Abstract
CYP2J2 metabolizes arachidonic acid to 20-HETE and EETs which play a critical role in the regulation of renal, pulmonary, cardiac and vascular function. However, the contribution of CYP2J2 to EET formation in the liver remains poorly characterized. Similarly, information is sparse regarding the extent and variability of CYP2J2 expression during human development. This investigation was undertaken to characterize the variability of CYP2J2 expression in fetal liver, heart, kidney, lung, intestine and brain and postnatal liver samples. CYP2J2 mRNA expression was measured using quantitative PCR, and immunoreactive CYP2J2 was examined using two anti-CYP2J2 antibodies. CYP2J2 mRNA was ubiquitously expressed in pre- and postnatal samples. Fetal hepatic mRNA expression varied 127-fold (1351 ± 717 transcripts/ng total RNA), but this variation was reduced to 8-fold after exclusion of four samples with extremely low levels of mRNA. Amounts of immunoreactive protein also varied substantially among samples without an apparent relationship with transcript number or genotype. Western blot analysis revealed a different protein pattern between prenatal and postnatal liver samples. DNA resequencing of selected subjects identified a single novel SNP (CYP2J2*10), which was found in only one subject and therefore did not explain the large variability in CYP2J2 protein content. In vitro expression suggests that the protein product of CYP2J2*10 confers reduced enzymatic activity. Aberrant splicing produces three minor transcripts which were present in all samples tested. Due to premature termination codons none encodes functional protein. The mechanisms leading to variable amounts of immunoreactive protein and distinct pre- and postnatal CYP2J2 protein patterns warrant further investigation.
Introduction
Arachidonic acid products 20-hydroxyeicosatetraenoic acid (20-HETE) and 5,6-, 8,9–11,12, and 14,15-epoxyeicosatrienoic acids (EETs) play a critical role in the regulation of renal, pulmonary and cardiovascular function. Recent studies have indicated that conversion of arachidonic acid to these metabolites is primarily catalyzed by cytochrome P450 (CYP) enzymes in the brain, lung, kidney, and peripheral vasculature. While several enzymes from different CYP families, among them members of the CYP1A, 2B, 2C, 2D, 2E, 2J, and 4A subfamilies, have been implicated in EET formation, CYP2J2 has emerged as one of the major extrahepatic enzymes in humans. However, its contribution to EET formation in the liver remains poorly characterized. CYP2J2 has also been reported to be expressed in endothelial cells where EETs, in addition to regulating vascular tone, act as anti-inflammatory mediators. The role of CYP-mediated metabolites of arachidonic acid, including those generated by CYP2J2, has been reviewed and summarized in great detail by Roman (Roman, 2002).
There is also evidence that CYP2J2 plays a role in carcinogenesis. Recent investigations have suggested that upregulation of this enzyme, likely through a c-Jun-responsive module, promotes the neoplastic phenotype of carcinoma cells and may be involved in the pathogenesis of a variety of human cancers (Jiang et al., 2005; Marden and Murray, 2005). However, CYP2J2 can also metabolize a number of xenobiotics including the antihistamine drugs ebastine (Hashizume et al., 2002) and astemizole (Matsumoto et al., 2002) contributing to their presystemic elimination in the small intestine. Taken together, CYP2J2 emerges as a crucial enzyme in a number of physiological functions.
The human CYP2J2 cDNA was first cloned and characterized by Wu et al. 1996 (Wu et al., 1996). In vitro expression demonstrated that it metabolized arachidonic acid predominantly via olefin epoxidation to EETs. Immunoblotting studies revealed that it was primarily expressed in heart and, to a lesser extent, in liver, ileum, jejunum, colon and kidney. Furthermore, interindividual variation in the expression of CYP2J2 mRNA in heart and liver was remarkably low. The expression of CYP2J2 in human tissues was subsequently investigated in more detail demonstrating that the enzyme was abundantly expressed in several tissues (Enayetallah et al., 2004). The CYP2J2 gene was eventually cloned and sequenced, and was found to consist of nine exons similar to most other members of the human CYP2 family (King et al., 2002). Currently, the Cytochrome P450 nomenclature committee has defined nine allelic CYP2J2 variants (http://www.cypalleles.ki.se/cyp2j2.htm). In vitro expression studies indicated reduced metabolic activity towards both arachidonic acid and linoleic acid for CYP2J2*2, *3 and *6, toward arachidonic acid for CYP2J2*4, while the CYP2J2*5 variant did not appear to have altered activity (King et al., 2002). CYP2J2*7 involves a G>T substitution in the regulatory region at position −76 (−50 of transcription start) which causes the loss of an Sp1 transcription factor binding site. Consequently, this leads to lower amounts of CYP2J2 protein and reduced levels of circulating CYP2J2 epoxygenase metabolites in vivo (King et al., 2002; Spiecker et al., 2004). In addition, two novel variants, CYP2J2*8 and *9, have recently been discovered in a Korean population, and in vitro expressed CYP2J2*8 was associated with severely compromised activity towards astemizole O-demethylation and ebastine hydroxylation (Lee et al., 2005). The limited allele frequency data available suggest that the majority of reduced-function alleles are rare and that the occurrence of some alleles may be restricted to certain populations (e.g. CYP2J2*3-*6 were only detected in individuals of African descent (King et al., 2002) and CYP2J2*8 and *9 were only detected in Koreans (Lee et al., 2005)). The sole variant that appears to be present in subjects of all ethnicities is CYP2J2*7 which has been found at frequencies between 0.026 and 0.18 (King et al., 2002; Lee et al., 2005; Wang et al., 2006).
Most studies on CYP2J2 have been performed with adult subjects or adult-derived tissues. Since CYP2J2 appears to play an important role in EET formation, has a wide tissue distribution and is highly conserved (i.e. low degree of polymorphic expression), one may speculate that it also carries out critical functions during human development. However, to our knowledge, there is limited information about prenatal CYP2J2 gene expression and function. To gain more insight into CYP2J2 expression during human development, we performed quantitative PCR and immunoblot analyses on a large panel of liver samples genotyped for CYP2J2 and compared expression levels to that in other tissues. We also detected and characterized alternative splice variants, and resequenced selected samples with high and low CYP2J2 protein levels.
Methods
Source of DNA and RNA
Tissue samples were obtained through NICHD-supported tissue retrieval programs, the University of Maryland Brain and Tissue Bank for Developmental Disorders, (Baltimore, MD), and the Central Laboratory for Human Embryology at the University of Washington (Seattle, WA). RNA from 76 liver specimens (n=58, prenatal; n=18, postnatal) and specimens of fetal heart (n=5), lung (n=10), kidney (n=9), gut (n=4), brain (n=3), testis (n=2) and adrenal (n=2) were used for the study. The use of these tissues was approved by the University of Missouri-Kansas City Pediatric Health Sciences Review Board. ‘Brain’ refers to cerebrum or cerebellum and was derived from midsections 5, 7, 9, 11 or 13 (all from the University of Maryland). The fetal samples of this study are essentially the same as described previously (Leeder et al., 2005). The post-mortem interval was <6 hours for all samples.
Genomic DNA (n=376) was derived from liver tissue (n=76), or discarded or anticoagulated blood obtained for routine clinical management of hospitalized patients (Caucasians, n=128; African Americans, n=128; American Asians, n=44). Ethnicity was self-reported or retrieved from data sheets accompanying the tissues. The protocol for sample collection and usage was approved by the University of Missouri-Kansas City Pediatric Health Sciences Review Board.
DNA isolation and genotyping for CYP2J2
DNA was isolated using DNeasy Tissue or QIAamp Blood DNA kits (Qiagen, Valencia, CA). Genotyping was performed by PCR-RFLP. Assays were designed de novo using GenBank accession AF272142 as reference sequence. Details including primer sequences and PCR reaction conditions are given in Table 1. PCR amplifications were performed with JumpStart REDTaq DNA polymerase (Sigma, St. Louis, MO) at 1.5 mM MgCl2 with the buffer provided or at optimized MgCl2 concentrations (Table 1). Fragment lengths, restriction enzymes used and restriction patterns for respective assays are shown in Table 2. Restriction enzymes and the 100 bp DNA ladder were purchased from New England Biolabs (Beverly, MA). Digested PCR fragments were separated on 3% agarose gels containing Synergel (Diversified Biotech, Boston, MA).
Table 1.
Primer sequences and PCR reaction conditions
| Primer name | Position AF272142 | Primer Sequence | PCR (bp) | Application | Anneal °C | PCR details |
|---|---|---|---|---|---|---|
| 10 F
11 R |
6007–6026
39147–39168 |
5′ GAA GAG CAG GAG GAC GTC TG
5′ GTC TTC TTA CTT TCC TTG CCC C |
1572 | full length cDNA | 59 | REDAccuTaq LA |
| 28 F
15 R |
6223–6240
21012–21033 |
5′ CAC CTG GAG GTT CAG CTG
5′ CTT GAA ATG AGG GTC AAA AGG C |
194, 357 | SV4 detection (see Fig 4) | 58 | REDTaq |
| 16 F
19 R |
24969–24988
38959–38976 |
5′ GGC CCT CTA CCC AGA AAT CC
5′ CTG TTC TCC GAG GCA TGC |
188, 376, 527 | SV2+3 detection (see Fig 4) | 66 | REDTaq |
| 14 F
15 R |
16807–16832
21012–21033 |
5′ CCC CTA TGC GAG AAC ATA TCT TTA AG
5′ CTT GAA ATG AGG GTC AAA AGG C |
209 | QRT-PCR | 61 | REDTaq |
| 3 F
4 R |
20426–20448
21075–21094 |
5′ GGA ATC AGA ATA CTG TAA TGT TG
5′ GGT ACT CAA AGC GTT CTC CG |
669 | *2 and *4 genotyping | 57 | REDTaq
2 mM MgCl2 |
| 3 F
5 R |
20426–20448
20703–20732 |
5′ GGA ATC AGA ATA CTG TAA TGT TG
5′ CTA TCT TCC ATT CCT AGC ACA GTG CTa tGC |
307 | *3 genotyping | 57 | REDTaq
3 mM MgCl2 |
| 6 F
7 R |
27698–27718
27921–27941 |
5′ GAA CCC TTC CCT CTT TAC CAG
5′ CCC TAA AAG ATG GGC TTG AAG |
244 | *5 genotyping | 60 | REDTaq |
| 8 F
9 R |
31609–31627
31831–31848 |
5′ CCC CTC TGC ATA GGT GTG C
5′ CCC TGG ACT CCC ACA CAC |
240 | *6 genotyping | 60 | REDTaq |
| 1 F
2 R |
5764–5782
6101–6118 |
5′ GAT TGA ACC GAA CAG AGG C
5′ GCA GAA AGG CGA CAG TGC |
355 | *7 genotyping | 60 | REDTaq
1 mM MgCl2 |
| 30 F
31 R |
1481–1507
6775–6796 |
5′ TTC ACA CTG AGC ATC TAG CAA TTT GTC
5′ CTG CAT CAA GAA GGG TCC CAG G |
5316 | seq template
upstream, exon 1 |
62 | REDAccuTaq LA |
| 34 F
37 R |
16168–16190
21565–21589 |
5′ AGG CAT GTA CCA CCA TAC CCA GC
5′ AGA GCA ACC AGC ACA GAT GAC TCT C |
5422 | seq template
exons 2, 3, 4 |
68 | Platinum Taq High Fidelity, 5% DMSO |
| 38 F
41 R |
22358–22384
28363–28389 |
5′ GCA TCT ATG CCC TAC TAC CAC ATT CCC
5′ ACA CCA GCA TTA ATA TGA TGT TGT CCC |
6032 | seq template
exons 5, 6, 7 |
60 | REDAccuTaq LA |
| 44 F
45 R |
31201–31224
39916–39940 |
5′ GGG GTG CCA TGT ACT AGA CAT TGG
5′ CAA GAA AAG ATC ACA CCC ACC ATC G |
8740 | seq template
exons 8 and 9 |
65 | REDAccuTaq LA |
| 75-F
55 R |
16782–16809
16992–17011 |
5′ CAA AAC TTT GGG AAC CGC CCC GTG ggC C
5′ AGG GAG TGA CTT CAA TCA GG |
230 | *10 Exon 2 SNP genotyping | 58 | REDTaq |
| 74 F
46 R |
5416–5443
5592–5611 |
5′AAC AAA TGT TAA TTA AGC ACT TAT cAT G
5′ TCT CGG TAG TCT CAT CTT GC |
196 | Upstream SNP genotyping | 52 | REDTaq |
The table gives the name, sequence and position (GenBank accession AF272142) of each primer. Primers are shown in pairs as they were used for different applications (cDNA amplification, quantitative PCR, genotyping or generation of sequencing template). The lengths of respective PCR amplicons are shown in base pairs (bp) and annealing temperatures for each reaction are given in ºC. Longer amplicons were generated with either the JumpStart REDAccuTaq LA DNA Polymerase Mix (REDAccuTaq LA) or Platinum Taq High Fidelity using their accompanying buffer. Shorter amplicons were amplified with JumpStart REDTaq DNA Polymerase (REDTaq) and the buffer provided. Some reactions were supplemented with MgCl2 or DMSO with final concentrations as indicated. For additional details, please see text.
Table 2.
PCR-RFLP-based CYP2J2 genotyping assays
| Allele | Enzyme | PCR in bp | Digest ºC | wild-type pattern in bp | variant pattern in bp |
|---|---|---|---|---|---|
| *2 | Hpy188I | 669 | 37 | 350+108+102+84+18+7 | 434+108+102+18+7 |
| *3 | NsiI | 307 | 37 | 280 + 27 | 141 + 139 + 27 |
| *4 | Tsp509I | 669 | 65 | 379 + 290 | 379 + 253 + 37 |
| *5 | MseI | 244 | 37 | 214 + 30 | 174 + 40+ 30 |
| *6 | Tsp509I | 240 | 65 | 157 + 83 | 240 |
| *7 | AluI | 355 | 37 | 301 + 54 | 165 + 136 + 54 |
| *10 | ApaI | 230 | 25 | 202 + 28 | 230 |
Primer sequences and other PCR details are given in Table 1. Each reaction contained 2.5U restriction enzyme and the appropriate accompanying restriction buffer that was diluted 20-fold to a final concentration 0.5x. Digestion reactions were incubated >6 hrs or overnight to ensure completeness.
RNA isolation and quantitative real-time (QRT)-PCR analysis
Total RNA was extracted from frozen tissue (20–35 mg) according to the Qiagen RNeasy protocol (Qiagen, Valencia, CA) with an on-column DNase I treatment. RNA quality was assured by agarose gel electrophoresis and quantity measured spectrophotometrically. QRT-PCR was performed with the QuantiTect SYBR green one-step RT-PCR kit from Qiagen on a DNA Engine Opticon 2 instrument (MJ Research, Boston, MA). Each 12 μl reaction contained 15 ng RNA and was carried out in triplicate. Primers and annealing temperatures are given in Table 1. Serial dilutions of a CYP2J2 amplicon served as standard to generate a calibration curve ranging from 102 to 107 molecules. Transcript numbers were calculated from log-linear regression analysis of the calibration curve. Data were normalized to 18S rRNA which was measured with the TaqMan Ribosomal RNA Control Reagent Kit (Applied Biosystems, Foster City, CA).
Immunodetection of CYP2J2 protein
Microsomal fractions were prepared from prenatal and postnatal liver as described previously (Leeder et al., 2005). Microsomal samples were heated to 80°C for 10 minutes. Microsomal proteins (4 μg per lane) were separated using the Novex NuPAGE System (10 well 4–12% gradient gels, XCell SureLock Mini-Cell apparatus, LDS sample buffer, sample reducing agent, antioxidant, MOPS SDS running buffer; Invitrogen, Carlsbad, CA) or the Criterion XT System (18 well 10% Criterion XT gels, Criterion Cell apparatus, XT sample buffer, reducing agent, MOPS running buffer; Bio-Rad, Hercules, CA). Proteins were separated at 150 or 180 volts for 3 hours when using the Novex and Criterion systems, respectively. Proteins were transferred to Hybond-C Extra Nitrocellulose (Amersham Biosciences, Piscataway, NJ) using a Bio-Rad Semi-dry transfer unit with transfer buffer consisting of 48 mM Tris, 39 mM glycine, 20% methanol, 0.0375% SDS at pH 9.2. Membranes were blocked overnight at room temperature on a rocker in 10 mM Tris, 150 mM NaCl, 0.2% Tween 20, pH 8 (TNT) containing 4% skim milk powder (M-TNT). Blocked membranes were incubated for 2 hours with CYP2J2 antibodies designated as anti-CYP2J2rec (Wu et al., 1996) or anti-CYP2J2pep3 (Seubert et al, 2004) diluted 1:50,000-fold in M-TNT and alternately washed in TNT and H2O (5 min X 3 TNT/H2O wash cycles). The anti-CYP2J2rec antibody was made against a partially purified preparation of human CYP2J2 protein and has been shown to cross-reacts with CYP2J proteins of multiple species, but not with non-CYP2J isoforms. The anti CYP2J2pep3 antibody was raised against a peptide common to all CYP2J isoforms (see (King et al., 2002) for details). To detect bound antibody, blots were incubated with horseradish peroxidase-conjugated donkey anti-rabbit antibody (1:50,000) (Amersham Biosciences, Piscataway, NJ) in M-TNT for 30 min, then alternately washed in TNT and H2O (5 min X 3 TNT/H2O wash cycles), incubated with ECL Advance chemiluminescense reagents according to the manufacturer’s protocol (Amersham Biosciences, Piscataway, NJ), and exposed to Hyperfilm ECL. Films were scanned using a flat-bed scanner. Heterologously expressed CYP2J2 protein (BD Gentest, San Jose, CA) was present on each membrane as a positive control. Each sample was run twice to ensure reproducibility.
cDNA synthesis
Reverse transcription was performed on 2 μg total RNA with an oligo (dT) primer using the Qiagen Omniscript RT kit. Reaction volumes were 20 μl. Subsequent CYP2J2 transcript-specific PCR reactions (full length or partial) were carried out on 0.4 μl cDNA in 8 μl reactions (Table 1).
Amplification, cloning and characterization of CYP2J2 transcripts
Full length CYP2J2 cDNA was amplified in a long-range PCR reaction to allow generation of fragments shorter and longer than the expected wild-type 1.57 kb transcript, should such transcripts exist. Subsequently, cDNA from two samples (one prenatal testis, one postnatal liver) containing potential variant transcripts was cloned with the pCR-XL-TOPO cloning kit designed for efficient cloning of larger amplicons (Invitrogen, Carlsbad, CA) and screened for insert length. Clones with inserts deviating in size from that of correctly spliced mRNA were further analyzed by DNA sequencing with DYEnamic ET dye terminator chemistry and a MegaBACE 500 capillary sequencer (Amersham Biosciences, Piscataway, NJ).
To detect differentially spliced variants in panels of cDNAs from various tissues, we designed primers amplifying the affected regions. The reaction for splice variants SV2 and SV3 generated three fragments identifying correctly spliced, exon 7A deleted and exon 7B retained transcripts, respectively, while the reaction for SV4 yielded two PCR products corresponding to correctly spliced and exon 2 deleted transcripts, respectively. Primers and other reaction details are given in Table 1.
CYP2J2 gene resequencing
Four long-range PCR products ranging from 5.3 to 8.7 kb in length and covering all exons as well as flanking intronic and the 3′and 5′-untranslated regions were generated (Table 1). The fragments were treated with Exo/SAP-IT (USB, Cleveland, OH) and subsequently sequenced with DYEnamic ET dye terminator chemistry and a MegaBACE 500 capillary sequencer using appropriately spaced CYP2J2 primers to cover all exons, at least 100 bp of flanking intronic, 550 bp upstream and 375 bp of downstream sequence, respectively.
Automated splice site analysis
The strengths of splice sites and splice enhancer binding sites were determined by automated analysis available at https://splice.cmh.edu using models based on the April 2003 release of the Human Genome Sequence (Build 33). According to the models, donor and acceptor splice sites are defined by an information content (Ri value) >0 bit (Rogan et al., 1998; Nalla and Rogan, 2005).
In vitro expression and characterization of CYP2J2*10
The CYP2J2 cDNA was expressed in the bacterial expression vector PCW+ (source pCWhum3A4HT, (Guryev et al., 2001)) after truncation of the N-terminus and addition of a hexahistidine tag to the C-terminus to facilitate purification (wild-type construct, CYP2J2.1). The P115L amino acid change was introduced into the wild-type construct using the primers: 5′-CCGCCCCGTGACCCTTATGCGAGAACATA 3′ and 3- TATGTTCTCGCATAAGGGTCACGGGGCGG-5′ (the changed nucleotide is underlined) with a QuickChange II XL site directed mutagenesis kit (Strategene, La Jolla, CA) according to the manufacturer’s instructions. The integrity of the variant construct was confirmed by DNA sequencing. Protein expression was performed as previously described (Cheesman et al., 2003) and also in the presence of chaperone proteins (GroES-GroEL) (Mitsuda and Iwasaki, 2006). Expressed CYP2J2 protein was affinity purified using a Ni-NTA agarose column from Qiagen (Valencia, CA) and eluted fractions were dialyzed against 100mM Potassium phosphate buffer, pH 7.4, containing 20% glycerol prior to storage in 15–20 μM aliquots at −80 °C. Heme content in these preparations was determined using the pyridine hemochromagen assay of sodium dithionite reduced heme (Smith, 1975). Details of the bacterial expression and purification systems will be described elsewhere (Jones et al., in preparation).
Statistical Analyses
Analyses were performed with the SPSS version 12.0 software package for Windows (SPSS Inc., Chicago, IL). A normal distribution was established by Q-Q-Plot for the number of transcripts/ng total RNA. Univariate analyses of variance were performed using log-transformed values for number of transcripts/ng total RNA. To assess whether expression levels differed among tissues, a mixed model analysis with a Bonferroni adjustment for multiple comparisons was performed. Linear regression analysis was performed with Microsoft Excel 2002. A p-value <0.05 was considered statistically significant.
Results
CYP2J2 genotyping
All subjects for which tissue was available (n=76) were genotyped for CYP2J2*2 through *7. No CYP2J2*2, *3, *4, *5, or *6 alleles were found in any of the samples. CYP2J2*7, however, was observed at a frequency of 0.099 in this ethnically mixed population (Caucasian, n=35; African American, n=25; other or unknown, n= 16).
Variability in CYP2J2 gene transcription
Expression of CYP2J2 mRNA is reported as transcripts per nanogram total RNA corrected for the expression of 18S rRNA as recommended by Koch et al. (Koch et al., 2002) and as applied previously to fetal liver CYP3A expression (Leeder et al., 2005). As shown in Fig. 1A, expression varied 127-fold in the panel of human fetal liver samples (n=58). Mean ± SD (range) expression was 1351 ± 717 transcripts/ng total RNA (23 to 2957 transcripts/ng total RNA). A subset of four samples had markedly reduced levels of CYP2J2 mRNA, of which three samples (CMM311, CMM32, CMM361) subsequently were found to be devoid of immunoreactive protein while the fourth sample, CMM1153, did reveal protein. These four samples were significantly different when compared to the remaining 54 samples (p=0.001). These samples have been previously reported to contain extremely low levels of CYP3A7 mRNA, immunoreactive protein and catalytic activity (Leeder et al., 2005). In the subset of 54 samples mRNA expression was normally distributed, and the level of expression was not affected by age, either estimated gestational age (r2 =0.047) or postnatal age (r2 =0.0003). When excluded from the analysis, variability in CYP2J2 mRNA expression decreased from 127-fold to 8-fold (n=54). Furthermore, there was no statistically significant difference between the pre- and postnatal groups regardless whether the four samples described above were included (p=0.86) or excluded (p=0.17) from the analysis. In the 18 postnatal samples (ranging from 4 days to 18 years of age), mRNA expression varied only 19-fold (mean ± SD, 1482 ± 1229 transcripts/ng total RNA, range: 253 to 4728 transcripts/ng total RNA). The lowest transcript number was observed in a sample from a 16 day-old infant that was homozygous for the CYP2J2*7 allele. We also noted that this sample exhibited some RNA degradation and it was not conclusive whether the low transcription level was due to the genotype or RNA quality. Exclusion of this sample decreased the variability to 15-fold; mean ± SD (range) expression was 1554 ± 1227 transcripts/ng total RNA (326 to 4728 transcripts/ng total RNA). Genotype, i.e. the presence of the CYP2J2*7 allele did not impact the number of transcripts/ng total RNA. As shown in Figure 2, there was no statistically significant difference between the two genotype groups in either the fetal (p=0.37) or postnatal group (p=0.67).
Figure 1. Expression of CYP2J2 mRNA in human tissues during development.
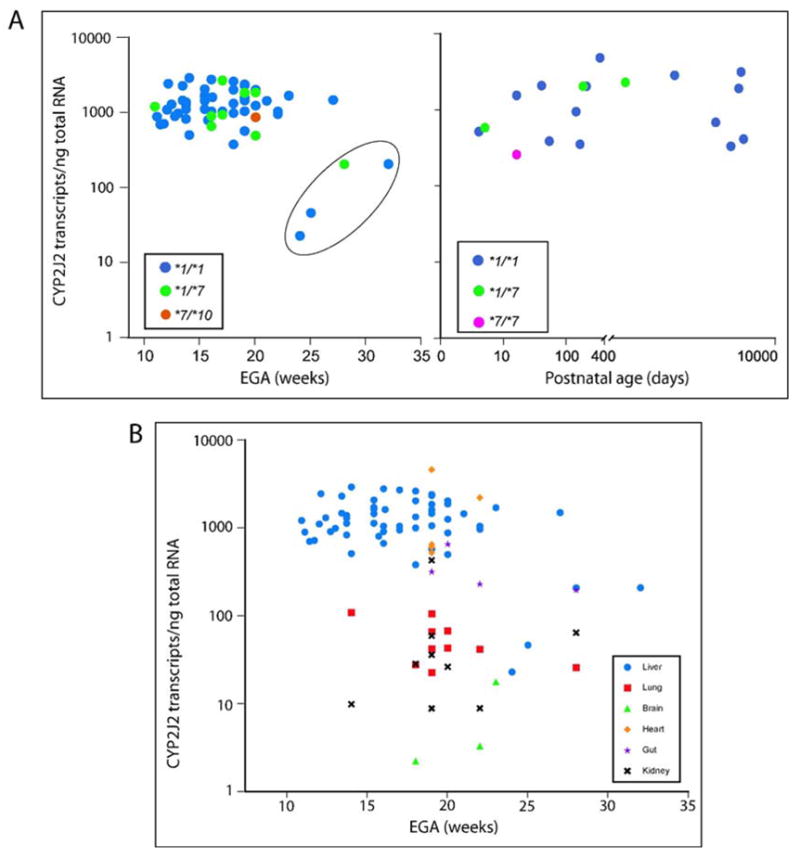
QRT-PCR was performed on 15 ng total RNA in triplicate reactions. Serial dilutions of a CYP2J2 amplicon served as a standard to generate a calibration curve ranging from 102 to 107 copies. Transcript numbers were calculated from log-linear regression analysis of the calibration curve and normalized for 18S ribosomal RNA content.
A: Expression in human fetal and postnatal liver. The genotype of each sample is as indicated in the inset. The 127 fold-variation within the fetal samples decreased to 8-fold upon exclusion of four samples with the lowest transcript numbers (circled). These samples were found to contain low levels of RNA, protein and activity in a previous study (Leeder et al., 2005). EGA, estimated gestational age. There is no significant difference between the pre-and postnatal groups (p=0.82; circled samples excluded, p=0.17). The circled samples are significantly different (p=0.001).
B: Expression in extra-hepatic tissues. Each tissue is coded by a symbol as given in the inset. Liver samples are shown for comparison. Heart and intestine are not significantly different from liver (p=1.0 and 0.14). Lung and kidney are comparable (p=1.0), but differ significantly from liver (p=0.001).
Figure 2. Relationship between CYP2J2 mRNA expression and genotype.
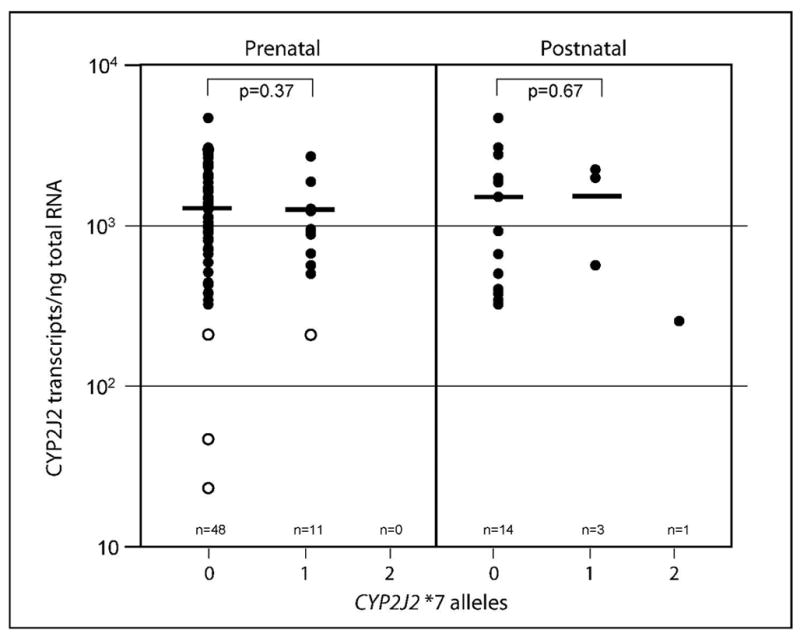
The figure shows the relationship between the number of transcripts/ng total RNA and the presence of zero (0), one (1) or two (2) CYP2J2*7 alleles. There was no significant difference between samples with zero and one CYP2J2*7 alleles regardless whether the four samples with low transcript numbers were included (p=0.71) or excluded (p=0.37). No CYP2J2*2-*6 allelic variants were found. The graph contains fetal and postnatal samples. The samples circled in Figure 1A are shown with open circles; the bar indicates the mean transcript number.
Smaller panels of extra-hepatic tissues were also investigated and demonstrated that CYP2J2 was ubiquitously expressed in the fetus (Fig 1B). QRT-PCR revealed the highest expression levels in prenatal heart (n=5, mean ± SD 1735 ± 1777 transcripts/ng total RNA) and intestine (n=4, mean ± SD 356 ± 213 transcripts/ng total RNA) followed by lung (n=10, mean ± SD 56 ± 32 transcripts/ng total RNA) and kidney (n=9, mean ± SD 75 ± 135 transcripts/ng total RNA). Lowest levels were observed in brain (n=3; 2, 8 and 18 transcripts/ng total RNA). Univariate analysis of variance revealed that mRNA expression was comparable between liver and heart (p=1.0) or intestine (p=0.15), but was significantly greater than that observed in lung ((p=0.001) and kidney (p=0.001). No statistically significant difference was observed between lung and kidney. The outcome did not change when all samples derived from the four subjects with low liver mRNA (circled in Figure 1A) were excluded from the analysis.
Variability in CYP2J2 immunoreactive protein
Immunoreactive protein content was assessed in 51 prenatal and 18 postnatal liver microsomal samples. Representative immunoblots illustrating the range of immunoreactive CYP2J2 content in samples ranging from 11.1 weeks of gestation to 17 years of life is shown in Fig 3A. Of note, the polyclonal anti-CYP2J2rec antibody recognized two predominant immunoreactive proteins in the postnatal samples and in one prenatal sample (CMM1153; 32 weeks EGA). Upon close inspection, some samples revealed an additional minor band. This band pattern has been observed previously in adult liver microsomes, but not in heart microsomes (Wu et al., 1996). In contrast, the lower molecular mass protein was not observed in all other fetal liver samples. The CYP2J2pep3 anti-peptide antibody consistently recognized two proteins in the fetal samples while the postnatal pattern consisted of multiple bands of variable intensities (Fig 2B). Of note, sample CMM1153 again exhibited a pattern resembling that observed in postnatal microsomes. These data suggested the presence of novel CYP2J2 isoform(s) or alternatively, the presence of an unrelated protein with an epitope(s) recognized by both CYP2J2 antibodies used.
Figure 3. CYP2J2 immunoreactive protein in fetal and postnatal liver.
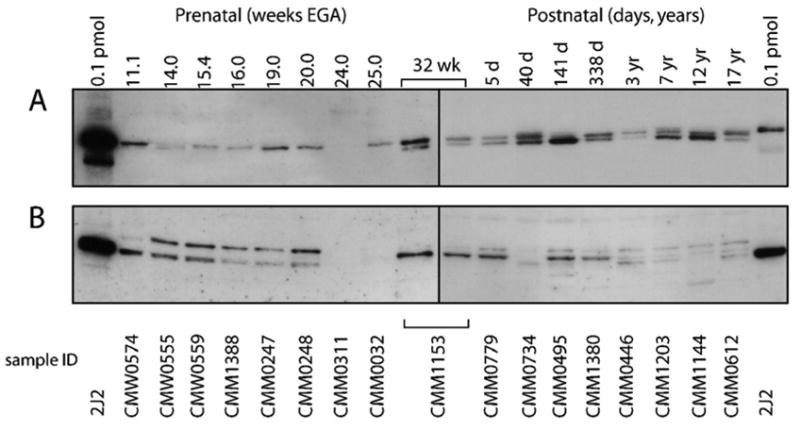
A representative set of samples ranging from 11.1 to 32 weeks EGA and 5 days to 17 years postnatal age are shown. Each lane contained 4 μg microsomal protein, the first and last lanes contained 0.1 pmol recombinant CYP2J2 protein as a positive control. Sample CMM1153, which exhibits a pattern that is more consistent with that observed in the postnatal samples, was loaded on both blots as a reference for band patterns. Membranes containing the fetal and postnatal samples were exposed for 30 and 10 seconds, respectively.
A: detection of immunoreactive protein with anti-CYP2J2rec
B: detection of immunoreactive protein with anti-CYP2J2pep3
Despite comparable CYP2J2 mRNA transcript levels in the prenatal and postnatal liver samples, the amount of immunodetectable protein was considerably lower in the fetus. As a consequence, a longer exposure time was needed to visualize immunoreactive CYP2J2 protein in prenatal samples compared to the postnatal samples (30 sec exposure vs. 10 sec, respectively). The extent of this phenomenon can be appreciated in Fig 3A and 3B by comparing the intensity of the signal for the 0.1 pmol standard of recombinant CYP2J2 protein included in each panel.
Considerable variability in protein content was also observed among the panel of prenatal samples. This finding is summarized in Figure 4. The amount of immunoreactive protein ranged from ‘low’ (including non-detectable) to ‘intermediate’ and ‘high’ (by visual inspection, categories used for descriptive purposes only). The ‘low’ protein bands approximated 0.006 pmol of CYP2J2 protein per μg microsomal protein, while the strongest bands in the ‘high’ protein category were between 0.012 and 0.02 pmol of CYP2J2 protein per μg microsomal protein. No relationship between transcript number and the amount of immunoreactive protein was apparent across the panel of 51 samples (not shown).
Figure 4. Variability of fetal CYP2J2 immunoreactive protein and relationship between protein content and mRNA expression.
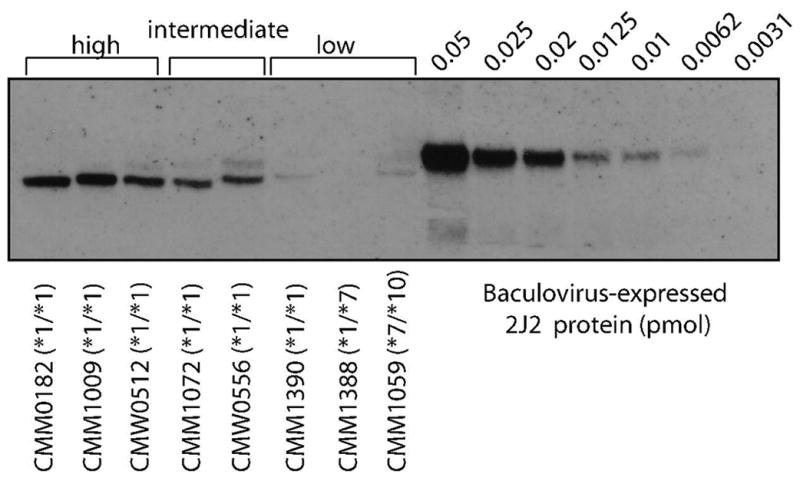
Each lane contained 4 μg microsomal protein. Anti-CYP2J2 antibody was used for panels A and B.
Immunoblot with representative samples demonstrating the range of immunoreactive protein observed. For descriptive purposes, ‘low’, ‘intermediate’ and ‘high’ protein categories were assigned by visual inspection. There was no relationship between genotype and amount of protein observed.
CYP2J2 splice variants and automated splice site analysis
Because mRNA expression appeared to be comparable in prenatal and postnatal liver and immunoreactive protein expression patterns were variable, we explored the possibility that developmentally regulated alternative splicing may lead to reduced expression of the canonical transcript and thus, translation into CYP2J2 protein in prenatal liver. Therefore, full-length cDNA was amplified from a panel of tissue samples. The canonical transcript, as well as amplification products that were smaller and larger than the correctly spliced transcript, were detected in all samples implying the presence of alternative splice products (Fig 5A). Cloning and sequencing the cDNA from two tissue samples (prenatal testis and postnatal liver) with appreciable levels of the variant transcripts revealed three novel splice variants (Fig 5B). Two variants, SV2 and SV4, had deletions of exon 2 and exon 7, respectively, while SV3 retained a cryptic exon. To discriminate the canonical exon 7 from the cryptic exon 7, they have been designated as exons 7A and 7B, respectively. Both exon deletions and the retention cause frame shifts and introduce premature stop codons.
Figure 5. Expression of CYP2J2 splice variants.
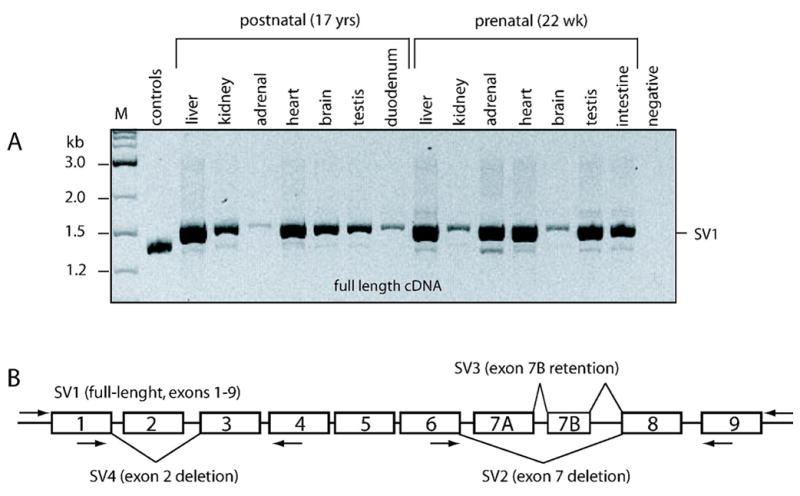
Two sets of tissues, one from a postnatal (17 year old) and one from a fetal (22 wks estimated gestational age) male, were utilized for alternate splice variant analysis. Each reaction contained cDNA corresponding to 40 ng total RNA.
A: Full-length cDNA (SV1) was the major amplification product in each tissue analyzed. Minor variants were visible across the panels. M, 1 kb ladder (A); control, amplification from a plasmid mix containing the exon 2 and exon 7 deletions.
B: Graphic display of alternate splice variants identified by cloning and sequencing. Primer locations are as indicated. Boxed depict exons. Transcript-specific PCR for the exon 2 deletion variant (SV4) did not produce any visible product (not shown).
Splice variants SV2 and SV3 were minor transcripts and difficult to visualize by gel electrophoresis, but appeared to be present in all tissues tested (not shown). In contrast, PCR amplification specific for the exon 2 deletion variant (SV4) did not reveal any visible product in a panel of tissues including the two subjects from which it was originally cloned (CMM684 and CMM1409), suggesting that it constitutes only a rare variant (not shown).
The strengths of the splice donor and acceptor sites were calculated with the information theory-based splice site analysis tool that we have successfully applied to the analysis of other CYP splice sites (Rogan et al., 2003; Gaedigk et al., 2005) (Table 3). Each canonical donor and acceptor splice site contained at least 3.7 bits of information. The exon 2 and exon 7A deletions could not be explained by the presence of weak splice acceptor or donor sites. However, the observed splice junctions of the cryptic exon 7B corresponded to strong 9.0 bit and 5.2 bit acceptor and donor sites, respectively, as predicted by splice site analysis.
Table 3.
Automated splice site analysis.
| Exon | Splice Acceptor Site | Position AF272142 | Splice Donor Site | Position AF272142 |
|---|---|---|---|---|
| Exon 1 | n/a | n/a | 6.9 | 6241 |
| Exon 2 | 6.0 | 16676 | 9.9 | 16840 |
| Exon 3 | 13.6 | 20464 | 8.7 | 20615 |
| Exon 4 | 14.6 | 21007 | 11.3 | 21169 |
| Exon 5 | 7.1 | 22815 | 10.6 | 22993 |
| Exon 6 | 3.7 | 24849 | 10.8 | 24992 |
| Exon 7A | 10.0 | 27718 | 8.7 | 27907 |
| Exon 7B | 9.0 | 28260 | 5.2 | 28412 |
| Exon 8 | 9.6 | 31673 | 10.6 | 31813 |
| Exon 9 | 13.0 | 38950 | n/a | n/a |
Splice acceptor and donor site strengths are given in bits. The current model is based on the Human Genome Sequence (Build 33) available at https://splice.cmh.edu. The position of the splice site is indicated in reference to GenBank accession AF272142 and as defined.
The variability in fetal liver CYP2J2 protein expression patterns was unlikely due to SV2, SV3 and SV4, since these splice variants were minor and rather ubiquitously present across tissues. Therefore, we searched for additional splice variants, which may have escaped our initial cloning/screening experiments. However, amplification of cDNA covering exons 2 through 9 and exons 2 through 7A did not reveal any additional splice products in samples classified as ‘high’ (n=3), ‘intermediate’ (n=2) and ‘low’ (n=3) protein expression. The samples used for this analysis corresponded to those shown in Figure 4.
Discovery and characterization of the novel CYP2J2*10 allele
A nonsynonymous C>T SNP was discovered at AF272142 g.16810 (cDNA position 344) when selected DNA samples with ‘low’ levels of immunoreactive protein were resequenced. This SNP was detected in a single subject of unknown ethnicity (CMM1059) and was confirmed by sequencing multiple PCR-generated templates as well as by PCR-RFLP genotyping. Subsequent genotyping of the tissue DNA panel (n=76 including the carrier) and an additional set of DNAs including 128 Caucasians, 128 African Americans and 44 American Asians (total of 600 alleles) did not reveal any other carrier, making this allele a rare event. The novel allele was designated as CYP2J2*10 by the P450 Nomenclature Committee at http://www.cypalleles.ki.se/cyp2j2.htm.
To assess the functional consequence of the P115L amino acid substitution, CYP2J2.10 protein was expressed in the presence and absence of chaperone proteins GroEL/GroES using a bacterial expression system. Based on protein content, expression with the chaperones yielded 432 and 345 nmol/L CYP2J2 wild-type and CYP2J2.10 protein, respectively. SDS-PAGE analysis with the CYP2J2pep3 antibody showed a protein band at 57 kDa that was identical in size to that derived from the wild-type cDNA and both expressed proteins. However, CYP2J2.10 appeared to be devoid of heme as judged by the lack of a P450 CO-difference spectrum at 450nm and the presence of a large absorbance peak at 280nm (not shown). Moreover, the ratio of free to protein-bound heme revealed only trace amounts of heme in the CYP2J2.10 preparation, while heme was readily incorporated into wild-type protein (ratios of 0.87 and 0.02 for wild-type and CYP2J2.10, respectively). These results strongly indicate that P115L affects proper heme binding or protein folding. Therefore, CYP2J2.10 protein activity is likely severely compromised. Preliminary results using terfenadine as a probe substrate for CYP2J2 activity corroborate our findings and further support our conclusion that CYP2J2.10 protein activity is likely severely compromised (Jones et al., in preparation). An alignment of CYP2 family members including human, rat and rabbit sequences showed that P115 is located within the substrate recognition site (SRS)-1 that proceeds the formation of the B’-helix, but it does not appear to be highly conserved. At this point in time, however, it remains speculative whether P115 is important for protein folding and/or heme binding.
Discussion
The findings of this study demonstrate that CYP2J2 mRNA is expressed in human fetal liver as early as 11 weeks EGA, expression levels are comparable to those observed in fetal heart, interindividual variability is relatively low and there is no apparent change in the level of expression after birth. Furthermore, transcript was also detected in every other extrahepatic tissue tested, albeit at lower levels. In contrast to the apparently tight regulation of gene transcription in fetal liver (Fig 1A), there were marked differences in the amount of immunoreactive protein present in this tissue (Fig 3 and 4). Taken together, this suggests that processes other than transcription regulate variability in CYP2J2 protein content. Unfortunately, to date there is no validated CYP2J2 probe substrate available that would allow specific determination of CYP2J2 catalytic activity. Certain antihistamine drugs, including ebastine and astemizole, are known to be metabolized by CYP2J2, but other P450s such as CYP3A4, CYP4F and CYP2D6 have been reported to contribute to the metabolism of these substrates (Hashizume et al., 2002; Matsumoto et al., 2002). While CYP3A4 and CYP2D6 are not expressed at appreciable amounts in fetal liver, their contribution to fetal ebastine and astemizole metabolism may be negligible. However, CYP3A5 and CYP3A7, which are present in fetal liver (Leeder et al., 2005) and CYP4F family members, which likely are also expressed prenatally, given their importance in fatty acid epoxide inactivation (Le Quere et al., 2004) may contribute to the overall activity measured towards ebastine and astemizole and therefore render these probes nonspecific. Efforts to overcome these challenges are currently underway.
We also detected different pre- and postnatal protein expression patterns. In the postnatal liver samples, the anti-CYP2J2 antibody recognized two or three distinct bands, which is similar to the observations of Wu et al. in adult liver using the same antibody (Wu et al., 1996). Interestingly, a predominant immunoreactive protein was detected in the fetal liver tissue samples, similar to the results reported by Wu et al. in adult heart. These authors speculated that adult liver contains additional CYP2J2 isoforms and/or other proteins that share immunochemical determinants with CYP2J2, but their nature remains elusive. Regardless, it appears that protein expression in adult heart resembles that in fetal liver and that there is a distinct pattern in postnatal liver. Differential protein patterns between pre-and postnatal liver were also observed with a second antibody, anti-CYP2J2pep3, providing further evidence that these developmental differences are real. On the other hand, Gu et al. found only a single immunoreactive band in fetal olfactory mucosa (n=8) and liver (n=6) as well as in their single adult control sample using an anti-rat Cyp2J4 antibody (Gu et al., 2000). We can only speculate at this point in time whether the anti-rat Cyp2J4 antibody is indeed more specific compared to the antibodies used in this study and others (Wu et al., 1996; Yu et al., 2000; King et al., 2002; Jiang et al., 2005) or fails to detect certain CYP2J2 postnatal liver-specific isoforms. Clearly, further studies are required to determine if the various proteins recognized by the CYP2J2 antibodies are indeed isoforms and if so, to characterize their functional properties in a developmental context.
The four samples with low CYP2J2 transcript numbers (Fig 1A) were derived from fetuses with severe congenital anomalies. These samples also presented with low CYP3A7 mRNA expression, protein content and activity. As described and discussed in detail by Leeder et al., 2005, these observations could not be explained by poor sample quality. Noteworthy is sample CMM1153, which demonstrated a pattern of immunoreactivity unlike the other three samples (Fig 1A and 3), but very similar to that observed in postnatal samples. This unique presentation of a ‘fetal’ sample may be explained by compromised viability due to porencephaly or other unknown factors.
In order to explain the variability in protein content, the liver panel was genotyped for CYP2J2*2 through *7 by PCR-RFLP. The only variant found was the more common CYP2J2*7 allele, which could not explain the variable amounts of protein. Therefore, we performed gene resequencing on eight subjects with high, intermediate and low protein levels, but no sequence variations explaining the variable amounts of protein were revealed. Only a single novel SNP at position AF272142 g.16810 was discovered in one subject of unknown ethnicity. Subsequent genotyping of the liver panel and additional Caucasian, African American and Asian samples totaling 600 chromosomes, did not reveal any additional carriers of this novel allele termed CYP2J2*10. The failure to detect any additional novel SNPs within the coding and 100bp of intronic sequences is consistent with previous studies which discovered few sequence variations at rather low frequencies (King et al., 2002; Spiecker et al., 2004; Lee et al., 2005). However, the presence of a SNP(s) located deep within an intron may explain low levels of protein. Such a scenario is the main cause of CYP3A5 poor metabolism, where a SNP in intron 3 creates a cryptic splice site that leads to a premature stop codon and nonfunctional truncated protein (Kuehl et al., 2001). Since resequencing focused on exons, flanking intron portions and 500 bp of promoter sequence, SNPs outside these regions eluded discovery in this investigation.
Gene reporter studies have demonstrated that the loss of an SP1 binding site on the CYP2J2*7 allele causes a reduction in transcription level (Spiecker et al., 2004), but the transcript number/ng total RNA was not significantly different between our homozygous and heterozygous groups, respectively (Fig. 2). At this point we can only speculate whether the functional allele compensates for the reduced expression of CYP2J2*7 in liver tissue, whether there are additional splice variants that are initiated from a different promoter or whether other factors are at play. Albeit significantly lower concentrations of EET metabolites in plasma have been attributed to the presence of the CYP2J2*7 allele (Spiecker et al., 2004), this in vivo phenotype reflects EET metabolism of all tissues where CYP2J2 is expressed. Clearly, additional studies on hepatic CYP2J2 activity are warranted.
To assess whether C>T (P115L) in CYP2J2*10 conveys any functional consequences, CYP2J2.10 protein was expressed in vitro in a bacterial system (Jones et al., in preparation). Since the majority of apoprotein was devoid of heme, one may speculate that P115L intervenes with heme incorporation in vivo and thereby compromises the amount of functional protein. CYP2J2*10 was found in a composite heterozygote with CYP2J2*7, which could explain the low level of immunoreactive protein in this particular sample. Yet, given the rarity of this allelic variant, it did not explain the observed variability and likely does not play a significant role in overall interindividual variation in CYP2J2 levels.
Next, in an effort to detect any aberrant mRNA splice variants, cDNA was amplified from the same subjects (representing high, intermediate and low protein) in whom the CYP2J2 gene was resequenced. Each revealed a major product corresponding to the correctly spliced mRNA and two minor products. Subsequent analysis on full-length cDNA on a set of tissues from one pre- and one postnatal subject did not reveal any additional tissue-specific splice products. These findings are also in agreement with the automated splice site analysis shown in Table 3 that calculated relative high bit values for almost all canonical splice sites. None of the three alternate splice variants that were ultimately identified (Fig 5B) encode functional protein due to frame shifts introduced by deletion and retention events. Again, this set of experiments failed to find any explanation for the variability in protein expression. There is no evidence in the literature suggesting that CYP2J2 transcripts include alternative exons at the 5′-end (i.e. other canonical exons 1), but it is conceivable that alternate first exon(s) exist. Such exons were, however, not captured by using primers directed to the known exon 1 sequence. Further characterization of splice events affecting the 5′-ends may ultimately discover additional splice variants that could account for the variable amounts of protein detected. Possibly, such variants may also encode the immunoreactive proteins identified in postnatal and adult liver, but not fetal liver and adult heart.
In summary, we provide an initial characterization of CYP2J2 mRNA and protein expression during human development. While mRNA expression appears to be tightly regulated, immunoreactive protein varies considerably. Furthermore, resequencing of the gene and characterization of splice variants did not provide any explanation for the variability seen on the protein level. Future investigations will be directed towards the development of specific probe substrates to permit assessment of CYP2J2 activity in liver microsomal preparations and more detailed characterizations of mRNA splicing in the 5′ and 3′-ends of CYP2J2 transcripts.
Acknowledgments
We thank Stephen D. Simon, PhD for statistical support.
Non-standard abbreviations
- ECL
enhanced chemiluminesence
- EET
epoxyeicosatrienoic acid
- EGA
estimated gestational age
- 20-HETE
20-hydroxyeicosatetraenoic acid
- PCR-RFLP
polymerase chain reaction-restriction fragment length polymorphism
- SV
splice variant
Footnotes
Financial support: This work was supported by RO1 ES10855-05 from the National Institute of Environmental Health Sciences and in part by the Intramural Research Program at the NIH, NIEHS (DCZ).
References
- Cheesman MJ, Baer BR, Zheng Y, Gillam E, Rettie AE. Rabbit CYP4B1 engineered for high-level expression in Escherichia coli: ligand stabilization and processing of the N-terminus and heme prosthetic group. Arch Biochem Biophys. 2003;416:17–24. doi: 10.1016/s0003-9861(03)00278-9. [DOI] [PubMed] [Google Scholar]
- Enayetallah AE, French RA, Thibodeau MS, Grant DF. Distribution of soluble epoxide hydrolase and of cytochrome P450 2C8, 2C9 and 2J2 in human tissues. J Histochem Cytochem. 2004;52:447–454. doi: 10.1177/002215540405200403. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Gaedigk R, Leeder JS. CYP2D7 splice variants in human liver and brain: Does CYP2D7 encode functional protein? Biochem Biophys Res Comm. 2005;336:1241–1250. doi: 10.1016/j.bbrc.2005.08.255. [DOI] [PubMed] [Google Scholar]
- Gu J, Su T, Chen Y, Zhang Q-Y, Ding X. Expression of biotransformation enzymes in human fetal olfactory mucosa: potential roles in developmental toxicity. Toxicol Appl Pharmacol. 2000;165:158–162. doi: 10.1006/taap.2000.8923. [DOI] [PubMed] [Google Scholar]
- Guryev O, Gilep A, Usanov S, Estabrook R. Interaction of Apo-cytochrome b5 with Cytochromes P4503A4 and P45017A: Relevance of Heme Transfer Reactions. Biochemistry. 2001;40:5018–5031. doi: 10.1021/bi002305w. [DOI] [PubMed] [Google Scholar]
- Hashizume T, Imaoka S, Mise M, Terauchi Y, Fuji T, Miyazaki H, Kamataki T, Funae Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J Pharmacol Exp Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chen C-L, Card JW, Yang S, Chen J-X, Fu X-N, Ning Y-G, Xiao X, Zeldin DC, Wang DW. Cytochrome P450 2J2 promotes the neoplastic phenotype of carcinoma cells and is up-regulated in human tumors. Cancer Res. 2005;65:4707–4715. doi: 10.1158/0008-5472.CAN-04-4173. [DOI] [PubMed] [Google Scholar]
- King LM, Ma X, Srettabunjong S, Graves J, NBradburry JA, Li LMS, Liao JK, Mohrenweiser H, Zeldin DC. Cloning of CYP2J2 gene and identification of functional polymorphisms. Mol Pharmacol. 2002;61:840–852. doi: 10.1124/mol.61.4.840. [DOI] [PubMed] [Google Scholar]
- Koch I, Weil R, Wolbold R, Brockmöller J, Hustert E, Burk O, Nuessler AC, Neuhaus P, Eichelbaum M, Zanger UM, Wojnowski L. Interindividual variabilty and tissue-specificity in the expression of cytochrome P450 3A mRNA. Drug Metab Dispos. 2002;30:1108–1114. doi: 10.1124/dmd.30.10.1108. [DOI] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nature Genetics. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- Le Quere V, Plee-Gauter E, Potin P, Madec S, Salaun JP. Human CYP4F3s are the main catalysts in the oxidation of fatty acid epoxides. J Lipid Res. 2004;45:1446–1458. doi: 10.1194/jlr.M300463-JLR200. [DOI] [PubMed] [Google Scholar]
- Lee SS, Jeong HE, Liu KH, Ryu JY, Moon T, Yoon CN, Oh SJ, Yun CH, Shin JG. Identification and functional characterization of novel CYP2J2 variants: G312R varaint causes loss of enzyme catalytic activity. Pharmacogenet Genomics. 2005;15:105–113. doi: 10.1097/01213011-200502000-00006. [DOI] [PubMed] [Google Scholar]
- Leeder JS, Gaedigk R, Marcucci KA, Gaedigk A, Vyhlidal CA, Schindel BP, Pearce RE. Variability of CYP3A7 expression in human fetal liver. J Pharmacol Exp Ther. 2005;314:626–635. doi: 10.1124/jpet.105.086504. [DOI] [PubMed] [Google Scholar]
- Marden NY, Murray M. Characterization of a c-Jun-responsive module in the 5′-flank of the human CYP2J2 gene that regulates transactivation. Biochem J. 2005;391:631–640. doi: 10.1042/BJ20050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Hirama T, Matsubara T, Nagata K, Yamazoe Y. Involvement of CYP2J2 on the intestinal first-pass metabolism of antihistamine drug, astemizole. Drug Metab Dispos. 2002;30:1240–1245. doi: 10.1124/dmd.30.11.1240. [DOI] [PubMed] [Google Scholar]
- Mitsuda M, Iwasaki M. Improvement in the expression of CYP2B6 by co-expression with molecular chaperones GroES/EL in Escherichia coli. Protein Expr Purif. 2006;46:401–405. doi: 10.1016/j.pep.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Nalla VK, Rogan PK. Automated splicing mutation analysis by information theory. Human Mutation. 2005;25:334–342. doi: 10.1002/humu.20151. [DOI] [PubMed] [Google Scholar]
- Rogan PK, Faux BM, Schneider TD. Information analysis of human splice site mutations. Human Mutation. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Rogan PK, Svojanovsky S, Leeder JS. Information theory-based analysis of CYP2C19, CYP2D6 and CYP3A5 splicing mutations. Pharmacogenetics. 2003;13:207–218. doi: 10.1097/00008571-200304000-00005. [DOI] [PubMed] [Google Scholar]
- Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- Smith K. Porphyrins and Metalloporphyrins. Elsevier Science; Amsterdam: 1975. [Google Scholar]
- Seubert J, Yang B, Bradbury A, Graves J, Miller L, Gabel S, Gooch R, Foley J, Newman J, Mao L, Rockman HA, Hammock BD, Murphy E, Zeldin DC. Enhanced postischemic functional recovery in CYP2J2 transgenic hearts involves mitochondrial ATP-sensitive K+ channels and p42/p44 MAPK pathway. Circ Res. 2004;95:506–514. doi: 10.1161/01.RES.0000139436.89654.c8. [DOI] [PubMed] [Google Scholar]
- Spiecker M, Darius H, Hankeln T, Soufi M, Sattler AM, Schaefer JR, Node K, Borgel J, Mugge A, Lindpaintner K, Huesing A, Maisch B, Zeldin DC, Liao JK. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 2004;110:2132–2136. doi: 10.1161/01.CIR.0000143832.91812.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jiang Y, Liu Y, Lin C, Cheng G, Chen X, Hao B, Tan W, Lin D, He F. CYP2J2*7 single nucleotide polymorphism in a Chinese population. Clin Chim Acta. 2006;365:125–128. doi: 10.1016/j.cca.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic epoxygenase highly expressed in heart. J Biol Chem. 1996;271:3460–3468. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- Yu Z, Huse LM, Adler P, Graham L, Ma J, Zeldin DC, Kroetz DL. Increased CYP2J2 expression and epoxyeicosatrienoic acid formation in spontanous hypertensive rat kidney. Mol Pharmacol. 2000;57:1011–1020. [PubMed] [Google Scholar]


