Abstract
A major pathway of eukaryotic messenger RNA (mRNA) turnover begins with deadenylation, followed by decapping and 5′ to 3′ exonucleolytic decay. We provide evidence that mRNA decapping and 5′ to 3′ degradation occur in discrete cytoplasmic foci in yeast, which we call processing bodies (P bodies). First, proteins that activate or catalyze decapping are concentrated in P bodies. Second, inhibiting mRNA turnover before decapping leads to loss of P bodies; however, inhibiting turnover at, or after, decapping, increases the abundance and size of P bodies. Finally, mRNA degradation intermediates are localized to P bodies. These results define the flux of mRNAs between polysomes and P bodies as a critical aspect of cytoplasmic mRNA metabolism and a possible site for regulation of mRNA degradation.
Messenger RNA turnover is an important step in regulating gene expression. In yeast, the major pathway of mRNA decay is initiated by shortening of the 3′ poly(adenosine) [poly(A)] tail, followed by removal of the cap by the decapping enzyme Dcp1p/Dcp2p, which in turn allows 5′ to 3′ exonucleolytic decay (1-8). Decapping is a key step in this pathway, because it permits the destruction of the mRNA and is a site of numerous control inputs (9).
Several observations suggest that decapping occurs when the mRNA undergoes a transition from a translationally competent messenger ribonucleoprotein (mRNP) to an mRNP state destined for decay. For example, the translation initiation factor eIF4E, which binds the cap structure, is an inhibitor of decapping both in vitro and in vivo (10, 11). Moreover, deadenylated mRNAs that interact with a complex of Lsm1-7 proteins, which activates decapping, are no longer bound by eIF4E or eIF4G (12).
The hypothesis that mRNAs enter a non-translating state after deadenylation and before decapping is analogous to the storage of mRNA in numerous biological contexts where deadenylated mRNAs are translationally repressed before their later activation. Consistent with a mechanistic similarity between decapping and mRNA storage, Dhh1p, which is an activator of decapping in yeast (13), has homologs that are required for the translational repression and storage of maternal mRNAs in Xenopus, Drosophila, and Caenorhabditis (14-16). Such stored mRNAs are often localized in discrete cytoplasmic granules, which represent accumulations of translationally repressed mRNAs (15). This analogy suggested the possibility that Dhh1p and other factors involved in mRNA decapping would be found in specific cytoplasmic sites in yeast.
To determine the localization of proteins involved in decay, we constructed green fluorescent protein (GFP) fusions of yeast mRNA decapping factors and determined their location in live cells (17). These fusion proteins are at the C terminal, include the full-length protein, and are functional (18). We observed that both subunits of the decapping enzyme, Dcp1p and Dcp2p, are strikingly concentrated in discrete cytoplasmic foci (Fig. 1, B and C). In addition, we observed that the decapping activators Lsm1p (19), Pat1p (19), and Dhh1p, are also all concentrated in similar foci (Fig. 1, D to F). In contrast, GFP alone (Fig. 1G) was distributed throughout the cell. The size and number of these foci vary between individual cells and can also be affected by the tagged protein examined and the growth conditions. Using Dhh1p as a marker, we observed 2.4 ± 1.4 foci per cell. These foci are cytoplasmic in comparison with live-cell DNA staining (20).
Fig. 1.
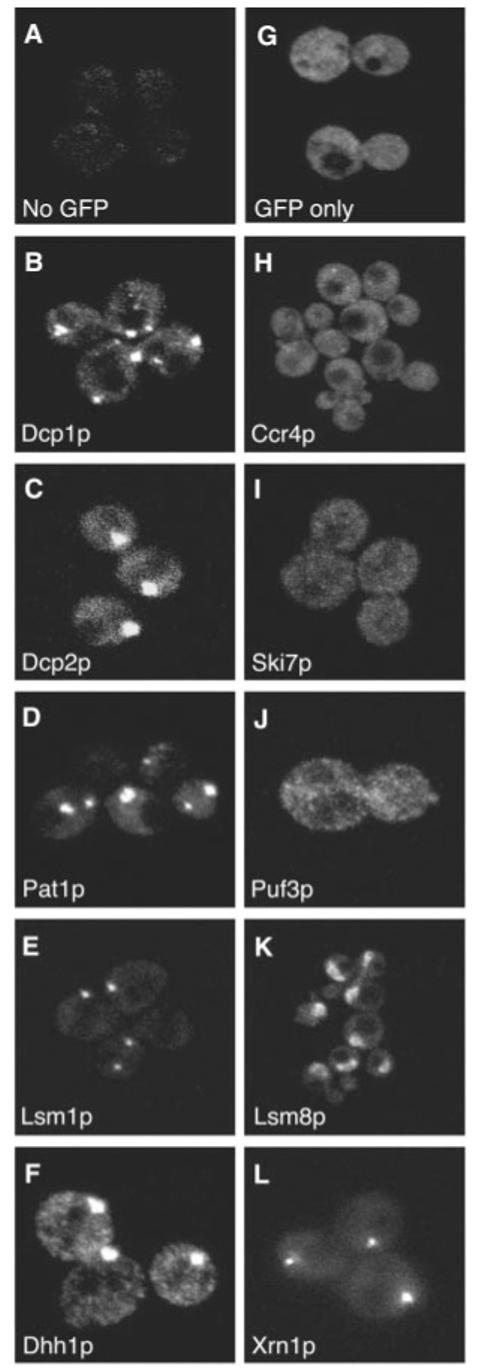
Decapping factors and Xrn1p localize to discrete foci in the cell. Proteins involved in mRNA decay were tagged with GFP following the PCR-based gene modification method described by Longtine et al. (17). (A) No GFP control. Cells expressing GFP tagged versions of factors involved in decapping are shown in (B to F) and factors involved in other aspects of mRNA turnover are shown in (H to L), and (G) shows untagged version of GFP only. The protein observed by GFP tagging is shown in the bottom left corner of each panel.
To determine the range of proteins concentrated in these sites, we tagged proteins involved in other steps of mRNA turnover and examined their distribution. We observed that Ccr4p, a subunit of the major cytoplasmic dead-enylase (21), Ski7p, a subunit of the cytoplasmic exosome (22) , and Puf3p, an mRNA specific 3′ UTR binding protein (23), are more uniformly distributed throughout the cell (Fig. 1, H to J). We do observe some small foci containing Ccr4p, which suggests that Ccr4p can transiently localize to the foci containing the decapping factors or is present in foci of its own. Lsm8p, which forms an Lsm2-8p complex associated with the U6 snRNA is not in foci and is localized to the nucleus (Fig. 1K). In contrast, the 5′-3′ exonuclease Xrn1p, which degrades mRNAs after decapping, is also localized to discrete foci (Fig. 1L).
Two observations indicate that the decapping factors and Xrn1p are in the same foci. First, these proteins copurify and coimmuno-precipitate in a variety of experiments (12, 13, 19, 24, 25). Second, we examined the localization of Dcp1p, Dhh1p, and Xrn1p tagged with GFP in cells containing red fluorescent protein (RFP)-tagged Lsm1p. Our results show that all of these proteins are present in the same foci (fig. S1). We refer to these structures as P bodies (for cytoplasmic processing bodies).
The P bodies might represent sites of storage of mRNA decay factors or may be specific sites wherein mRNAs are decapped and degraded 5′ to 3′. If P bodies are dynamic sites of decapping and exonucleolysis, then inhibiting mRNA turnover before decapping should reduce the number of the P bodies, whereas inhibiting decay at, or after decapping should increase the number of P bodies. To test this possibility, we used Dhh1p as a marker for P bodies and examined its distribution in various mutants that affect mRNA turnover.
We observed that strains blocked at the actual step of decapping (dcp1Δ) or 5′ to 3′ degradation (xrn1Δ) showed dramatic increases in the size and abundance of P bodies (compare Fig. 2A with Fig. 2, B and C). The results of Western analysis show that the level of Dhh1 p-GFP does not significantly differ in these strains, which indicates that the changes in the P bodies are not due to differences in Dhh1p-GFP abundance (fig. S2). In contrast, in a ccr4Δ strain, which is deficient in deadenylation, P bodies are markedly reduced, but not altogether absent (Fig. 2D). The fact that some small P bodies are observed is consistent with the observation that some decapping still occurs in a ccr4Δ strain (21). However, because ccr4Δ strains show a reduction in Dhh1p-GFP expression (fig. S2), we cannot rule out that part of the reduction in P bodies in a ccr4Δ strain is due to decreased Dhh1p-GFP. Because storage sites for mRNA decapping and exonucleolysis factors would be expected to increase when dead-enylation is blocked, these results are inconsistent with P bodies being storage sites for mRNA decay factors. Moreover, the decrease in P bodies when deadenylation is inhibited, and the increase in P bodies when the decapping or 5′ to 3′ exonucleolytic digestion is inhibited, is consistent with P bodies being specific sites of mRNA decay. Given this, we suggest that P bodies are sites of mRNA decapping and 5′ to 3′ exonucleolysis and that the size of P bodies reflects the flux of mRNAs undergoing decapping. This interpretation is also supported by the observation that treatment of cells with cycloheximide, which rapidly inhibits decapping, presumably by trapping mRNAs on polysomes (26), caused a loss of P bodies within 10 minutes (fig. S3).
Fig. 2.
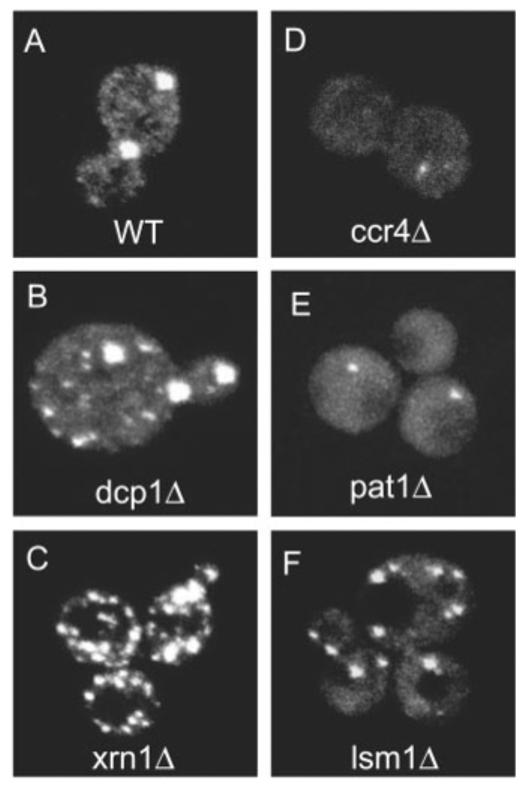
Alteration of mRNA decay affects the size and the number of P bodies. Using Dhh1p-GFP as a marker for P bodies, P bodies were observed in strains which are deleted for mRNA decay factors. (A) shows P bodies in WT cells, (B to F) show P bodies in dcp1Δ, xrn1Δ, ccr4Δ, pat1Δ, and lsm1Δ cells, respectively. Average number of P bodies in dcp1Δ and xrn1Δ strains per cell were 6.25 ± 3.7 and 12.7 ± 7.8, respectively, which, compared with wild-type strains in a Student's t test, gave P values of ≪0.001.
We also examined P-body formation in strains deleted for various activators of decapping. In a pat1Δ strain, the size of the foci decreases (Fig. 2E), whereas in an lsm1Δ strain, the number of foci increases (Fig. 2F). This suggests that these proteins differ in the mechanism by which they activate decapping. This is consistent with the observation that Pat1p coimmunoprecipitates with mRNA that is associated with translation initiation factors, whereas Lsm1p is only associated with mRNAs that are not bound to translation initiation factors (12).
If P bodies are sites of decay, then mRNAs should be transiently associated with these sites just before and during degradation. To specifically examine the subcellular location of mRNAs in the process of degradation, we introduced a poly(G) tract in the 3′ UTR of the unstable MFA2 mRNA. This forms a block to the exonuclease Xrn1p and leads to the accumulation of an intermediate in the decay process (2). To observe the localization of this intermediate, we introduced into the mRNA downstream of the poly(G) tract binding sites for the MS2 bacteriophage coat protein and also expressed a MS2-GFP fusion protein. The interaction of the MS2-GFP protein with the MS2 sites on the RNA allows the mRNA to be localized in the cells (27).
An important result was that RNA decay fragments are seen in discrete foci (Fig. 3A, top panel on the left). In contrast, in strains expressing MFA2 transcripts lacking the MS2 sites, no foci could be seen (Fig. 3A, middle panel on the left). This indicates that these foci represent MS2-GFP protein bound to RNA and not free MS2-GFP protein. Moreover, strains expressing an MFA2 transcript without a poly(G) tract inserted to trap the decay intermediate also did not show discrete accumulation of MS2-GFP (Fig. 3A, bottom panel on the left). This indicated that the RNA seen in the foci is the decay intermediate (Fig. 3A). In addition, strains expressing a PGK1 mRNA with a poly(G) and MS2 binding sites in the 3′ UTR also accumulate an mRNA decay intermediate (2), and that intermediate is localized to foci (Fig. 3C). To determine that the RNA-containing foci are P bodies, we showed that the MFA2 decay intermediate colocalized with Lsm1p-RFP (Fig. 3B). This indicates that mRNAs in the process of decay are associated with P bodies.
Fig. 3.
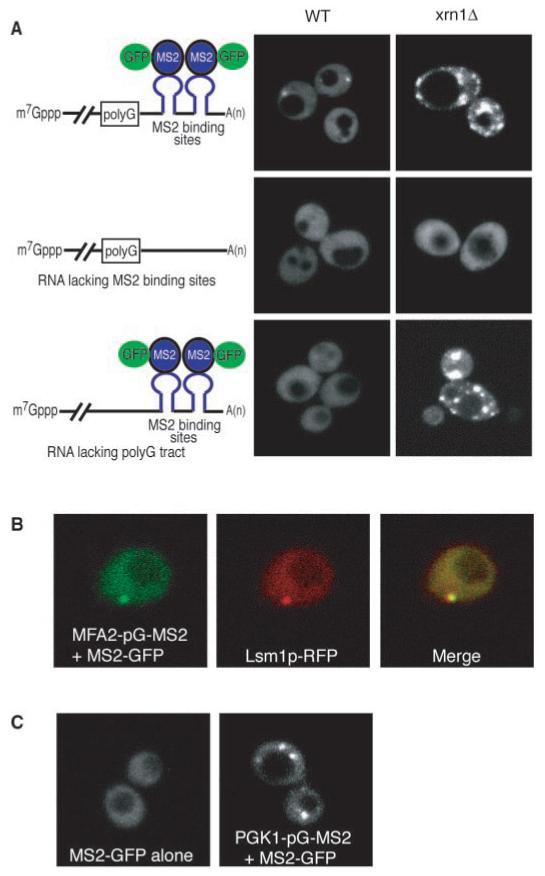
RNA decay intermediates are localized to P bodies. (A) WT (left column) and xrn1Δ (right column) cells are shown expressing the MFA2 mRNA with poly(G) and MS2 sites (top row), or the MFA2 mRNA with only a poly(G) tract (middle row), or the MFA2 mRNA with only the MS2 sites (bottom row). The schematic diagram of the reporter RNA expressed from the plasmid and its interaction with the MS2-GFP fusion protein is shown on the left. For simplicity, only a single protein is shown bound to a stem loop; however, because MS2 coat protein binds as a dimer, at least two MS2-GFP molecules are bound per site. (B) Left, RNA; middle, Lsm1p-RFP; and right, the merge generated by Adobe Photoshop. (C) WT cells expressing only the MS2-GFP fusion protein (left) or with a PGK1 reporter mRNA with a poly(G) tract and MS2 binding sites (right).
We also trapped intermediates of mRNA decay by removing the Xrn1p exonuclease, which leads to the accumulation of deadenylated decapped mRNAs (4, 8). Thus, mRNA decay intermediates should accumulate in these cells independent of the poly(G) tract. We observed that the MFA2 mRNA [with or without an inserted poly(G) tract] accumulated in P bodies in the xrn1Δ cells (Fig. 3A, top and bottom panels on the right). Consistent with the observation that P-body number increases in xrn1Δ cells (Fig. 2), we observed that in xrn1Δ strains the number of RNA-containing foci also increased.
From the observations that mRNA decapping factors and decay intermediates are concentrated in P bodies, and that the size and number of P bodies is affected by altering mRNA decay, we define P bodies as sites of mRNA decapping and 5′ to 3′ exonucleolytic decay. However, since these mRNA decay factors are not exclusively in P bodies, it is possible that decapping and 5′ to 3′ degradation may also occur outside of P bodies. Moreover, because recent results indicate that Dcp1p, Dcp2p, Lsm1p, and Xrn1p are found in discrete cytoplasmic foci in mammalian cells (28-30), we suggest that equivalent P bodies are present in mammalian cells and, presumably, also function as specific sites of mRNA degradation.
From these and prior observations, we present a model for the dynamic relationship between translation and decapping. In this model, mRNAs exist in two distinct functional states, one in polysomes and one sequestered into a nontranslating pool, which would be the pool present in P bodies and the direct substrate for mRNA decapping. Transitions between these two pools would be dictated by the translation status of the mRNA. For example, trapping the mRNAs in polysomes with cycloheximide inhibits decapping and leads to loss of P bodies within 10 min after treatment with the drug (fig. S3). This suggests that the polysome and P body pool of mRNAs are distinct. This is supported by the fact that factors involved in translation are not seen in distinct foci under these conditions (31). In contrast, inhibition of translation initiation, either in cis or in trans (5, 10, 32) leads to an increase in decapping rate, presumably by increasing the flux of mRNA through P bodies.
The occurrence of decay in P bodies adds to a growing number of examples where mRNA molecules are sequestered into specific subcytoplasmic compartments (33, 34) and suggests that the assembly, disassembly, and transitions between such compartments will be important control points in mRNA metabolism.
An exciting issue is whether P bodies are sites of additional events in cytoplasmic mRNA biology. Because components of P bodies are also utilized for maternal mRNA storage, we anticipate that P bodies, or closely related structures, will be sites of mRNA storage for later translation activation. In this case, storage could be achieved by simply inhibiting the decapping reaction. This predicts that future analysis of P bodies and stored mRNAs, either maternal or in somatic cells such as neurons, may reveal an underlying conservation of biochemical function. Moreover, because both mRNA storage and decapping involve transitions between a translation competent and a translation repressed state, we speculate that other transitions of mRNA molecules into and out of translation might occur within P bodies.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/300/5620/805/
DC1
Materials and Methods
Figs. S1 to S3
Table S1
References
References and Notes
- 1.Shyu AB, Belasco JG, Greenberg ME. Genes Dev. 1991;5:221. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 2.Decker CJ, Parker R. Genes Dev. 1993;7:1632. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 3.Muhlrad D, Parker R. Genes Dev. 1992;6:2100. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 4.Muhlrad D, Decker CJ, Parker R. Genes Dev. 1994;8:855. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 5.Muhlrad D, Decker CJ, Parker R. Mol Cell Biol. 1995;15:2145. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beelman CA, et al. Nature. 1996;382:642. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 7.Dunckley T, Parker R. EMBO J. 1999;18:5411. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CL, Stevens A. Mol Cell Biol. 1993;13:4826. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucker M, Parker R. Annu Rev Biochem. 2000;69:571. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz DC, Parker R. Mol Cell Biol. 1999;19:5247. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz DC, Parker R. Mol Cell Biol. 2000;20:7933. [Google Scholar]
- 12.Tharun S, Parker R. Mol Cell. 2001;8:1075. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 13.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. RNA. 2001;7:1717. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ladomery M, Wade E, Sommerville J. Nucleic Acids Res. 1997;25:965. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Development. 2001;128:3233. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 16.Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. Development. 2001;128:3221. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- 17.Longtine MS, et al. Yeast. 1998;14:953. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18. The GFP-tagged fusions of decay factors were shown to be functional in vivo for mRNA decay by comparing the ratio of the full length to the decay fragment at steady state of PGK1pG and MFA2pG reporter mRNAs.
- 19.Tharun S, et al. Nature. 2000;404:515. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 20.Sheth U, Parker R. unpublished observations. [Google Scholar]
- 21.Tucker M, et al. Cell. 2001;104:377. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 22.van Hoof A, Staples RR, Baker RE, Parker R. Mol Cell Biol. 2000;20:8230. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivas W, Parker R. EMBO J. 2000;19:6602. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnerot C, Boeck R, Lapeyre B. Mol Cell Biol. 2000;20:5939. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. EMBO J. 2000;19:1661. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beelman C, Parker R. J Biol Chem. 1994;269:9687. [PubMed] [Google Scholar]
- 27.Bertrand E, et al. Mol Cell. 1998;2:437. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 28.Dijk E. van, et al. EMBO J. 2002;21:6915. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lykke-Andersen J. Mol Cell Biol. 2002;22:8114. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. RNA. 2002;8:1489. [PMC free article] [PubMed] [Google Scholar]
- 31.Brengues M, Parker R. unpublished observation. [Google Scholar]
- 32.LaGrandeur T, Parker R. RNA. 1999;5:4420. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson P, Kedersha N. J Cell Sci. 2002;115:3227. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 34.Kedersha N, et al. Mol Biol Cell. 2002;13:195. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. We thank M. A. V. Sanchez for the MFA2-pG-MS2 plasmid, P. Hilleren for the PGK1-pG-MS2 plasmid, K. Bloom for the MS2-GFP plasmid, A. van Hoof for the strain expressing SKI7-GFP, R. Tsein for the mRFP plasmid, H. Fares for experimental assistance, J. M. Coller for helpful discussions, and the members of the Parker Lab for their advice. Supported by Howard Hughes Medical Institute and NIH (GM45443)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


