlc2 expression [12] indicates all cardiomyocytes (magenta), and anti-fluorescein immunohistochemistry indicates labeled progeny (blue). (F,G) Schematics depicting territories containing atrial CPs (yellow) and ventricular CPs (red) within the LMZ (F; [21]) and the ALPM (G; J.J.S. and D.Y., unpublished data). (H–J) Schematics depicting the position of atrial (yellow) and ventricular (red) cardiomyocytes within the cardiac cone (H), the heart tube (I), and the cardiac chambers (J), as determined by expression patterns of amhc and vmhc [12, 13].
The wild-type fate map provides valuable insight into the organization of CPs at stages before they are recognizable with molecular markers; the strength of fate mapping as an experimental strategy becomes even more apparent when it is applied to the analysis of embryos exhibiting abnormal cardiomyocyte populations. For example, as mentioned above, one-eyed pinhead mutants display a more striking deficiency of ventricular cardiomyocytes than of atrial cardiomyocytes [16], but this observation alone does not demonstrate whether Nodal signaling promotes ventricular fate assignment, differentiation, growth, or survival. The relative orientation of ventricular and atrial CPs in the late blastula (Figure 1F) raises the possibility that chamber fate assignment could be influenced by differential exposure to Nodal ligands that are secreted from the embryonic margin. To test this hypothesis, alterations to the CP fate map were examined in embryos with reduced levels of Nodal signaling [21]. In contrast to the wild-type fate map, the distribution of CPs shifted toward the margin when Nodal signaling was reduced. In wild-type embryos, the blastomeres closest to the margin never become atrial CPs; in embryos with reduced Nodal signaling, both ACPs and VCPs were found adjacent to the margin. This fate transformation indicates that Nodal signaling promotes the assignment of ventricular fate in CPs derived from the most marginal blastomeres.
In addition to detecting alterations in patterning of the heart fields, fate mapping can be a powerful strategy for monitoring the number of CPs and their production of cardiomyocytes. For instance, the observation that reduction of RA signaling results in an increased number of cardiomyocytes suggested that RA signaling could repress the formation of CPs; alternatively, RA could restrict the number of cardiomyocytes produced by individual CPs. To distinguish between these potential roles of RA, Keegan et al. constructed a CP fate map in embryos treated with a RA receptor antagonist [19]. This analysis revealed that a loss of RA signaling causes a fate transformation that increases the number of CPs within the LMZ, presumably at the expense of other mesendodermal cell types. Although there were more CPs in antagonist-treated embryos, the typical CP appeared to produce a normal number of cardiomyocytes; the average cardiac contribution per CP was comparable (~4–5 cardiomyocytes) to that observed in the wild-type fate map. Thus, RA signaling plays a critical role in restricting the specification of CP identity.
It is clear that several pathways act to induce, limit, and organize cardiac specification within the zebrafish heart fields. Fate mapping of CPs can yield significant insights by providing both visible and quantifiable data for the assessment of CP density, productivity, and organization. As investigators continue to uncover genes that affect the production of ventricular and atrial myocardium, high-resolution fate mapping in the late blastula (Figure 1A,E) and also at subsequent stages (Figure 1B,G) will remain an indispensable tool for defining the impact of these genes on cardiac specification. Ultimately, extensions of these studies should reveal the full landscape of signals responsible for creating the terrain depicted by the CP fate map.
3. Defining cardiac cytoarchitecture: subcellular mechanics of morphogenesis
Once cardiac specification is complete, substantial morphogenetic changes are necessary to create the three-dimensional form of the functional heart [23]. First, as myocardial differentiation progresses, the bilateral populations of cardiomyocytes move medially, eventually merging at the embryonic midline. Through a process called cardiac fusion, they form a ring of cells, referred to as the cardiac cone (Figure 1C,H), that encircles the endocardial precursors. Next, the cardiac cone extends into a linear tube: the cone’s axis gradually lengthens and shifts from a dorsal-ventral plane to an anterior-posterior plane (Figure 1C,D,H,I). Once extension is complete, the heart tube is a muscular cylinder lined with endocardium, and the inner and outer circumferences of the cone have become the arterial and venous apertures of the tube (Figure 1D,I). Next, localized bulges emerge from the walls of the heart tube: this process, called ballooning, creates the characteristic curvatures of the ventricle and the atrium (Figure 1E,J). Chambers are further demarcated by the distinctive constriction of the atrioventricular (AV) canal; morphogenetic changes also occur inside this portion of the heart, where endocardial cushions (ECs) form and are then remodeled into AV valve leaflets [24]. Altogether, cardiac morphogenesis is a dynamic and multifaceted process that must be carefully regulated to insure proper cardiac structure.
Many zebrafish mutations disrupt cardiac morphology, and analysis of these mutants has indicated several important regulators of discrete morphogenetic processes. In the most extreme of these mutant phenotypes, the bilateral populations of cardiomyocytes never travel to the midline, resulting in two separate hearts forming in lateral positions, a condition known as cardia bifida [23]. Analysis of cardia bifida mutants has revealed key requirements for myocardial migration. Mutations in the transcription factor genes casanova (sox32), bonnie and clyde, and faust (gata5) and the Nodal coreceptor gene one-eyed pinhead all disrupt endoderm specification and secondarily cause cardia bifida, suggesting that the endoderm provides an important signal or substrate utilized by migrating cardiomyocytes [17, 25–27]. The molecular mechanisms of endodermal-myocardial interactions remain mysterious, although it is intriguing to consider a potential role for sphingolipid signaling, since the sphingosine-1-phosphate receptor gene miles apart (edg5) plays a cell non-autonomous role in promoting myocardial migration [28]. In addition to putative interactions with the endoderm, migrating cardiomyocytes require interactions with components of the extracellular matrix, particularly Fibronectin, as demonstrated by the cardia bifida phenotype of natter (fibronectin) mutants [29].
Another class of zebrafish mutations permit myocardial migration to the midline but then affect the morphology of the heart tube or cardiac chambers. Mutations in the genes heart and soul (prcki), snakehead (atp1a1a.1), and nagie oko (mpp5) hinder heart tube extension: mutant hearts resemble arrested cardiac cones or stunted heart tubes [12, 30–34]. The PRKCi, Atp1a1a.1, and Mpp5 proteins are all known to exhibit apicobasally polarized localization within epithelial cells, suggesting the importance of cell polarity to the process of tube extension.
Mutations in the heart of glass (heg), santa (krit1), and valentine (ccm2) genes do not seem to influence the shape of the heart tube, but they do cause severe distortion of chamber shape: the mutant hearts have thin-walled, extremely dilated chambers [35, 36]. All three of these genes are expressed in endothelial cells, with heg clearly being expressed in the endocardium, suggesting that endocardial-myocardial signaling plays a key role in regulating chamber wall thickening. Epigenetic influences, such as the biomechanical forces created by blood flow, also contribute to the formation of chamber shape. The zebrafish locus weak atrium encodes an atrium-specific myosin heavy chain (amhc) that is required for atrial contractility [13]. In addition to atrial defects and the consequent reduction in blood flow, weak atrium mutants also display ventricular defects: the mutant ventricle acquires an unusually small shape without characteristic chamber curvatures. Since amhc is expressed only in the atrium [13], this ventricular phenotype represents a secondary consequence of atrial dysfunction, presumably reflecting an impact of normal hemodynamics on chamber ballooning.
AV valve morphogenesis, like chamber morphogenesis, involves interplay between multiple signals, including those generated by biomechanical forces. Several zebrafish mutations interfere with the formation of ECs in the AV canal. For example, jekyll (udp-glucose dehydrogenase) mutants fail to form ECs [37]. UDP-glucose dehydrogenase (Ugdh) produces substrates used in the modification of extracellular matrix proteins that are known to facilitate Wnt and Fgf signaling [38]. Perhaps Ugdh is important for the control of Wnt signaling during EC formation: constitutive activation of Wnt signaling via mutation of the tumor suppressor gene apc leads to excessive EC formation beyond the boundaries of the AV canal, and overexpression of apc inhibits EC formation [39]. Other studies employing pharmacological inhibitors have suggested that Calcineurin/NFAT signaling promotes EC formation, while Notch signaling inhibits EC formation [24, 40]. Additionally, a number of lines of evidence demonstrate that cardiac function plays a key role in the induction of ECs. Mutation of genes that are important for cardiac contractility, including the cardiac actin gene cardiofunk (actc1) and the cardiac troponin T gene silent heart (tnnt2), prevents EC formation, suggesting that shear stress, produced by blood flow, or a stretch response, produced by contraction, could be required to initiate valve development [24, 41]. The notion of shear stress influencing valve morphogenesis has also been suggested by Hove et al., who demonstrated that implanting beads to obstruct blood flow at either the inflow or outflow end of the heart can inhibit EC formation [42].
Altogether, studies of zebrafish mutations affecting cardiac morphogenesis indicate the vast complexity of its regulation. To reveal the precise way that each pathway regulates a particular morphogenetic process, it is essential to understand the effects that specific genes exert on the dynamic changes to cardiac cytoarchitecture that underlie morphogenesis. How do the shapes and sizes of individual cells dictate the dimensions of the entire organ? How do cell-cell interactions contribute to morphological rearrangements? How do individual cell movements establish larger patterns of tissue movement? The optical accessibility of the zebrafish provides unparalleled opportunities for resolving the mechanisms of morphogenesis on a cellular and subcellular level. Visualization of myocardial and endocardial cells has been greatly enhanced by transgenes that express gfp in cardiomyocytes (e.g. Tg(cmlc2:egfp))[43] or endothelial cells (e.g. Tg(flk1:egfp)) [24]. In concert with other molecular markers, these transgenes have been instrumental in revealing how key regulatory genes effect changes in cellular polarity, interactions, and morphology that, in turn, control specific steps of morphogenesis.
Any morphogenetic rearrangement of embryonic tissues is likely to begin with changes in the cytoarchitecture of individual cells. For example, Trinh and Stainier have demonstrated that migrating cardiomyocytes begin to form a polarized epithelium as they approach the embryonic midline [29]. By examining the localization of markers of apicobasal polarity in embryos expressing Tg(cmlc2:egfp) (Figure 2), they determined the dynamics of polarity acquisition in differentiating cardiomyocytes. Intriguingly, mutations that disrupt apicobasal polarity, such as natter and hands off, also inhibit myocardial migration; these data reveal previously unappreciated roles of Fibronectin and Hand2 in the establishment of apicobasal polarity and suggest that formation of a polarized epithelium is a requirement for the coordination of myocardial migration [29, 44]. Myocardial epithelial polarity also seems to be integral to the process of heart tube extension. Both PRKCi and Mpp5 are components of apically localized protein complexes, and analysis of prkci and mpp5 mutants demonstrates that both genes are required cell-autonomously to insure the polarity and integrity of the myocardial epithelium [34]. As Atp1a1a.1 is basolaterally localized in chick cardiomyocytes [45], it is interesting to speculate that its function during heart tube extension could also be related to myocardial apicobasal polarity [32]. Together, these data point to the importance of organization and coherence of the myocardium during the formation of the cardiac cone and its transformation into the heart tube.
Figure 2.
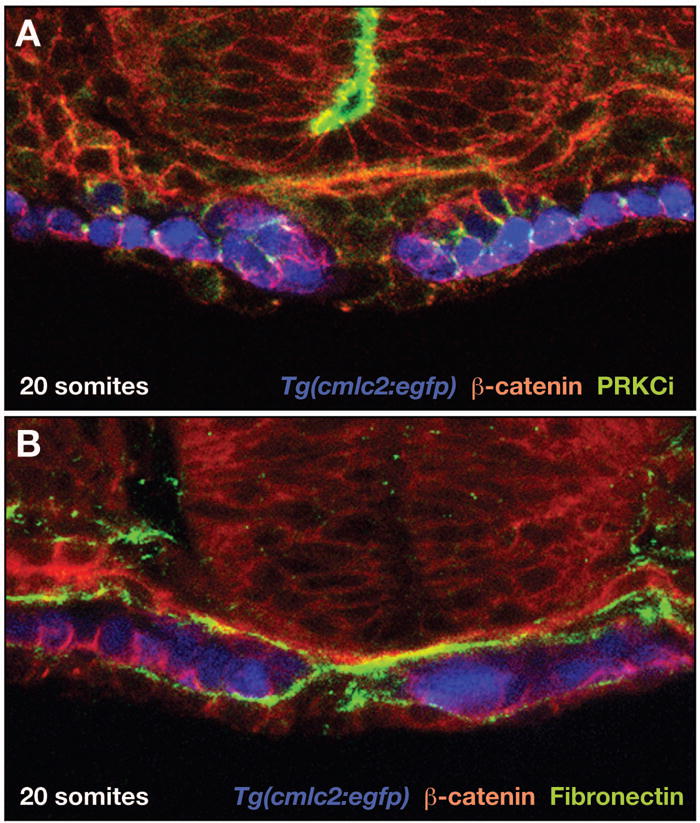
Examination of cardiac cytoarchitecture demonstrates that the migrating myocardium is a polarized epithelium. (A,B) Confocal images of transverse sections, dorsal to the top, of Tg(cmlc2:egfp) [43] embryos at the 20-somite stage, as in [29]. Expression of Tg(cmlc2:egfp) (pseudocolored blue) indicates bilateral populations of cardiomyocytes in the process of cardiac fusion. Immunohistochemistry for β-catenin (red), PRKCi (green, A), and Fibronectin (green, B) indicates protein localization. (A) At this stage, cardiomyocytes exhibit clear apicobasal polarity, with β-catenin localized basolaterally and PRKCi localized apicolaterally. (B) Fibronectin deposition is evident adjacent to the basal surface of the myocardial epithelia. Images provided by L. Trinh and D. Stainier.
Detailed examination of cytoarchitecture has also shed light on the mechanisms regulating AV valve formation. Using Tg(flk1:egfp) and Tg(tie2:egfp) to visualize endocardium, Beis et al. determined that EC formation is preceded by striking changes in cell shapes and protein localization within the endocardium of the AV canal [24]. Specifically, AV endocardial cells, previously squamous in appearance, become more cuboidal and begin to exhibit lateral localization of the adhesion molecule Dm-grasp. Examination of this early aspect of AV endocardial differentiation in embryos with valve defects has demonstrated distinct roles for the implicated pathways. In silent heart mutants, AV endocardium remains squamous and does not express Dm-grasp, indicating the importance of cardiac function for the initiation of endocardial differentiation [24]. In contrast, Calcineurin signaling is not required for initial differentiation of the AV canal endocardium, but it is important for the subsequent epithelial to mesenchymal transformation that creates ECs [24, 40]. Notch signaling, on the other hand, seems to be responsible for the spatial restriction of AV canal differentiation [24]. In embryos treated with the Notch pathway inhibitor DAPT, ventricular endocardium undergoes differentiation similar to that normally observed only in the AV canal.
Characterization of cardiac cell shape changes and their subcellular patterning adds depth to our understanding of the forces that drive cardiac morphogenesis; mutant analysis complements this approach by bridging the gap between gene function and cellular activities. It will be interesting to see how future studies apply cytoarchitectural analysis to other aspects of cardiac morphogenesis, such as cardiac ballooning or the interactions of myocardial and endocardial cells during tube assembly. Furthermore, future work will add a new point of view of morphogenesis by employing transgenes for timelapse imaging of cardiac cell rearrangements. Eventually, cytoarchitectural and timelapse approaches to morphogenesis are likely to merge, once the reagents are available for monitoring the dynamics of subcellular protein localization while morphogenesis is underway.
4. High-speed imaging: quantification of cardiac function
The embryonic zebrafish heart drives circulation through rhythmic, serial contractions of the ventricle and atrium [10]. Cardiomyocyte contraction requires the assembly of sarcomeres, large protein complexes that generate contractile force, and their organization into myofibrillar arrays. Sarcomeres contract in response to cellular depolarization and remain refractory to additional contractile cues until repolarization has occurred. Cardiomyocyte depolarization and repolarization require the coordinated release of Ca2+, Na+, and K+ ions intracellularly and extracellularly. Although the physiology of a mature cardiomyocyte is well characterized, less is known about how developing cardiomyocytes acquire and coordinate their contractile properties. How is sarcomere assembly regulated? What sets the initial embryonic heart rate? How does the contractile pattern of the primitive heart tube transition into that of a two-chambered organ? Zebrafish mutations causing contractility defects have the potential to uncover the genetic pathways that control the development of cardiac function.
Numerous zebrafish mutations interfere with cardiac function, causing the heart to appear noncontractile or poorly contractile. Several of these mutations disrupt genes encoding components of the sarcomere, including pickwick (titin) [46], tell tale heart (cardiac myosin light chain 2) [47], weak atrium (amhc) [13], cardiofunk (actc1) [41], and silent heart (tnnt2) [48], and lead to problems with sarcomere assembly or maintenance. Other mutations in this category affect genes encoding components of the electrical conduction system that controls cardiomyocyte depolarization and repolarization. For example, the island beat (cacna1c) locus encodes an α 1c subunit of a L-type calcium channel; island beat mutants exhibit an abnormally shaped and noncontractile ventricle as well as atrial fibrillation, demonstrating chamber-specific roles for this mechanism of calcium transport [49]. Like island beat, mutation of tremblor also causes ventricular noncontractility and atrial arrhythmia; this locus encodes a sodium/calcium exchanger, Slc8a1a, also known as Ncx1 [50, 51]. The breakdance mutation causes an arrhythmia reminiscent of that observed in human long QT syndrome; fittingly, the breakdance gene encodes a rapidly activating, delayed rectifier K+ channel (kcnh2), defects in which are also causative for the human syndrome [52]. Analysis of contractility mutants also suggests that endocardial-myocardial signaling may be required to maintain contractility. Mutation of dead beat causes a progressive loss of ventricular contractility; this locus encodes PLCγ1, a component of the vascular endothelial growth factor (VEGF) signaling pathway [53]. Mosaic analyses demonstrate that dead beat is required cell-autonomously for maintenance of ventricular contractility, thereby demonstrating a previously unappreciated role for VEGF signaling in cardiomyocytes. Manipulation of VEGF-PLCγ1 signaling in rat cardiomyocytes in culture affects their calcium cycling, suggesting a similar function for VEGF in the embryonic heart [53].
It is easy to classify the phenotypes of contractility mutants by qualitative features, such as noncontracility, reduced contractility, or arrhythmia. However, these categories oversimplify the diversity of functional phenotypes, which can reflect distinctions between the roles of specific components of the contractile apparatus or conduction system. Poor contractility, for example, could represent a number of different problems, including defective sarcomere assembly, ineffective sarcomere function, or reduced calcium cycling; these defects could be present throughout the heart or only in a specific chamber or region. To distinguish between possible roles for regulators of cardiac function, it is essential to use techniques appropriate for the quantification of functional parameters. New high-speed imaging technologies make it possible to resolve several useful metrics of cardiac contraction, blood flow, and cardiac conduction.
High-speed imaging is not necessary for the measurement of certain indicators of cardiac function in the zebrafish embryo: capturing images at normal video rate, it is straightforward to generate simple diagnostics like heart rate and ventricular shortening [32]. However, far more information can be extracted from high-speed analysis. Recent advancements in confocal microscopy allow the fast and sensitive imaging of fluorescent blood cells, endocardium, and myocardium when they are in motion; sophisticated computational tools can then reconstruct the optical sections to create four-dimensional depictions of the beating heart [54]. These data sets facilitate calculation of a variety of relevant metrics of cardiac function. For example, measurement of blood cell velocities and chamber volumes has demonstrated the existence of significant shear forces inside the heart at the stages when such forces are proposed to trigger EC formation [42]. Reconstruction of the waves of endocardial and myocardial movement and the patterns of blood cell displacement within the contracting heart tube have shown that the fluid dynamics of the heart tube resemble a suction pump, challenging the notion that the heart tube drives circulation via peristalsis (Figure 3) [55]. Examination of temporal changes in chamber volumes, cardiac cushion movements, and patterns of blood flow have documented the maturation of the AV valve into a functional apparatus for preventing backwards flow [54]. This quantitative characterization of the maturation of cardiac function in wild-type embryos provides a powerful framework for the future analysis of mutant phenotypes, although it is important to note that analysis of irregular contractions may require refinement of computational algorithms that are based on assumptions of the rhythmic activity expected in wild-type embryos [54].
Figure 3.
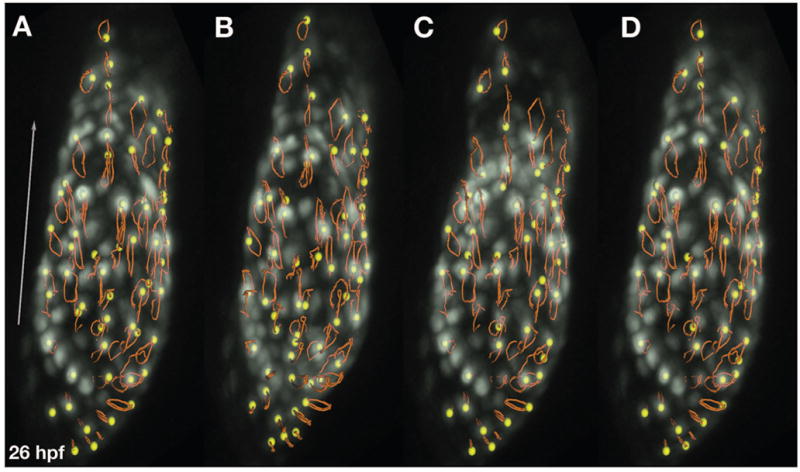
High-speed imaging facilitates the tracking of cardiomyocyte trajectories during cardiac contraction. (A–D) Series of 3D renderings documenting cardiac contraction in a Tg(cmlc2:egfp) [43] embryo at 26 hpf. The trajectories (orange tracks) of individual cardiomyocytes (yellow balls) are indicated. Trajectory patterns vary in different regions of the heart, indicating local distinctions in cardiomyocyte behavior during the heartbeat. As in [55], data were acquired by A. Forouhar using a Zeiss LSM 5 LIVE fast confocal microscope at 151 frames per second, reconstructed into 3D volumes via programs written by M. Liebling, and rendered using Imaris (Bitplane A.G.). The arrow shows the general direction of blood flow and is 100 microns long. Images provided by M. Liebling and M. Dickinson.
Tracking dynamic cell movements provides a clear readout of cardiac function; however, the origins of functional deficiencies lie within the cells. Sarcomere status can be carefully inspected via electron microscopy, but other techniques are necessary to monitor the cardiac conduction system. Electrocardiograms can be used to examine cardiac voltages in the zebrafish embryo, pacing techniques can test the electromechanical responsiveness of embryonic hearts, and whole-cell voltage-clamp recordings of dissociated cardiomyocytes can indicate the status of particular currents in individual cells [49]. It is also desirable to monitor conduction in context, tracking electrical impulses in four dimensions in vivo to resolve regional differences in conduction and changes over time. High-speed imaging of calcium indicators allows visualization of a wave of myocardial depolarization as it progresses from pole to pole. Application of this technique to the analysis of tremblor mutants revealed an absence of wave-like depolarization; instead, mutant embryos exhibit high and static levels of calcium within the ventricle and abnormal, sporadic calcium transients in the atrium [50, 51]. These observations correspond well to the tremblor contractility phenotype and to the established role of Ncx1 in calcium extrusion. Monitoring the progress of the calcium wavefront also makes it possible to track the maturation of cardiac conduction during the process of chamber emergence. Combining calcium imaging and cardiac pacing, Milan et al. have shown that AV canal myocardium acquires special characteristics around 40 hours post fertilization (hpf), so as to slow conduction velocities between the atrium and ventricle [56]. Examination of conduction velocities in cloche mutants, which lack endocardium, demonstrated that endocardial-myocardial signaling is necessary to induce these conductive properties in the AV canal; in contrast, examination of silent heart mutants indicated that cardiac contractions and blood flow are dispensable for this aspect of AV canal development. Finally, antisense morpholino-mediated knockdown of notch1b and neuregulin, both of which are expressed in the endocardium, inhibited deceleration of conduction at the AV junction, implicating these genes in the functional maturation of the AV canal.
Overall, functional studies suggest a high degree of cross-species conservation of fundamental regulatory mechanisms. Extensions of this work will undoubtedly apply high-speed imaging techniques to the analysis of many other essential and conserved genes. Furthermore, it will be fruitful to employ high-speed imaging to the characterization of small molecules influencing cardiac function. Using automated microscopy, automated videography, and automated computational analysis, Burns et al. have created a high-throughput assay that sensitively and accurately monitors heart rate in Tg(cmlc2:gfp) embryos [57]. This strategy can be used to rapidly screen large libraries of small molecules for their effects on contractility. The imaging techniques described above can then be used to define the ways that particular compounds affect contraction and conduction, potentially accelerating the pace of drug discovery.
5. Future prospects: image is everything
Taken together, recent studies of cardiac development in the zebrafish embryo emphasize the importance of high-resolution imaging techniques for the analysis of essential regulatory mechanisms. Rather than simply assigning a gene a role in a general process, such as cardiomyocyte production or heart tube assembly, current techniques can reveal more mechanistic specifics of gene function. It is possible to track fate decisions made by individual cells, to follow subcellular localization of proteins that are integral to tissue movement and structure, and to monitor contractile and conductive contributions to the functional output of the organ. The next generation of experiments promise to add substantial breadth and depth to this body of work. Of course, today’s techniques will be applied to additional interesting genes, since there are a number of intriguing mutations in the collection that remain unexplored. Meanwhile, many new mutations are being generated, especially using newly refined genetic techniques, such as insertional mutagenesis and TILLING [58, 59]. Furthermore, the continued development of imaging technologies should rapidly expand the horizons of the field. For example, it will be particularly exciting to employ new fluorescent proteins with dynamic properties, such as the photoconvertible protein Kaede, for tracking cell movements and rearrangements [60]. Moreover, as imaging techniques facilitate a deep understanding of zebrafish cardiac development, it will be important to extend these studies across species to examine the degree of conservation of the relevant developmental mechanisms. Assuming that the fundamental aspects of the key regulatory pathways are broadly conserved, high-resolution studies of zebrafish cardiac specification, morphogenesis, and function should shed considerable light on the cellular and subcellular mechanisms of mammalian cardiogenesis and the etiology of congenital heart disease.
Acknowledgments
We thank L. Trinh, D. Stainier, M. Liebling, and M. Dickinson for generously providing images and input for Figures 2 and 3. Research in the Yelon lab is supported by grants from the National Institutes of Health, the American Heart Association, and the March of Dimes.
Abbreviations
- (CP)
cardiac progenitor
- (RA)
retinoic acid
- (LMZ)
lateral marginal zone
- (ALPM)
anterior lateral plate mesoderm
- (hpf)
hours post fertilization
- (AV)
atrioventricular
- (ECs)
endocardial cushions
- (VEGF)
vascular endothelial growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yelon D, Weinstein BM, Fishman MC. In: Cardiovascular system, in Pattern Formation. Zebrafish L, Solnica-Krezel, editors. Springer-Verlag; Heidelberg: 2002. pp. 298–321. [Google Scholar]
- 2.Trinh LA, Stainier DY. Cardiac development. Methods Cell Biol. 2004;76:455–73. doi: 10.1016/s0091-679x(04)76020-3. [DOI] [PubMed] [Google Scholar]
- 3.Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2(12):956–66. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 4.Pelster B, Burggren WW. Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio) Circ Res. 1996;79(2):358–62. doi: 10.1161/01.res.79.2.358. [DOI] [PubMed] [Google Scholar]
- 5.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 7.Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–92. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 8.Chen JN, Haffter P, Odenthal J, Vogelsang E, Brand M, van Eeden FJ, et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development. 1996;123:293–302. doi: 10.1242/dev.123.1.293. [DOI] [PubMed] [Google Scholar]
- 9.Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–99. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Warren KS, Wu JC, Pinet F, Fishman MC. The genetic basis of cardiac function: dissection by zebrafish (Danio rerio) screens. Philos Trans R Soc Lond B Biol Sci. 2000;355(1399):939–44. doi: 10.1098/rstb.2000.0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu N, Sedmera D, Yost HJ, Clark EB. Structure and function of the developing zebrafish heart. Anat Rec. 2000;260(2):148–57. doi: 10.1002/1097-0185(20001001)260:2<148::AID-AR50>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214(1):23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- 13.Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130(24):6121–9. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- 14.Yelon D, Feldman JL, Keegan BR. Genetic regulation of cardiac patterning in zebrafish. Cold Spring Harb Symp Quant Biol. 2002;67:19–25. doi: 10.1101/sqb.2002.67.19. [DOI] [PubMed] [Google Scholar]
- 15.Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127(2):225–35. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- 16.Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Dev Biol. 2001;234(2):330–8. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- 17.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13(22):2983–95. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, et al. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127(12):2573–82. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- 19.Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307(5707):247–9. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- 20.Kozlowski DJ, Murakami T, Ho RK, Weinberg ES. Regional cell movement and tissue patterning in the zebrafish embryo revealed by fate mapping with caged fluorescein. Biochem Cell Biol. 1997;75(5):551–62. [PubMed] [Google Scholar]
- 21.Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131(13):3081–91. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- 22.Stainier DY, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development. 1993;119(1):31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- 23.Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13(6):507–13. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- 24.Beis D, Bartman T, Jin SW, Scott IC, D’Amico LA, Ober EA, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14(10):1279–89. [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, Yelon D, et al. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15(12):1493–505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyrieras N, Strahle U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGF-beta-related signaling. Curr Biol. 1998;8(13):783–6. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- 28.Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406(6792):192–5. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- 29.Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6(3):371–82. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 30.Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, et al. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11(19):1492–502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- 31.Peterson RT, Mably JD, Chen JN, Fishman MC. Convergence of distinct pathways to heart patterning revealed by the small molecule concentramide and the mutation heart-and-soul. Curr Biol. 2001;11(19):1481–91. doi: 10.1016/s0960-9822(01)00482-1. [DOI] [PubMed] [Google Scholar]
- 32.Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, et al. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130(25):6165–73. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- 33.Yuan S, Joseph EM. The small heart mutation reveals novel roles of Na+/K+-ATPase in maintaining ventricular cardiomyocyte morphology and viability in zebrafish. Circ Res. 2004;95(6):595–603. doi: 10.1161/01.RES.0000141529.48143.6e. [DOI] [PubMed] [Google Scholar]
- 34.Rohr S, Bit-Avragim N, Abdelilah-Seyfried S. Heart and soul/PRKCi and nagie oko/Mpp5 regulate myocardial coherence and remodeling during cardiac morphogenesis. Development. 2006;133(1):107–15. doi: 10.1242/dev.02182. [DOI] [PubMed] [Google Scholar]
- 35.Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13(24):2138–47. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 36.Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133(16):3139–46. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 37.Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293(5535):1670–3. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- 38.Lander AD, Selleck SB. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol. 2000;148(2):227–32. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, et al. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature. 2003;425(6958):633–7. doi: 10.1038/nature02028. [DOI] [PubMed] [Google Scholar]
- 40.Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, et al. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118(5):649–63. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Bartman T, Walsh EC, Wen KK, McKane M, Ren J, Alexander J, et al. Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2004;2(5):E129. doi: 10.1371/journal.pbio.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421(6919):172–7. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- 43.Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228(1):30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- 44.Trinh LA, Yelon D, Stainier DY. Hand2 regulates epithelial formation during myocardial differentiation. Curr Biol. 2005;15:441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- 45.Linask KK. N-cadherin localization in early heart development and polar expression of Na+,K(+)-ATPase, and integrin during pericardial coelom formation and epithelialization of the differentiating myocardium. Dev Biol. 1992;151(1):213–24. doi: 10.1016/0012-1606(92)90228-9. [DOI] [PubMed] [Google Scholar]
- 46.Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, et al. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30(2):205–9. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- 47.Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, et al. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99(3):323–31. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- 48.Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31(1):106–10. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 49.Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1(2):265–75. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 50.Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, et al. Mutation in sodium-calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci U S A. 2005;102(49):17699–704. doi: 10.1073/pnas.0502679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, et al. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A. 2005;102(49):17705–10. doi: 10.1073/pnas.0502683102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langheinrich U, Vacun G, Wagner T. Zebrafish embryos express an orthologue of HERG and are sensitive toward a range of QT-prolonging drugs inducing severe arrhythmia. Toxicol Appl Pharmacol. 2003;193(3):370–82. doi: 10.1016/j.taap.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Rottbauer W, Just S, Wessels G, Trano N, Most P, Katus HA, et al. VEGF-PLCgamma1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 2005;19(13):1624–34. doi: 10.1101/gad.1319405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liebling M, Forouhar AS, Wolleschensky R, Zimmermann B, Ankerhold R, Fraser SE, et al. Rapid three-dimensional imaging and analysis of the beating embryonic heart reveals functional changes during development. Dev Dyn. 2006 doi: 10.1002/dvdy.20926. [DOI] [PubMed] [Google Scholar]
- 55.Forouhar AS, Liebling M, Hickerson A, Nasiraei-Moghaddam A, Tsai HJ, Hove JR, et al. The embryonic vertebrate heart tube is a dynamic suction pump. Science. 2006;312(5774):751–3. doi: 10.1126/science.1123775. [DOI] [PubMed] [Google Scholar]
- 56.Milan DJ, Giokas AC, Serluca FC, Peterson RT, MacRae CA. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development. 2006;133(6):1125–32. doi: 10.1242/dev.02279. [DOI] [PubMed] [Google Scholar]
- 57.Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1(5):263–4. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- 58.Amsterdam A, Hopkins N. Retroviral-mediated insertional mutagenesis in zebrafish. Methods Cell Biol. 2004;77:3–20. doi: 10.1016/s0091-679x(04)77001-6. [DOI] [PubMed] [Google Scholar]
- 59.Stemple DL. TILLING--a high-throughput harvest for functional genomics. Nat Rev Genet. 2004;5(2):145–50. doi: 10.1038/nrg1273. [DOI] [PubMed] [Google Scholar]
- 60.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44(3):136–42. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]


