Abstract
The synthesis of phospholipids in the yeast Saccharomyces cerevisiae is regulated by zinc, an essential mineral required for growth and metabolism. Cells depleted of zinc contain increased levels of phosphatidylinositol and decreased levels of phosphatidylethanolamine. In addition to the major phospholipids, the levels of the minor phospholipids phosphatidate and diacylglycerol pyrophosphate decrease in the vacuole membrane of zinc-depleted cells. Alterations in phosphatidylinositol and phosphatidylethanolamine can be ascribed to an increase in PIS1-encoded phosphatidylinositol synthase activity and to decreases in the activities of CDP-diacylglycerol pathway enzymes including the CHO1-encoded phosphatidylserine synthase, respectively. Alterations in the minor vacuole membrane phospholipids are due to the induction of the DPP1-encoded diacylglycerol pyrophosphate phosphatase. These changes in the activities of phospholipid biosynthetic enzymes result from differential regulation of gene expression at the level of transcription. Under zinc-deplete conditions, the positive transcription factor Zap1p stimulates the expression of the DPP1 and PIS1 genes through the cis-acting element UASZRE. In contrast, the negative regulatory protein Opi1p, which is involved in inositol-mediated regulation of phospholipid synthesis, represses the expression of the CHO1 gene through the cis-acting element UASINO. Regulation of phospholipid synthesis may provide an important mechanism by which cells cope with the stress of zinc depletion, given the roles that phospholipids play in the structure and function of cellular membranes.
Keywords: Phospholipid synthesis, Phosphatidylinositol synthase, Phosphatidylserine synthase, Transcriptional regulation, Yeast, Zinc
1. Introduction
The yeast Saccharomyces cerevisiae serves as a eukaryotic model organism to study the regulation of phospholipid synthesis [1–6]. Almost all of the structural genes responsible for phospholipid synthesis have been identified in S. cerevisiae [1–8], and many of the phospholipid biosynthetic enzymes have been purified from the organism [1–6]. The characterization of these genes along with their encoded enzymes has significantly advanced the understanding of phospholipid synthesis and its regulation in eukaryotes.
Phospholipids play diverse roles that are essential for growth and metabolism. It is well known that phospholipids govern membrane-associated functions such as enzyme catalysis, receptor-mediated signaling, and solute transport [9,10]. In addition, phospholipids are precursors for the synthesis of macromolecules [11–15], serve as molecular chaperons [16,17], serve in protein modification for membrane association [18], and are reservoirs of second messengers [19]. Thus, as shown in S. cerevisiae, the activities of phospholipid biosynthetic enzymes are regulated to cope with a variety of stress conditions (e.g., nutrient depletion) [6,20–23,23,24]. In this review, we will discuss how phospholipid synthesis in S. cerevisiae is regulated in response to zinc depletion.
2. Phospholipid biosynthetic pathways in S. cerevisiae
The major phospholipids found in the membranes of S. cerevisiae include phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylserine (PS) [1–3,6,25]. Mitochondrial membranes also contain phosphatidylglycerol (PG) and cardiolipin (CL) [1–3,6,25]. The most common fatty acids esterified to the glycerophosphate backbone of the phospholipids include palmitic acid, palmitoleic acid, stearic acid, and oleic acid [25–27].
Phospholipid synthesis in S. cerevisiae occurs by complementary pathways common to those found in mammalian cells (Fig. 1) [1,2,6,28,29]. One exception is that in yeast PS is synthesized from CDP-diacylglycerol (CDP-DG) and serine, whereas in mammalian cells, PS is synthesized from PE or PC by an exchange reaction with serine [1,2,6]. In yeast, PS, PE, and PC are synthesized from phosphatidate (PA) via the CDP-DG pathway (Fig. 1). The energy-rich liponucleotide CDP-DG is synthesized from PA and CTP by the CDS1-encoded CDP-DG synthase [30,31]. CDP-DG may then donate its phosphatidyl moiety to serine to form PS [32] in the reaction catalyzed by the CHO1-encoded PS synthase [33–35]. PS is dephosphorylated to PE by the PSD1- [36,37] and PSD2-encoded [38] PS decarboxylase enzymes. PE is then converted to PC by the three-step AdoMet-dependent methylation reactions [39]. The first methylation reaction is catalyzed by the CHO2-encoded PE methyltransferase [40,41] and the last two methylation reactions are catalyzed by the OPI3-encoded phospholipid methyltransferase [40,42].
Fig. 1.
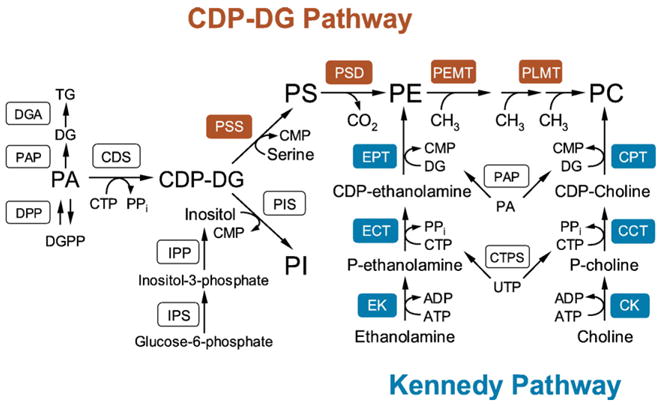
Pathways for the synthesis of the major phospholipids in S. cerevisiae. The pathways shown for the synthesis of phospholipids include the relevant steps discussed in the review. The CDP-DG pathway enzymes (PSS, PS synthase; PSD, PS decarboxylase; PEMT, PE methyltransferase; and PLMT, phospholipid methyltransferase) are highlighted in red. The Kennedy pathway enzymes (EK, ethanolamine kinase; ECT, phosphoethanolamine cytidylyltransferase; EPT, ethanolaminephosphotransferase; CK, choline kinase; CCT, phosphocholine cytidylyltransferase; CPT, cholinephosphotransferase) are highlighted in blue. IPS, inositol phosphate synthase; IPP, inositol phosphate phosphatase; PIS, PI synthase; DPP, DGPP phosphatase; PAP, PA phosphatase; DGA, DG acyltransferase; CTPS, CTP synthetase.
PE and PC are also synthesized via the CDP-ethanolamine and CDP-choline branches of the Kennedy pathway (Fig. 1). Ethanolamine and choline are phosphorylated with ATP to form phosphoethanolamine and phosphocholine, respectively, by the EKI1-encoded ethanolamine kinase [43] and the CKI1-encoded choline kinase [44]. Phosphoethanolamine and phosphocholine are activated with CTP to form CDP-ethanolamine and CDP-choline, respectively, by the ECT1-encoded phosphoethanolamine cytidylyltransferase [45] and the PCT1-encoded phosphocholine cytidylyltransferase [46]. CDP-ethanolamine and CDP-choline react with diacylglycerol (DG) to form PE and PC, respectively, in the reaction catalyzed by the EPT1-encoded ethanolamine phosphotransferase [47,48] and the CPT1-encoded choline phosphotransferase [49,50].
The PIS1-encoded PI synthase [51,52] catalyzes the formation of PI by displacement of CMP from CDP-DG with inositol [53]. The inositol used in this reaction is derived from glucose-6-phosphate via the reactions catalyzed by the INO1-encoded inositol-3-phosphate synthase [54,55] and the INM1-encoded inositol-3-phosphate phosphatase [56]. In the CL pathway (not shown in Fig. 1), PGP is formed from CDP-DG by displacement of CMP with glycerol-3-phosphate in the reaction catalyzed by the PGS1-encoded phosphatidylglycerophosphate (PGP) synthase [57]. PGP is dephosphorylated to PG by a phosphatase. The CRD1-encoded CL synthase [58–60] catalyzes the reaction between PG and CDP-DG to generate CL. The CTP required for the synthesis of the activated, energy-rich intermediates (CDP-DG, CDP-ethanolamine, and CDP-choline) is derived from UTP by the URA7- [61] and URA8-encoded [62] CTP synthetase enzymes. The DG used for the synthesis of PE and PC via the Kennedy pathway is derived from PA by the PAH1-encoded Mg2+-dependent PA phosphatase, and it is also used as a substrate for the synthesis of triacylglycerol [63].
The CDP-DG pathway is primarily used for the synthesis of PE and PC when cells are grown in the absence of ethanolamine and choline [1,2,6,64,65]. Yet, the Kennedy pathway contributes to the synthesis of PE and PC in this growth condition [5,43,66]. For example, the PC synthesized via the CDP-DG pathway is constantly hydrolyzed to choline and PA [66,67] by the SPO14-encoded [68,69] phospholipase D. The choline is incorporated back into PC via the CDP-choline branch of the Kennedy pathway, and the PA is converted to other phospholipids via the intermediates CDP-DG and DG [2,3,6]. Analysis of mutants in S. cerevisiae [6,70,71] as well as in mammalian cells [72,73] indicates that the physiological roles of PC synthesized via the two pathways are different.
The Kennedy pathway plays a critical role in phospholipid synthesis when enzymes in the CDP-DG pathway are defective [1,2,4,6]. The cho2 opi3 mutant deficient in the methylation of PE requires choline for growth and synthesizes PC via the CDP-choline branch of the Kennedy pathway [40–42,74]. The cho1 and psd1 psd2 mutants deficient in the synthesis of PS [75,76] and PE [38,77], respectively, can synthesize PC when supplemented with ethanolamine. The ethanolamine is incorporated into PE via the CDP-ethanolamine branch of the Kennedy pathway, and the PE is subsequently methylated to form PC. The Kennedy pathway mutants (e.g., cki1 eki1, cpt1 ept1) defective in both the CDP-choline and CDP-ethanolamine branches can synthesize PC only via the CDP-DG pathway [43,78,79]. However, unlike the CDP-DG pathway mutants [38,40–42,74–77], the Kennedy pathway mutants do not exhibit any auxotrophic requirements [43,79]. These mutants have an essentially normal complement of phospholipids including PC [43,79].
3. Zinc-mediated regulation of the DPP1-encoded DGPP phosphatase
Zinc is an essential nutrient required for the growth and metabolism of S. cerevisiae and higher eukaryotes [80]. Zinc is a cofactor for key metabolic enzymes such as alcohol dehydrogenase, carbonic anhydrase, proteases, RNA polymerases, and superoxide dismutase [80]; it is also a structural component of a diverse set of proteins such as chaperons, lipid binding proteins, and transcription factors [81,82]. Accordingly, an insufficient amount of zinc is detrimental to organisms. Zinc deficiency in rats is associated with oxidative damage to DNA, lipids, and proteins [83]; in humans, it is manifested by defects in appetite, cognitive function, embryonic development, epithelial integrity, and immune function [84].
In the genome-wide analysis of gene expression in the S. cerevisiae cell depleted of zinc, DPP1 was identified as one of the most highly induced genes, and this regulation is coordinated with the regulation of genes responsible for zinc transport [85] (see below). The DPP1 gene encodes diacylglycerol pyrophosphate (DGPP) phosphatase that is associated with the vacuole membrane [24,86–88]. This enzyme catalyzes the removal of the β-phosphate from DGPP to form PA, followed by the dephosphorylation of PA to form DG [86]. Since DGPP is a preferred substrate to PA, the DPP1-encoded DGPP phosphatase does not dephosphorylate PA in the presence of relatively low levels of DGPP [86]. The DGPP molecule was originally identified in plants as the product of the PA kinase [89]. Research with plants under a variety of stress conditions indicates that DGPP may function as a signaling molecule [90,91,91–93]. The accumulation of DGPP is transient and coincides with a rise in the level of PA [91,93].
The regulation of DPP1 expression in zinc-depleted cells is mediated by the zinc-sensing and zinc-inducible transcription factor Zap1p [94–97], which binds to the cis-acting element UASZRE (zinc-responsive element) in the DPP1 promoter [24,85]. Analysis of mutants lacking zinc transporters in the plasma membrane and in the vacuole membrane indicates that DPP1 expression is sensitive to the cytosolic level of zinc [24]. The regulation of DGPP phosphatase expression correlates with the metabolism of DGPP and PA in the vacuole membrane [98]. When cells are grown in zinc-rich conditions, DGPP and PA account for 0.6 mol% and 1.4 mol% of the total phospholipids in vacuole membranes. Under zinc-depleted conditions, however, the amounts of DGPP and PA are decreased to an undetectable level and 0.3 mol%, respectively [98].
The DPP1 gene is not essential, and dpp1Δ mutant cells do not exhibit any distinct phenotypes under various growth conditions [87], including fluctuations in zinc supplementation [99]. Thus, the role of DGPP phosphatase during zinc depletion would have to complement other mechanisms that respond to this stress. Although the function of DGPP phosphatase in yeast is still unclear, we speculate that the enzyme controls the levels of DGPP and PA in vacuolar membranes, which in turn mediates the cellular functions occurring in response to zinc depletion.
4. Zinc-mediated regulation of phospholipid synthesis
Zinc depletion has more global effects on phospholipid synthesis in the cell. In addition to the changes in DGPP and PA, it results in a reduction in the level of PE and an increase in the level of PI in the vacuole membrane [98]. The cellular levels of PE and PI are also altered similarly in response to zinc depletion [23]. These changes in the major phospholipids are independent of the regulation of the DPP1-encoded DGPP phosphatase activity [100].
In zinc-depleted cells, the activity levels of all enzymes in the CDP-DG pathway (PS synthase, PS decarboxylase, PE methyl-transferase, and phospholipid methyltransferase) are reduced, while the activity of PI synthase is elevated [23]. Thus, the decrease in the cellular PE content correlates with the decreases in the activities of PS synthase and PS decarboxylase, and the increase in the cellular PI content correlates with the increase in the activity of PI synthase. Although the activities of the phospholipid methyltransferase enzymes are also reduced in zinc-depleted cells, these changes do not have a major effect on PC content [23]. Stimulation of the Kennedy pathway for phospholipid synthesis appears to compensate for the decrease in activities of the CDP-DG pathway enzymes. Recent studies have shown that the EKI1-encoded ethanolamine kinase is induced in zinc-depleted cells, and this regulation is mediated in part by Zap1p [101].
The coordinate regulation of the PI synthase and PS synthase enzymes, which compete for CDP-DG (Fig. 1), is part of an overall mechanism by which the synthesis of PI is coordinately regulated with the synthesis of PC [1,2,4–6,102,103]. The induction of PI synthase expression may represent one mechanism by which cells cope with zinc depletion, given that PI is a precursor to inositol-containing lipid molecules (sphingolipids, phosphoinositides, and glycosylphosphatidylinositol anchors) that are essential to growth and metabolism [2,13,104–114]. The repression of PS synthase activity alleviates the competition both enzymes have for CDP-DG [115]. These important phospholipid biosynthetic enzymes are regulated differently at the level of transcription in zinc-depleted cells.
5. Zinc-mediated regulation of the PIS1-encoded PI synthase
The increase in the activity of PI synthase in zinc-depleted cells results from the regulation of the PIS1 expression at the level of transcription, as shown by an increase in the levels of PIS1 mRNA, its encoded protein Pis1p, and the β-galactosidase activity driven by a PPIS1-lacZ reporter gene [116]. Like DPP1, the regulation of PIS1 transcription is mediated by Zap1p [116]. A model for the transcriptional regulation of PIS1 by Zap1p in response to zinc depletion is shown in Fig. 2 (left). The promoter of the PIS1 gene contains a sequence that shares homology with a consensus UASZRE (ACCTTNAAGGT) [116]. Electrophoretic mobility shift assays with DNA probes containing the putative UASZRE sequence and purified recombinant Zap1p show that the sequence in the PIS1 promoter is required for Zap1p binding in vitro [116]. Moreover, mutations in sequence to a nonconsensus UASZRE abolish Zap1p-DNA interactions in vitro and the induction of PIS1 gene expression in response to zinc depletion [116]. In contrast to DPP1, the PIS1 gene was not identified in the genome-wide analysis of gene expression that revealed 46 direct Zap1p target genes that are highly induced by zinc depletion [85]. This is attributed to the relatively modest level of PIS1 induction (~2-fold) when compared with the >10-fold inductions of other Zap1p target genes (e.g., DPP1) [24,85]. Notwithstanding, the 2-fold induction of the PIS1 gene in response to zinc depletion correlates with the ~2-fold increase in the PI content of yeast cells depleted of zinc [23].
Fig. 2.
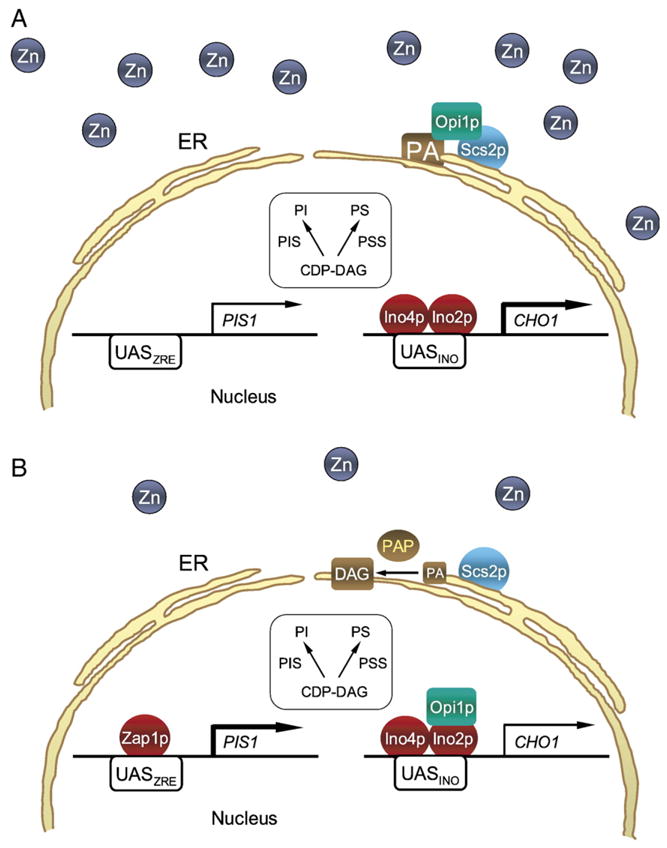
Models for the transcriptional regulation of PIS1 and CHO1 by zinc depletion in the absence of inositol. (A) PIS1 (left) and CHO1 (right) are expressed at some level when cells are grown in a zinc-rich medium (depicted by numerous zinc atoms outside the nucleus). Maximum expression of CHO1 (indicated by the bold arrow) is dependent on the interaction of the Ino2p–Ino4p complex with the UASINO element in the gene promoter. Under this growth condition, the repressor Opi1p is associated with the nuclear/ER membrane through interactions with PA and Scs2p. (B) when zinc is limiting (depicted by a reduced number of zinc atoms outside the nucleus), the Zap1p transcription factor is induced and binds to the UASZRE in the PIS1 gene promoter to increase transcription (indicated by the bold arrow). Transcription of CHO1 is attenuated in zinc-depleted cells by the interaction of Opi1p with Ino2p (indicated by the thin arrow). Dissociation of Opi1p from the nuclear/ER membrane and its translocation into the nucleus are caused by a decrease in PA concentration. An increase in Mg2+-dependent PA phosphatase (PAP) activity may be responsible for the decrease in PA concentration.
6. Zinc-mediated regulation of the CHO1-encoded PS synthase
The expression of CHO1-encoded PS synthase is also controlled at the level of transcription in zinc-depleted cells [23]. In contrast to PI synthase, zinc depletion results in the repression of CHO1 expression, resulting in decreased levels of the PS synthase mRNA, protein, and activity [23]. The downregulation of PS synthase expression and the lack of a UASZRE in the CHO1 promoter indicate that the transcription factor Zap1p does not directly control the expression of PS synthase. Moreover, an indirect effect of Zap1p on the expression of PS synthase is ruled out because a zap1Δ mutation does not affect the zinc-mediated regulation of the enzyme [23]. Instead, the repression of PS synthase by zinc depletion is mediated through the UASINO element in the CHO1 promoter and by the phospholipid synthesis regulatory proteins Opi1p, Ino2p, and Ino4p [23]. This conclusion is supported by the observations that mutations in the UASINO element abolish the zinc-mediated regulation of CHO1 expression, and that regulation PS synthase expression by zinc depletion is lost in ino2Δ, ino4Δ, and opi1Δ mutants [23].
Ino2p, Ino4p, and Opi1p play an important role in the inositol-mediated regulation of CHO1 and other UASINO-containing genes involved in phospholipid synthesis [4–6,117–119]. Inositol is an essential nutrient that can be synthesized in S. cerevisiae by the INO1-encoded inositol-3-phosphate synthase (Fig. 1). The essential nature of inositol stems from the fact that it is the water-soluble precursor for the synthesis of PI and other inositol-containing lipids [2,13,107,110–114]. Ino2p [120] and Ino4p [121] are positive regulatory proteins, whereas Opi1p [122] is a negative regulatory protein. The UASINO element contains a consensus-binding site (CANNTG) for an Ino2p-Ino4p heterodimer, which is required for maximum expression of the co-regulated UASINO-containing genes [4–6,123–125]. The CHO1 and other UASINO-containing genes are maximally expressed when inositol is absent from the growth medium, but these genes are repressed when inositol is supplemented to the growth medium. The coordinate repression of UASINO-containing genes by inositol requires the ongoing synthesis of PC [79,126], and is enhanced by the inclusion of choline in the growth medium [1,2,4–6]. According to the model for this regulation, which is based on a recent paper by Loewen et al. [127], Opi1p is associated with the nuclear/endoplasmic reticulum (ER) membrane through interactions with the integral membrane protein Scs2p (a VAP homolog) [128] and with PA [127] when cells are grown without inositol. Upon inositol supplementation, the level of PA reduces due to the utilization of CDP-DG and increased synthesis of PI. The decrease in the PA level results in loss of Opi1p association with the nuclear/ER membrane, followed by its translocation into the nucleus [127]. In the nucleus, Opi1p mediates repression of the co-regulated phospholipid biosynthetic genes through the UASINO element [129], but not by direct interaction [130]. Instead, Opi1p inhibits transcriptional activation by binding to DNA-bound Ino2p [131]. Interestingly, the repression of CHO1 by zinc depletion occurs through this regulatory circuit in the absence of inositol supplementation. A model for the zinc-mediated regulation of CHO1 transcription is shown in Fig. 2 (right).
The zinc-mediated repression of CHO1 may be explained if the INO2 gene is repressed and/or the OPI1 gene is induced by zinc depletion. However, data indicate that the expression of the INO2 and OPI1 genes is not significantly affected by zinc depletion [132]. Moreover, Ino2p and Ino4p are not zinc-containing proteins, and thus a reduction in their function is not likely a direct consequence of zinc depletion. The key aspect common to the repression of CHO1 (and other UASINO-containing genes) by inositol supplementation [1–6] and by zinc depletion [23] is the negative regulatory protein Opi1p. The concentration of PA in the nuclear/ER membrane plays a central role in the localization and regulation of Opi1p repressor function [127] (Fig. 2, right). PA levels in S. cerevisiae are governed by its de novo synthesis from glycerol-3-phosphate, its utilization by the CDP-DG synthase (CDP-DG pathway) and Mg2+-dependent PA phosphatase (Kennedy pathway) enzymes required for phospholipid synthesis, and the phospholipase D-mediated hydrolysis of PC [6]. As indicated above, the decrease in PA concentration and resultant translocation of Opi1p from the nuclear/ER membrane to the nucleus in response to inositol supplementation is attributed to an increase in PI synthesis and utilization of CDP-DG [127]. The increase in PI synthase expression and PI synthesis in response to zinc depletion, which occurs in the absence of inositol supplementation [23,116], is consistent with this model. It is unknown what effect zinc depletion has on the cellular concentration of inositol, which might affect the synthesis of PI. It is known however, that inositol supplementation results in the induction of Mg2+-dependent PA phosphatase activity [133], and preliminary studies have shown that this activity is also induced in response to zinc depletion [132]. These observations suggest that regulation of Mg2+-dependent PA phosphatase also contributes to the reduction of PA in the nuclear/ER membrane, the translocation of Opi1p into the nucleus, and repression of CHO1 and other UASINO-containing genes. The availability of the pah1Δ mutant defective in Mg2+-dependent PA phosphatase [63] will permit studies to examine this question.
7. Perspectives
The zinc-mediated regulation of phospholipid synthesis clearly impacts on membrane phospholipid composition (i.e., changes in the levels of PE and PI) [23,98]. Clues for the physiological relevance of this regulation might stem from the roles that specific phospholipids play in the structure and function of cellular membranes. Both PE and PI play a role in the modification of proteins for attachment to membranes. For example, PE is used directly for covalent modification and membrane attachment of Apg8p, a protein essential to the process of autophagy occurring in response to nutrient limitation [134–137], and indeed zinc depletion results in an elevation of Apg8p-PE [23]. PE is also used for the glycosylphosphatidylinositol modification of proteins for membrane attachment [12]. The glycosylphosphatidylinositol anchor is attached to proteins through the amine group of phosphoethanolamine that is derived from PE [12]. Interestingly, zinc depletion [85] induces the expression of MCD4 that encodes one of the enzymes responsible for the transfer of the phosphoethanolamine moiety of PE to make the anchor [138]. The importance of PE for Apg8p modification and for glycosylphosphatidylinositol anchor synthesis in response to zinc depletion warrants further examination. As indicated above, PI is also used for the synthesis of glycosylphosphatidylinositol anchor and for the synthesis of polyphosphoinositides and sphingolipids. The increase in PI content may be important for the synthesis of these molecules. Additional work is necessary to address these questions.
It is also noteworthy that the zinc-mediated regulation of phospholipid synthesis occurs in a coordinate manner with the control of zinc homeostasis. In S. cerevisiae, the cellular levels of zinc are controlled by zinc transporters in the plasma membrane (Zrt1p, Zrt2p, Fet4p) [139–141] and in the membranes of the vacuole (Zrt3p, Cot1p, Zrc1p) [142–145], endoplasmic reticulum (Msc2p, Zrg17p) [82,146], and mitochondria (Mrs3p, Mrs4p) [147]. The expression of these transporters is largely regulated at the transcriptional level to maintain zinc homeostasis [148]. For example, the expression of the high affinity zinc transporter Zrt1p is induced for increased zinc uptake when the cellular level of zinc is limiting, whereas the expression of Zrt1p is repressed to attenuate zinc uptake when the cellular level of zinc is high [139]. Like DPP1 and PIS1, the activation of ZRT1 expression is dependent on the transcription factor Zap1p and the cis-acting element UASZRE [94,95,139]. The fact that the zinc transporters are located within the phospholipid bilayer of cellular membranes raises the question as to whether changes in phospholipid composition in response to zinc depletion might regulate their function. Several reports have shown that PE plays a major role in transporter function. For example, PE is required for amino acid transporter function in S. cerevisiae [149,150], and PE content in Escherichia coli is required for function of the γ-aminobutyric acid [151], lactose [16,152], and phenylalanine [153] transporters. The availability of mutants (e.g., eki1, psd1, psd2) defective in PE synthesis should facilitate studies to address the importance of changing PE content for zinc transport function in S. cerevisiae.
Acknowledgments
We acknowledge our colleagues for their valuable contributions to the understanding of phospholipid synthesis regulation by zinc. This work was supported by United States Public Health Service Grant GM-28140 from the National Institutes of Health.
Abbreviations
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- PA
phosphatidate
- CDP-DG
CDP-diacylglycerol
- DGPP
diacylglycerol pyrophosphate
- UASZRE
upstream activating sequence zinc-responsive element
- UASINO
upstream activating sequence inositol-responsive element
References
- 1.Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Ann Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 2.Paltauf F, Kohlwein SD, Henry SA. In: Regulation and Compartmentalization of Lipid Synthesis in Yeast. Jones EW, Pringle JR, Broach JR, editors. Cold Spring Harbor; New York: 1992. pp. 415–500. [Google Scholar]
- 3.Carman GM, Zeimetz GM. Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271:13293–13296. doi: 10.1074/jbc.271.23.13293. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg ML, Lopes JM. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry SA, Patton-Vogt JL. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 6.Carman GM, Henry SA. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 7.Zaremberg V, McMaster CR. Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J Biol Chem. 2002;277:39035–39044. doi: 10.1074/jbc.M207753200. [DOI] [PubMed] [Google Scholar]
- 8.Zheng Z, Zou J. The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J Biol Chem. 2001;276:41710–41716. doi: 10.1074/jbc.M104749200. [DOI] [PubMed] [Google Scholar]
- 9.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Ann Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 10.Dowhan W, Mileykovskaya E, Bogdanov M. Diversity and versatility of lipid–protein interactions revealed by molecular genetic approaches. Biochim Biophys Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker GW, Lester RL. Biosynthesis of phosphoinositol-containing sphingolipids from phosphatidylinositol by a membrane preparation from Saccharomyces cerevisiae. J Bacteriol. 1980;142:747–754. doi: 10.1128/jb.142.3.747-754.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon AK, Stevens VL. Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J Biol Chem. 1992;267:15277–15280. [PubMed] [Google Scholar]
- 13.Lester RL, Dickson RC. Sphingolipids with inositolphosphate-containing head groups. Adv Lipid Res. 1993;26:253–274. [PubMed] [Google Scholar]
- 14.Fankhauser C, Homans SW, Thomas-Oates JE, McConville MJ, Desponds C, Conzelmann A, Ferguson MA. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J Biol Chem. 1993;268:26365–26374. [PubMed] [Google Scholar]
- 15.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 17.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 18.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 19.Exton JH. Phosphatidylcholine breakdown and signal transduction. Biochim Biophys Acta. 1994;1212:26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 20.Becker GW, Lester RL. Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J Biol Chem. 1977;252:8684–8691. [PubMed] [Google Scholar]
- 21.Griac P, Henry SA. The yeast inositol-sensitive upstream activating sequence, UASINO, responds to nitrogen limitation and general nutrient availability. Nucleic Acids Res. 1999;27:2043–2050. doi: 10.1093/nar/27.9.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homann MJ, Poole MA, Gaynor PM, Ho CT, Carman GM. Effect of growth phase on phospholipid biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1987;169:533–539. doi: 10.1128/jb.169.2.533-539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwanyshyn WM, Han GS, Carman GM. Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J Biol Chem. 2004;279:21976–21983. doi: 10.1074/jbc.M402047200. [DOI] [PubMed] [Google Scholar]
- 24.Han GS, Johnston CN, Chen X, Athenstaedt K, Daum G, Carman GM. Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J Biol Chem. 2001;276:10126–10133. doi: 10.1074/jbc.M011421200. [DOI] [PubMed] [Google Scholar]
- 25.Rattray JB, Schibeci A, Kidby DK. Lipids of yeast. Bacteriol Rev. 1975;39:197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bossie MA, Martin CE. Nutritional regulation of yeast delta-9 fatty acid desaturase activity. J Bacteriol. 1989;171:6409–6413. doi: 10.1128/jb.171.12.6409-6413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonough VM, Stukey JE, Martin CE. Specificity of unsaturated fatty acid-regulated expression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1992;267:5931–5936. [PubMed] [Google Scholar]
- 28.Voelker DR. New perspectives on the regulation of intermembrane glycerophospholipid traffic. J Lipid Res. 2003;44:441–449. doi: 10.1194/jlr.R200020-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Birner R, Daum G. Biogenesis and cellular dynamics of aminoglycer-ophospholipids. Int Rev Cyt. 2003;225:273–323. doi: 10.1016/s0074-7696(05)25007-6. [DOI] [PubMed] [Google Scholar]
- 30.Carter JR, Kennedy EP. Enzymatic synthesis of cytidine diphosphate diglyceride. J Lipid Res. 1966;7:678–683. [PubMed] [Google Scholar]
- 31.Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- 32.Kanfer JN, Kennedy EP. Metabolism and function of bacterial lipids. II. Biosynthesis of phospholipids in Escherichia coli. J Biol Chem. 1964;239:1720–1726. [PubMed] [Google Scholar]
- 33.Letts VA, Klig LS, Bae-Lee M, Carman GM, Henry SA. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc Natl Acad Sci U S A. 1983;80:7279–7283. doi: 10.1073/pnas.80.23.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikawa J, Tsukagoshi Y, Kodaki T, Yamashita S. Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase. Eur J Biochem. 1987;167:7–12. doi: 10.1111/j.1432-1033.1987.tb13297.x. [DOI] [PubMed] [Google Scholar]
- 35.Kiyono K, Miura K, Kushima Y, Hikiji T, Fukushima M, Shibuya I, Ohta A. Primary structure and product characterization of the Saccharomyces cerevisiae CHO1 gene that encodes phosphatidylserine synthase. J Biochem. 1987;102:1089–1100. doi: 10.1093/oxfordjournals.jbchem.a122147. [DOI] [PubMed] [Google Scholar]
- 36.Clancey CJ, Chang SC, Dowhan W. Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem. 1993;268:24580–24590. [PubMed] [Google Scholar]
- 37.Trotter PJ, Pedretti J, Voelker DR. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem. 1993;268:21416–21424. [PubMed] [Google Scholar]
- 38.Trotter PJ, Pedretti J, Yates R, Voelker DR. Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem. 1995;270:6071–6080. doi: 10.1074/jbc.270.11.6071. [DOI] [PubMed] [Google Scholar]
- 39.Bremer J, Greenberg DM. Methyl transferring enzyme system of microsomes in the biosynthesis of lecithin (phosphatidylcholine) Biochim Biophys Acta. 1961;46:205–216. [Google Scholar]
- 40.Kodaki T, Yamashita S. Yeast phosphatidylethanolamine methylation pathway: cloning and characterization of two distinct methyltransferase genes. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- 41.Summers EF, Letts VA, McGraw P, Henry SA. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGraw P, Henry SA. Mutations in the Saccharomyces cerevisiae OPI3 gene: effects on phospholipid methylation, growth, and cross pathway regulation of phospholipid synthesis. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim K, Kim KH, Storey MK, Voelker DR, Carman GM. Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase. J Biol Chem. 1999;274:14857–14866. doi: 10.1074/jbc.274.21.14857. [DOI] [PubMed] [Google Scholar]
- 44.Hosaka K, Kodaki T, Yamashita S. Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli. J Biol Chem. 1989;264:2053–2059. [PubMed] [Google Scholar]
- 45.Min-Seok R, Kawamata Y, Nakamura H, Ohta A, Takagi M. Isolation and characterization of ECT1 gene encoding CTP:phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae. J Biochem. 1996;120:1040–1047. doi: 10.1093/oxfordjournals.jbchem.a021497. [DOI] [PubMed] [Google Scholar]
- 46.Tsukagoshi Y, Nikawa J, Yamashita S. Molecular cloning and characterization of the gene encoding cholinephosphate cytidylyltransferase in Saccharomyces cerevisiae. Eur J Biochem. 1987;169:477–486. doi: 10.1111/j.1432-1033.1987.tb13635.x. [DOI] [PubMed] [Google Scholar]
- 47.Hjelmstad RH, Bell RM. The sn-1,2-diacylglycerol ethanolaminephosphotransferase of Saccharomyces cerevisiae. Isolation of mutants and cloning of the EPT1 gene. J Biol Chem. 1988;263:19748–19757. [PubMed] [Google Scholar]
- 48.Hjelmstad RH, Bell RM. sn-1,2-diacylglycerol choline- and ethano-laminephosphotransferases in Saccharomyces cerevisiae. Nucleotide sequence of the EPT1 gene and comparison of the CPT1 and EPT1 gene products. J Biol Chem. 1991;266:5094–5103. [PubMed] [Google Scholar]
- 49.Hjelmstad RH, Bell RM. Mutants of Saccharomyces cerevisiae defective in sn-1,2-diacylglycerol cholinephosphotransferase: isolation, characterization, and cloning of the CPT1 gene. J Biol Chem. 1987;262:3909–3917. [PubMed] [Google Scholar]
- 50.Hjelmstad RH, Bell RM. The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene. J Biol Chem. 1990;265:1755–1764. [PubMed] [Google Scholar]
- 51.Nikawa J, Yamashita S. Molecular cloning of the gene encoding CDP-diacylglycerol-inositol 3-phosphatidyl transferase in Saccharomyces cerevisiae. Eur J Biochem. 1984;143:251–256. doi: 10.1111/j.1432-1033.1984.tb08366.x. [DOI] [PubMed] [Google Scholar]
- 52.Nikawa J, Kodaki T, Yamashita S. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987;262:4876–4881. [PubMed] [Google Scholar]
- 53.Paulus H, Kennedy EP. The enzymatic synthesis of inositol monophosphatide. J Biol Chem. 1960;235:1303–1311. [PubMed] [Google Scholar]
- 54.Klig LS, Henry SA. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc Natl Acad Sci U S A. 1984;81:3816–3820. doi: 10.1073/pnas.81.12.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dean-Johnson M, Henry SA. Biosynthesis of inositol in yeast. Primary structure of myo-inositol-1-phosphate synthase (EC 5.5.1.4) and functional analysis of its structural gene, the INO1 locus. J Biol Chem. 1989;264:1274–1283. [PubMed] [Google Scholar]
- 56.Murray M, Greenberg ML. Expression of yeast INM1 encoding inositol monophosphatase is regulated by inositol, carbon source and growth stage and is decreased by lithium and valproate. Mol Microbiol. 2000;36:651–661. doi: 10.1046/j.1365-2958.2000.01886.x. [DOI] [PubMed] [Google Scholar]
- 57.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 58.Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- 59.Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol Microbiol. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- 60.Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Clslp) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- 61.Ozier-Kalogeropoulos O, Fasiolo F, Adeline MT, Collin J, Lacroute F. Cloning, sequencing and characterization of the Saccharomyces cerevisiae URA7 gene encoding CTP synthetase. Mol Gen Genet. 1991;231:7–16. doi: 10.1007/BF00293815. [DOI] [PubMed] [Google Scholar]
- 62.Ozier-Kalogeropoulos O, Adeline MT, Yang WL, Carman GM, Lacroute F. Use of synthetic lethal mutants to clone and characterize a novel CTP synthetase gene in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:431–439. doi: 10.1007/BF00281793. [DOI] [PubMed] [Google Scholar]
- 63.Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henry SA. In: Membrane Lipids of Yeast: Biochemical and Genetic Studies. Strathern JN, Jones EW, Broach JR, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1982. pp. 101–158. [Google Scholar]
- 65.Carman GM. Phosphatidylcholine Metabolism. In: Vance DE, editor. Saccharomyces Cerevisiae. CRC Press; Boca Raton, FL: 1989. pp. 165–183. [Google Scholar]
- 66.Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- 67.Xie ZG, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht J, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci U S A. 1998;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci U S A. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waksman M, Eli Y, Liscovitch M, Gerst JE. Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem. 1996;271:2361–2364. doi: 10.1074/jbc.271.5.2361. [DOI] [PubMed] [Google Scholar]
- 70.Dowd SR, Bier ME, Patton-Vogt JL. Turnover of phosphatidylcholine in Saccharomyces cerevisiae. The role of the CDP-choline pathway. J Biol Chem. 2001;276:3756–3763. doi: 10.1074/jbc.M003694200. [DOI] [PubMed] [Google Scholar]
- 71.Boumann HA, Damen MJ, Versluis C, Heck AJ, de Kruijff B, de Kroon AI. The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry. 2003;42:3054–3059. doi: 10.1021/bi026801r. [DOI] [PubMed] [Google Scholar]
- 72.Noga AA, Vance DE. A gender-specific role for phosphatidylethanolamine N-methyltransferase-derived phosphatidylcholine in the regulation of plasma high density and very low density lipoproteins in mice. J Biol Chem. 2003;278:21851–21859. doi: 10.1074/jbc.M301982200. [DOI] [PubMed] [Google Scholar]
- 73.Noga AA, Vance DE. Insights into the requirement of phosphatidylcholine synthesis for liver function in mice. J Lipid Res. 2003;44:1998–2005. doi: 10.1194/jlr.M300226-JLR200. [DOI] [PubMed] [Google Scholar]
- 74.Kodaki T, Yamashita S. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption. Eur J Biochem. 1989;185:243–251. doi: 10.1111/j.1432-1033.1989.tb15109.x. [DOI] [PubMed] [Google Scholar]
- 75.Atkinson K, Fogel S, Henry SA. Yeast mutant defective in phosphatidylserine synthesis. J Biol Chem. 1980;255:6653–6661. [PubMed] [Google Scholar]
- 76.Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. Yeast mutants auxotropic for choline or ethanolamine. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trotter PJ, Voelker DR. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- 78.McMaster CR, Bell RM. Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases. J Biol Chem. 1994;269:28010–28016. [PubMed] [Google Scholar]
- 79.Morash SC, McMaster CR, Hjelmstad RH, Bell RM. Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J Biol Chem. 1994;269:28769–28776. [PubMed] [Google Scholar]
- 80.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 81.Schwabe JW, Klug A. Zinc mining for protein domains [news; comment] Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 82.Ellis CD, Wang F, MacDiarmid CW, Clark S, Lyons T, Eide DJ. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J Cell Biol. 2004;166:325–335. doi: 10.1083/jcb.200401157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oteiza PL, Olin KL, Fraga CG, Keen CL. Oxidant defense systems in testes from zinc-deficient rats. Proc Soc Exp Biol Med. 1996;213:85–91. doi: 10.3181/00379727-213-44040. [DOI] [PubMed] [Google Scholar]
- 84.Walsh CT, Sandstead HH, Prasad AS, Newberne PM, Fraker PJ. Zinc: health effects and research priorities for the 1990s. Environ Health Perspect. 1994;102:5–46. doi: 10.1289/ehp.941025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad SciUS A. 2000;97:7957–7962. doi: 10.1073/pnas.97.14.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu WI, Liu Y, Riedel B, Wissing JB, Fischl AS, Carman GM. Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:1868–1876. doi: 10.1074/jbc.271.4.1868. [DOI] [PubMed] [Google Scholar]
- 87.Toke DA, Bennett WL, Dillon DA, Chen X, Oshiro J, Ostrander DB, Wu W-I, Cremesti A, Voelker DR, Fischl AS. Carman, Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J Biol Chem. 1998;273:3278–3284. doi: 10.1074/jbc.273.6.3278. [DOI] [PubMed] [Google Scholar]
- 88.Oshiro J, Han GS, Carman GM. Diacylglycerol pyrophosphate phosphatase in Saccarhomyces cerevisiae. Biochim Biophys Acta. 2003;1635:1–9. doi: 10.1016/j.bbalip.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Wissing JB, Behrbohm H. Diacylglycerol pyrophosphate, a novel phospholipid compound. FEBS Lett. 1993;315:95–99. doi: 10.1016/0014-5793(93)81141-l. [DOI] [PubMed] [Google Scholar]
- 90.Munnik T, de Vrije T, Irvine RF, Musgrave A. Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J Biol Chem. 1996;271:15708–15715. doi: 10.1074/jbc.271.26.15708. [DOI] [PubMed] [Google Scholar]
- 91.Munnik T, Meijer HJG, Ter Riet B, Hirt H, Frank W, Bartels D, Musgrave A. Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 2000;22:147–154. doi: 10.1046/j.1365-313x.2000.00725.x. [DOI] [PubMed] [Google Scholar]
- 92.Den Hartog M, Musgrave A, Munnik T. Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J. 2001;25:55–65. doi: 10.1046/j.1365-313x.2001.00931.x. [DOI] [PubMed] [Google Scholar]
- 93.Van der Luit AH, Piatti T, Van Doorn A, Musgrave A, Felix G, Boller T, Munnik T. Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 2000;123:1507–1515. doi: 10.1104/pp.123.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem. 1998;273:28713–28720. doi: 10.1074/jbc.273.44.28713. [DOI] [PubMed] [Google Scholar]
- 96.Bird AJ, Zhao H, Luo H, Jensen LT, Srinivasan C, Evans-Galea M, Winge DR, Eide DJ. A dual role for zinc fingers in both DNA binding and zinc sensing by the Zap1 transcriptional activator. EMBO J. 2000;19:3704–3713. doi: 10.1093/emboj/19.14.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bird AJ, McCall K, Kramer M, Blankman E, Winge DR, Eide DJ. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 2003;22:5137–5146. doi: 10.1093/emboj/cdg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Han GS, Johnston CN, Carman GM. Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J Biol Chem. 2004;279:5338–5345. doi: 10.1074/jbc.M311779200. [DOI] [PubMed] [Google Scholar]
- 99.Yuan DS. Zinc-regulated genes in Saccharomyces cerevisiae revealed by transposon tagging. Genetics. 2000;156:45–58. doi: 10.1093/genetics/156.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnston CN. The Effects of Zinc and the DPP1-Encoded Diacylglycerol Pyrophosphate Phosphatase on the Vacuolar Phospholipid Composition in Saccharomyces Cerevisiae. Rutgers University; 2002. [Google Scholar]
- 101.Kersting MC, Carman GM. Regulation of the Saccharomyces cerevisiae EKI1-encoded ethanolamine kinase by zinc depletion. J Biol Chem. 2006;281:13110–13116. doi: 10.1074/jbc.M601612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 103.Kersting MC, Choi HS, Carman GM. Regulation of the yeast EKI1-encoded ethanolamine kinase by inositol and choline. J Biol Chem. 2004;279:35353–35359. doi: 10.1074/jbc.M405704200. [DOI] [PubMed] [Google Scholar]
- 104.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Ann Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 105.Balla T. Phosphatidylinositol 4-kinases. Biochim Biophys Acta. 1998;1436:69–85. doi: 10.1016/s0005-2760(98)00134-9. [DOI] [PubMed] [Google Scholar]
- 106.Gehrmann T, Heilmayer LG., Jr Phosphatidylinositol 4-kinases. Eur J Biochem. 1998;253:357–370. doi: 10.1046/j.1432-1327.1998.2530357.x. [DOI] [PubMed] [Google Scholar]
- 107.Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- 108.Leidich SD, Drapp DA, Orlean P. A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J Biol Chem. 1994;269:10193–10196. [PubMed] [Google Scholar]
- 109.Leidich SD, Orlean P. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J Biol Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- 110.White MJ, Lopes JM, Henry SA. Inositol metabolism in yeasts. Adv Microb Physiol. 1991;32:1–51. doi: 10.1016/s0065-2911(08)60004-1. [DOI] [PubMed] [Google Scholar]
- 111.Downes CP, Macphee CH. myo-Inositol metabolites as cellular signals. Eur J Biochem. 1990;193:1–18. doi: 10.1111/j.1432-1033.1990.tb19297.x. [DOI] [PubMed] [Google Scholar]
- 112.Divecha N, Irvine RF. Phospholipid signaling. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- 113.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 114.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 115.Kelley MJ, Bailis AM, Henry SA, Carman GM. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- 116.Han SH, Han GS, Iwanyshyn WM, Carman GM. Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J Biol Chem. 2005;280:29017–29024. doi: 10.1074/jbc.M505881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bailis AM, Lopes JM, Kohlwein SD, Henry SA. Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:1411–1418. doi: 10.1093/nar/20.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bailis AM, Poole MA, Carman GM, Henry SA. The membrane-associated enzyme phosphatidylserine synthase of yeast is regulated at the level of mRNA abundance. Mol Cell Biol. 1987;7:167–176. doi: 10.1128/mcb.7.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Klig LS, Homann MJ, Carman GM, Henry SA. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant. J Bacteriol. 1985;162:1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nikoloff DM, McGraw P, Henry SA. The INO2 gene of Saccharomyces cerevisiae encodes a helix–loop–helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res. 1992;20:3253. doi: 10.1093/nar/20.12.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoshizaki DK, Hill JE, Henry SA. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- 122.White MJ, Hirsch JP, Henry SA. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- 123.Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Loewy BS, Henry SA. The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes. Mol Cell Biol. 1984;4:2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix–loop–helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gaynor PM, Gill T, Toutenhoofd S, Summers EF, McGraw P, Homann MJ, Henry SA, Carman GM. Regulation of phosphatidylethanolamine methyltransferase and phospholipid methyltransferase by phospholipid precursors in Saccharomyces cerevisiae. Biochim Biophys Acta. 1991;1090:326–332. doi: 10.1016/0167-4781(91)90197-t. [DOI] [PubMed] [Google Scholar]
- 127.Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 128.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. Embo J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid synthesis. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 130.Wagner C, Blank M, Strohmann B, Schüller HJ. Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast. 1999;15:843–854. doi: 10.1002/(SICI)1097-0061(199907)15:10A<843::AID-YEA424>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 131.Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol Microbiol. 2001;41:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- 132.Iwanyshyn WM. Regulation of Phospholipid Synthesis Saccharomyces Cerevisiae. Vol. 2005. Zinc: Rutgers University; [DOI] [PubMed] [Google Scholar]
- 133.Morlock KR, Lin YP, Carman GM. Regulation of phosphatidate phosphatase activity by inositol in Saccharomyces cerevisiae. J Bacteriol. 1988;170:3561–3566. doi: 10.1128/jb.170.8.3561-3566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Komatsu M, Tanida I, Ueno T, Ohsumi M, Ohsumi Y, Kominami E. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J Biol Chem. 2001;276:9846–9854. doi: 10.1074/jbc.M007737200. [DOI] [PubMed] [Google Scholar]
- 136.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. Embo J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gaynor EC, Mondesert G, Grimme SJ, Reed SI, Orlean P, Emr SD. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol Biol Cell. 1999;10:627–648. doi: 10.1091/mbc.10.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci US A. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 141.Waters BM, Eide DJ. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem. 2002;277:33749–33757. doi: 10.1074/jbc.M206214200. [DOI] [PubMed] [Google Scholar]
- 142.MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. Embo J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.MacDiarmid CW, Milanick MA, Eide DJ. Induction of the ZRC1 Metal Tolerance Gene in Zinc-limited Yeast Confers Resistance to Zinc Shock. J Biol Chem. 2003;278:15065–15072. doi: 10.1074/jbc.M300568200. [DOI] [PubMed] [Google Scholar]
- 144.Miyabe S, Izawa S, Inoue Y. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2001;282:79–83. doi: 10.1006/bbrc.2001.4522. [DOI] [PubMed] [Google Scholar]
- 145.Devirgiliis C, Murgia C, Danscher G, Perozzi G. Exchangeable zinc ions transiently accumulate in a vesicular compartment in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;323:58–64. doi: 10.1016/j.bbrc.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 146.Ellis CD, MacDiarmid CW, Eide DJ. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J Biol Chem. 2005;280:28811–28818. doi: 10.1074/jbc.M505500200. [DOI] [PubMed] [Google Scholar]
- 147.Muhlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J Biol Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- 148.Eide DJ. Multiple Regulatory Mechanisms Maintain Zinc Homeostasis in Saccharomyces cerevisiae. J Nutr. 2003;133:1532S–1535S. doi: 10.1093/jn/133.5.1532S. [DOI] [PubMed] [Google Scholar]
- 149.Opekarova M, Robl I, Tanner W. Phosphatidyl ethanolamine is essential for targeting the arginine transporter Can1p to the plasma membrane of yeast. Biochim Biophys Acta. 2002;1564:9–13. doi: 10.1016/s0005-2736(02)00455-8. [DOI] [PubMed] [Google Scholar]
- 150.Robl I, Grassl R, Tanner W, Opekarova M. Construction of phosphatidylethanolamine-less strain of Saccharomyces cerevisiae. Effect on amino acid transport. Yeast. 2001;18:251–260. doi: 10.1002/1097-0061(200102)18:3<251::AID-YEA667>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 151.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the gamma-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 152.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 153.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]


