Summary
Flk1 is the major receptor for VEGF on endothelial cells. During embryogenesis, flk1 is required for both vasculogenesis and angiogenesis and abnormally elevated flk1 expression is often associated with pathological conditions in adults. While the biological function of flk1 has been studied extensively, very little is known about how the flk1 gene is regulated at the transcriptional level. Our transgenic study led to the identification of a flk1 endothelial enhancer positioned approximately 5 kb upstream of the flk1 translation initiation site. Binding sites for FoxH1, scl, ets and gata factors are found in the zebrafish flk1 endothelial enhancer, as well as in upstream sequences of mouse flk1 and human kdr genes, suggesting that the regulatory machinery for flk1/kdr is conserved from fish to mammals. The roles of scl, ets and gata factors in hemangioblasts have been well defined, but the significance of FoxH1 in vessel formation has not been explored previously. Here we show that FoxH1 binds to the flk1 endothelial enhancer in vitro and functions as a repressor for flk1 transcription in cultured cells. Consistent with these findings, the expression level of flk1 is elevated in embryos lacking both maternal and zygotic FoxH1. We further show that overexpression of FoxH1 has a negative effect on vascular formation that can be counteracted by the down-regulation of smad2 activity in zebrafish embryos. Taken together, our data provide the first evidence that flk1 is a direct target of FoxH1 and that FoxH1 is involved in vessel formation in zebrafish.
Keywords: angiogenesis, blood vessel, FoxH1, flk1, zebrafish
Introduction
A functional vascular system is required for the proper development of vertebrate embryos and the survival of adults. A number of signaling pathways, including VEGF and TGFβ, have been shown to be involved in vascular formation, but the VEGF pathway is the only one known to be involved in both vasculogenesis (the formation of vessels from hemangioblasts) and angiogenesis (the sprouting and remodeling of existing vessels) (for review, see (Roman and Weinstein, 2000).
flk1 (also known as KDR and VEGFR2) is an endothelial-specific receptor of vascular endothelial growth factor (VEGF). During embryogenesis, flk1 expression is first detected in hemangioblasts, the common precursor of the endothelial and blood lineages, and remains active in endothelial cells during vascular formation. Consistent with the importance of flk1 in vessel formation, mouse embryos lacking flk1 do not have cells from endothelial or hematopoietic lineages (Shalaby et al., 1995). In zebrafish, embryos lacking the activity of the VEGF receptor, flk1/kdra, display a partial loss of segmental arteries (Habeck et al., 2002; Covassin et al., 2006). This mild phenotype is likely to be the result of the compensatory effect of a second flk1 homologue, kdrb. In fact, while kdrb morphants do not display apparent abnormalities in blood vessel formation, embryos lacking both flk1/kdra and kdrb show a complete loss of segmental arteries (Covassin et al., 2006).
The expression of flk1 in endothelial cells is tightly regulated. During embryogenesis, flk1 expression is down-regulated after the vasculature is established. In adults, significant flk1 expression is only seen in tissues undergoing active angiogenesis, such as the mammary glands during pregnancy. Abnormal elevation of flk1 expression is often associated with pathological conditions, such as neovasculorization in tumorigenesis (for review, see (Robinson and Stringer, 2001). The precise regulatory circuit that regulates the expression of flk1 is not well understood. However, the limited information available from studies in the mouse model has revealed critical roles for scl, ets and gata factors in flk1 regulation. Consensus binding sequences of these transcription factors are found in the mouse flk1 enhancer and in the human KDR promoter (Patterson et al., 1995; Kappel et al., 2000) and mutations in these transcription factor binding sites abolish endothelial expression of the flk1 gene in the mouse model (Kappel et al., 2000; Elvert et al., 2003). The regulatory relationship between flk1 and scl has also been studied in the zebrafish. Overexpression of scl increases flk1 expression in wild type embryos and can induce flk1 expression in the cloche mutant, which lacks endothelial and blood cells (Liao et al., 1998; Liao et al., 2000), demonstrating that scl is an important transcription factor that regulates flk1 gene expression. In addition, a recent study shows that overexpression of Etsrp, an endothelial-specific Ets-1 related protein, induces ectopic flk1 expression in wild type and clo mutant embryos (Pham et al., 2006; Sumanas and Lin, 2006). These findings suggest that the regulatory mechanisms of flk1 expression may be conserved among vertebrates.
FoxH1 (also known as Fast1) is a forkhead transcription factor that binds to Smad2/3 and mediates TGFβ signaling (for review, see (Attisano et al., 2001). Gene expression analysis showed that FoxH1 is expressed primarily during early developmental stages. Consistent with this finding, overexpression of FoxH1 induces the expression of a broad range of genes downstream of activin and leads to abnormal axis formation in Xenopus (Watanabe and Whitman, 1999). Furthermore, mouse and zebrafish embryos carrying mutations in FoxH1 develop gastrulation defects (Pogoda et al., 2000; Sirotkin et al., 2000; Hoodless et al., 2001; Yamamoto et al., 2001). The roles for FoxH1 at later developmental stages or in adults are not as clear. However, the findings that FoxH1−/− mouse embryos do not have the outflow tract or the right ventricle reveal a role of FoxH1 in the formation of the anterior heart field (von Both et al., 2004). Furthermore, the well-documented effects of TGFβ signaling on the proliferation, differentiation, migration and even survival of endothelial cells (for review, see (Goumans and Mummery, 2000; Goumans et al., 2003), suggest a role for FoxH1 in the regulation of endothelial cells.
To further understand how the flk1 gene is regulated at the transcriptional level in zebrafish, we systematically studied the regulatory elements of flk1/kdra (hereafter referred to as flk1). We identified a ∼6.4kb genomic sequence upstream of the translation initiation site of zebrafish flk1 that drives GFP expression in the zebrafish vasculature, resembling the endogenous flk1 expression pattern. Deletion analysis revealed that a ∼800 bp DNA fragment positioned approximately 4.3kb upstream of the flk1 translation initiation site is sufficient to drive flk1 expression in endothelial cells. Consensus sequences of transcription factor binding sites including scl, ets and gata factors are present in this 800bp minimal flk1 enhancer suggesting conserved regulatory machinery from fish to mammals. In addition, we provide evidence that FoxH1 is part of the regulatory machinery for flk1 gene expression in zebrafish. We found multiple FoxH1 binding sites in the upstream regulatory regions of zebrafish, mouse and human flk1 genes. By gel shift assay, we show that FoxH1 binds to the flk1 regulatory element. We further show that FoxH1 can repress the activity of the flk1 minimal enhancer in HEK293T cells. Consistent with this finding, the expression level of flk1 is moderately elevated in zebrafish embryos lacking both maternal and zygotic FoxH1 and overexpression of FoxH1 disrupts the formation of zebrafish vasculature. Moreover, we showed that flk1 gene expression is up-regulated in smad2 morphants and that such changes in flk1 expression levels could be counteracted by overexpression of FoxH1, indicating that the negative effects of FoxH1 on flk1 gene expression is smad2 dependent. These data suggest that FoxH1 functions as a negative modulator of flk1 gene expression and indicate interplay between TGFβ and VEGF signaling pathways in embryonic vascular formation.
Materials and Methods
Zebrafish husbandry and the generation of TG(flk1:GFP)la116 transgenic lines
The AB strain was used for this study. Adult fish and embryos were maintained as previously described (Westerfield, 2000).
A ∼6.4 kb genomic fragment upstream of the zebrafish flk1 gene (from −6410 to −1 upstream of flk1 translational initiation site) was cloned into pKS-GM2r vector 5' to green fluorescent protein (GFP) (kind gift from S. Lin) to create the flk(−6.4)-GFP construct. Approximately 50pg of linearized flk(−6.4)-GFP DNA was microinjected into zebrafish embryos at the 1-cell stage. The injected embryos were raised to adulthood and screened for founder fish with germline integration. Images of GFP expression patterns of TG(flk1:GFP)la116 embryos were acquired using a Zeiss SV-11 epifluorescence microscope or an Olympus IX70 confocal microscope equipped with a 10X objective.
Deletion constructs of flk(−6.4)-GFP
Serial deletion constructs of flk(−6.4)-GFP were generated using the ERASE-A-BASE system (Promega). Sequencing analysis showed that flk(−5.7)-GFP, flk(−5.0)-GFP, flk(−4.3)-GFP, flk(−3.5)-GFP and flk(−1.5)-GFP have sequences upstream of flk1 translational initiation site starting from −5680, −5016, −4353, −3565 and −1503, respectively. DNA fragments corresponding to sequences from −5045 to −3543, −4353 to −3543, and −5045 to −4326 were amplified by PCR using the following primers: −5045F, 5′-CCGCGGTTGTATGAAGTTTCTGTGTGAC-3′; −4353F, 5′-CCGCGGTCACCTTCTGCTAGTTAAAACC-3′; −4326R, 5′-GCGGCCGCGAATGAGGTTTTAACTAGCAGA-3′; −3543R, 5′-GCGGCCGCAATCCAAAGTAATTGATCCCTG-3′. These DNA fragments were then cloned into flk(−1.5)-GFP to create flk(−5.0,−3.5/−1.5)-GFP, flk(−5.0,−4.3/−1.5)-GFP and flk(−4.3,−3.5/−1.5)-GFP.
Approximately 50pg of each of these constructs were microinjected into zebrafish embryos at the 1-cell stage. The injected embryos were screened for transient expression of GFP using a Zeiss SV-11 epifluorescence microscope after one day of development.
mRNA and morpholino injection
mRNA encoding FoxH1 (kind gift from D. Meyer) (Pogoda et al., 2000), FAST-Eng and FAST-VP16 (kind gifts from M. Whittman) (Watanabe and Whitman, 1999) were synthesized using mMESSAGE mMACHINE (Ambion) and injected into TG(flk1:GFP)la116 embryos at the 1- to 4-cell stage.
An morpholino modified oligonucleotide specifically targeting the translation initiation site of smad2, smad2MO, was synthesized (GAGTGAAAGGCAAGATGGAGGACAT) (Genetools). 0.8 ng of smad2 morpholino were injected into TG(flk1:GFP)la116 embryos at the 1- to 4-cell stage.
The vasculature of the FoxH1 mRNA or smad2MO injected embryos were analyzed at between the 18-somite stage and 24 hpf (hour post fertilization). Embryos were photographed using a Zeiss SV-11 epifluorescence microscope and Axiocam digital camera and the intensity of GFP signals was quantified by the NIH ImageJ program. The p value was calculated by the student's t-Test.
GST fusion proteins
The zebrafish FoxH1 cDNA (from D. Meyer) was cloned into the SmaI site of pGEX-2TK (GE Healthcare) to create the pGEX-2TK-FoxH1 construct. pGEX-2TK-FoxH1 and pGEX-2TK were transformed into BL21 (Stratagene). The FoxH1-GST and GST proteins were purified using Bulk GST Purification Module (GE Healthcare). Protein concentrations were determined using the DC Protein Assay (Bio-Rad).
Electrophoretic Mobility Shift Assays
Oligonucleotides corresponding to flk1 upstream sequences from −3870 to −3844 were synthesized. Sequences of the upper strands are as follows: flk-FoxH1, 5′-CCGAATATTGTGTATTCGAGAAATATC-3′, and flk-FoxH1-mt, 5′-CCGAATATTcTGTATTCGAGAAATATC-3′ (the FoxH1 binding sites are underlined and the lowercase letter represents the substituted nucleotide). Oligonucleotides were annealed in 20mM Tris pH8.0, 1mM EDTA pH8.0 and 50mM NaCl.
3μg of purified FoxH1-GST and GST proteins were incubated with 32P end-labeled probes for 20min at room temperature in binding buffer containing 4% glycerol, 1mM MgCl2, 0.5mM EDTA, 0.5mM DTT, 50mM NaCl, 10mM Tris-HCl (pH 7.5) and 0.05mg/ml poly(dI-dC). Protein-DNA complexes were separated on a 6% nondenaturating polyacrylamide gel (37.5:1 acrylamide/bis-acrylamide) in 0.5X TBE. Unlabeled flk-FoxH1 or flk-FoxH1-mt were used as competitors at 50-, 100-, or 200-fold molar excess of labeled probes.
Luciferase Assay
FoxH1 cDNA was subcloned into pSG5 (Stratagene) to create pSG5-FoxH1 expression vector and upstream sequences of flk1 from −532 to −371 containing predicted TATA box and transcription starting site were amplified by PCR using flkP-F (5′-ACTGAGCGGCCGCTAGCCTGAATAAGTAGATAGC-3′) and flkP-R (5′-CGGGATCCTACCTCTGACTTTTCTACTGG-3′). The amplified DNA fragment was then cloned into a luciferase reporter vector, pGL3-basic (Promega), to create the pGL3-flkP construct. Upstream sequences of flk1 from −5045 to −3543 was amplified by PCR using the −5045F and −3543R primers, and cloned into pGL3-flkP to create pGL3-flk(−5.0, −3.5)P. The Gs in all three FoxH1 consensus binding sequences were substituted with Cs using QuikChange II Site-Directed Mutagenesis Kit (Stratagene) to create the pGL3-flk(−5.0, −3.5)P-mt construct. Primers used for site-directed mutagenesis are as follows: mt1-F; 5′-ATAATAATAAAGAGGCGTATTTATACATCTGTGCCAGG-3′, mt1-R; 5′-CCTGGCACAGATGTATAAATACGCCTCTTTATTATTAT-3′, mt2-F; 5′-GCTAGGCCGAATATTCTGTATTCGAGAAATATC-3′, mt2-R; 5′-GATATTTCTCGAATACAGAATATTCGGCCTAGC-3′, mt3-F; 5′-CATTAAATACCAATGAAGATCTGTAATCTAAAATTAT-3′ and mt3-R; 5′-ATAATTTTAGATTACAGATCTTCATTGGTATTTAATG-3′.
HEK293T cells were plated on 24-well plates at a density of 50,000 cells per well and transfected using Lipofectamine™ and Plus™ reagents (Invitrogen) with 0, 100 or 500 ng of pSG5-FoxH1, 10ng of luciferase reporter plasmid (pGL3-flk(−5.0, −3.5)P or pGL3-flk(−5.0, −3.5)P-mt), 2ng of pRL-CMV and various amounts of pSG5 plasmid to keep the total amount of DNA at 512ng for each transfection. Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega) and normalized with Renilla Luciferase activity for transfection efficiency.
Quantitative RT-PCR
Total RNA was extracted from the un-injected control and 100pg FoxH1 mRNA injected embryos at the 18-somite stage using RNAwiz (Ambion) and the first strand cDNA was synthesized using the SuperScript™ first-strand synthesis system for RT-PCR (Invitrogen). The relative flk1 expression level was determined by quantitative PCR using DNA Engine Opticon (MJ Research). β-actin was used as the internal control for normalization. Primers used: β-actin forward primer, 5′-CTGTCTTCCCATCCATCGTGGGTC-3′; β-actin reverse primer, 5′-CTCCATATCATCCCAGTTGGTGACA-3′. flk1 forward primer, 5′-GAGAACGGAACCAACAAGATCCACGAG-3′; flk1 reverse primer, 5′-CCCTCCAGCAGAACTGACTCCTTAC-3′.
Results and Discussion
Identification of an endothelial-specific regulatory element of the flk1 gene
As the first step toward understanding the regulation of flk1 at the transcriptional level, we isolated a ∼6.4kb DNA fragment upstream of the translation initiation site of the zebrafish flk1 gene by long-range PCR. We created a reporter construct, flk(−6.4)-GFP, using this 6.4kb flk1 upstream DNA fragment to drive GFP expression.
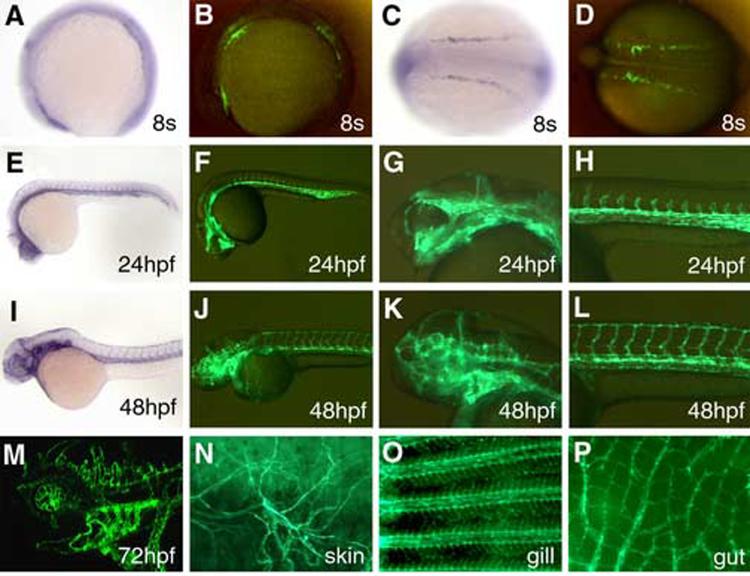
We generated a transgenic zebrafish line TG(flk1:GFP)la116 that carries a germline-integrated flk(−6.4)-GFP transgene and found that the GFP expression pattern in TG(flk1:GFP)la116 resembled the endogenous flk1 expression pattern. GFP signals can be observed in TG(flk1:GFP)la116 embryos as early as the 6-somite stage. At the 8-somite stage, GFP signals are detected in three patches of cells in TG(flk1:GFP)la116 embryos (Fig 1B,D), similar to the endogenous flk1 expression pattern (Fig.1A,C). The similarities between the endogenous flk1 expression pattern and GFP expression pattern in TG(flk1:GFP)la116 embryos continue as development proceeds. GFP expression is detected in the dorsal aorta, cardinal vein, intersomitic vessels and endothelial cells in the brain by 24 hours post fertilization (hpf), and in paired dorsal longitudinal anastomotic vessels (DLAV) by 48 hpf (Fig.1E-L). By three days of development, endothelial cells in the pharyngeal arches can be clearly detected by GFP expression in TG(flk1:GFP)la116 embryos (Fig.1M). This pattern is distinct from that observed in TG(fli:EGFP)y1 embryos where both the aortic arches and the mesenchyme of the forming jaw are GFP positive (Lawson and Weinstein, 2002). Furthermore, the GFP expression patterns in TG(flk1:GFP)la116 embryos are consistent with the other two independent transgenic lines using the same 6.4kb regulatory element (Cross et al., 2003; Beis et al., 2005; Jin et al., 2005), indicating that the ∼6.4kb flk1 upstream regulatory element is sufficient to drive gene expression in endothelial cells and that this endothelial expression pattern is not an artifact of the integration site of the flk(−6.4)-GFP transgene.
Fig.1.
GFP expression in TG(flk1:GFP)la116 embryos. (A-D) GFP signals driven by the flk(6.4)-GFP transgene can be detected in TG(flk1:GFP)la116 embryos as early as the 8-somite stage. flk1 expression is detected in three patches of cells by in situ hybridization (A). A similar pattern is seen in TG(flk1:GFP)la116 embryos (B). Lateral views are shown in panels A and B and the dorsal views are shown in panels C and D. Anterior to the left. (E-H) GFP expression pattern of TG(flk1:GFP)la116 embryos at 24hpf (F) resembles the flk1 pattern detected by in situ hybridization (E). Higher magnification images of the head and trunk are shown in G and H, respectively. (I-L) GFP expression pattern of TG(flk1:GFP)la116 embryos at 48hpf (J) resembles the flk1 pattern detected by in situ hybridization (I). Higher magnification images of the head and trunk are shown in K and L, respectively. (M) Confocal image of GFP expression in endothelial cells in the brain and brachial arches in 3-day-old TG(flk1:GFP)la116 embryos. (N-P) GFP expression remains active in adult TG(flk1:GFP)la116 fish. Images show GFP signals in endothelial cells on the skin (N), in the gill (O) and gut (P).
In addition, we found that while the spatial GFP expression of TG(flk1:GFP)la116 resembles the endogenous flk1 expression pattern revealed by the in situ hybridization, the temporal regulation is different. In zebrafish, the expression of flk1 in endothelial cells is greatly reduced after 3 days of development (Fouquet et al., 1997; Liao et al., 1997). However, the endothelial GFP signal remains strong in TG(flk1:GFP)la116 throughout embryogenesis and persists into adulthood (Fig.1N-P), indicating that the regulatory sequences responsible for down-regulating flk1 is not present or is not intact in the ∼6.4kb flk1 upstream DNA fragment.
Identification of the minimal flk1 endothelial enhancer
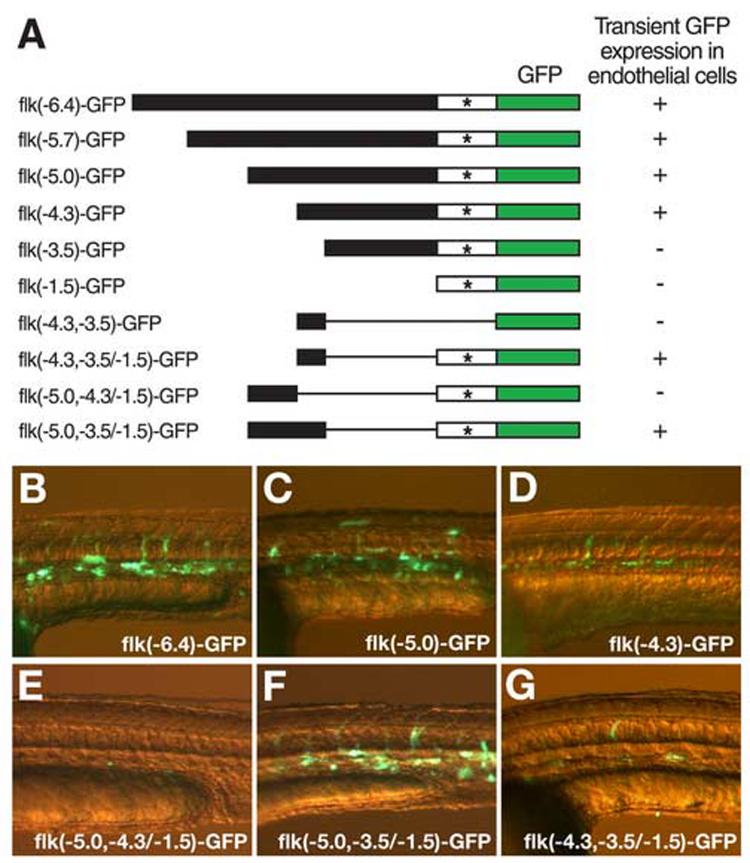
To identify critical cis-regulatory elements for flk1 expression in endothelial cells within the ∼6.4kb flk1 regulatory element, we made a series of deletion variants of flk(−6.4)-GFP. The flk(−5.7)-GFP, flk(−5.0)-GFP, flk(−4.3)-GFP, flk(−3.5)-GFP and flk(−1.5)-GFP deletion variants have the GFP reporter gene under the control of the ∼5.7kb, 5.0kb, 4.3kb, 3.5kb, and 1.5kb DNA fragments upstream from the flk1 translation initiation site, respectively. We injected these deletion constructs into zebrafish embryos at the 1-cell stage to evaluate the ability of these constructs to drive GFP expression in the zebrafish embryonic vasculature. We found that of the 196 embryos injected with flk(−6.4)-GFP, 158 (80.6%) had strong GFP expression in endothelial cells (Fig.2A,B). Similar results were observed in embryos injected with flk(−5.7)-GFP, flk(−5.0)-GFP and flk(−4.3)-GFP (52.0%, n=125, 52.4%, n= 145 and 51%, n=200, respectively) (Fig.2A, C, D). However, none of the embryos injected with the flk(−3.5)-GFP or flk(−1.5)-GFP constructs had GFP expression in endothelial cells (n=181 and 130, respectively) (Fig.2A). These data suggest that critical regulatory elements for flk1 endothelial expression are located at least 3.5 kb upstream of the flk1 translational initiation site.
Fig.2.
Deletion analysis of the flk1 regulatory region identifies a highly conserved element that is necessary for endothelial expression. (A) Schematic diagram of the deletion constructs of flk1-GFP reporter. Linearized DNA of each construct was injected in wild type zebrafish embryos at the 1-cell stage. GFP expression in injected embryos was analyzed after 1 day of development. The transient endothelial expression directed by each construct is summarized by a plus (endothelial expression) or a minus (no detectable endothelial expression) to the right of the line representing each construct. * marks the transcription initiation site of flk1. (B-G) Transient GFP expression in endothelial cells of 1-day-old embryos injected with flk(−6.4)-GFP (B), flk(−5.0)-GFP (C), flk(−4.3)-GFP (D), flk(−5.0,−4.3/−1.5)-GFP (E), flk(−5.0,−3.5/−1.5)-GFP (F) or flk(−4.3,−3.5/−1.5)-GFP (G).
Since flk(−4.3)-GFP was capable of driving GFP expression in endothelial cells but flk(−3.5)-GFP could not, we focused our analysis on the region of −4353 to −3543 upstream of flk1. We did not detect any GFP expression in embryos injected with the flk(−4.3,−3.5)-GFP construct, indicating that this fragment alone is not sufficient to drive gene expression in vivo (Fig.2A). One possible cause is that this construct lacks a functional promoter. The TATA box and the predicted transcriptional initiation site of flk1 are located within the 1.5kb fragment immediately upstream of the flk1 translation initiation site. Therefore, we created three additional constructs with various lengths of flk1 5' sequences from −5kb to −3.5kb fused to flk(−1.5)-GFP, flk(−5.0,−3.5/−1.5)-GFP, flk(−4.3,−3.5/−1.5)-GFP and flk(−5.0,−4.3/−1.5)-GFP. We injected these constructs into wild type zebrafish embryos at the 1-cell stage and found that while no GFP signals were detected in embryos injected with flk(−1.5)-GFP or flk(−5.0,−4.3/−1.5)-GFP (n=130 and n=176 , respectively), transient GFP expression in endothelial cells was driven by flk(−5.0,−3.5/−1.5)-GFP and flk(−4.3,−3.5/−1.5)-GFP constructs (68.8%, n= 218 and 53.9%, n=293, respectively) (Fig.2A, E-G). These findings indicate that the DNA fragment of −4353 to −3543 has the information required to drive flk1 expression in endothelial cells.
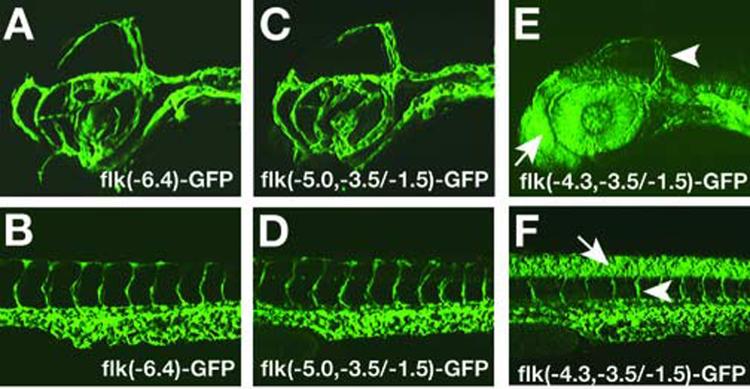
We established stable transgenic lines carrying germline-integrated flk(−5.0,−3.5/−1.5)-GFP or flk(−4.3,−3.5/−1.5)-GFP transgenes to analyze their GFP expression patterns in detail. We found that endothelial cells are marked by GFP expression in embryos carrying flk(−5.0,−3.5/−1.5)-GFP or flk(−4.3,−3.5/−1.5)-GFP transgenes, consistent with the findings obtained from transient expression analysis that sequences from −4353 to −3543 have all critical information to drive gene expression in endothelial cells . However, when analyzed closely, we found that while the GFP expression pattern of flk(−5.0,−3.5/−1.5)-GFP transgenic embryos is identical to that of TG(flk1:GFP)la116 embryos, flk(−4.3,−3.5/−1.5)-GFP transgenic embryos have GFP expression in the brain, the eyes and the neural tube in addition to endothelial cells (Fig.3). These observations, together with our finding that flk(−5.0,−4.3/−1.5)-GFP does not drive GFP expression in zebrafish embryos (Fig.2A,E), indicate that while the DNA fragment from −4353 to −3543 is sufficient to drive flk1 expression in endothelial cells, sequences from −5045 to −4325 are required to restrict flk1 gene expression to the endothelial cells.
Fig.3.
Transgenic analysis of the flk1 regulatory region identifies a 1.5kb minimal endothelial specific enhancer. (A-B) GFP expression patterns in the brain (A) and trunk (B) of 2-day-old TG(flk1:GFP)la116 embryos. (C-D) GFP expression patterns of 2-dayold embryos carrying germ line integrated flk(−5.0,−3.5/1.5)-GPF transgene resembles the patterns observed in TG(flk1:GFP)la116. (E-F) Embryos carrying germ line integrated flk(−4.3,−3.5/1.5)-GFP have GFP expression in endothelial cells (arrowhead) as well as neural tissues. Arrows point to GFP positive cells in forebrain in E and to GFP positive cells in neural tube in F.
Conserved regulatory control of flk1
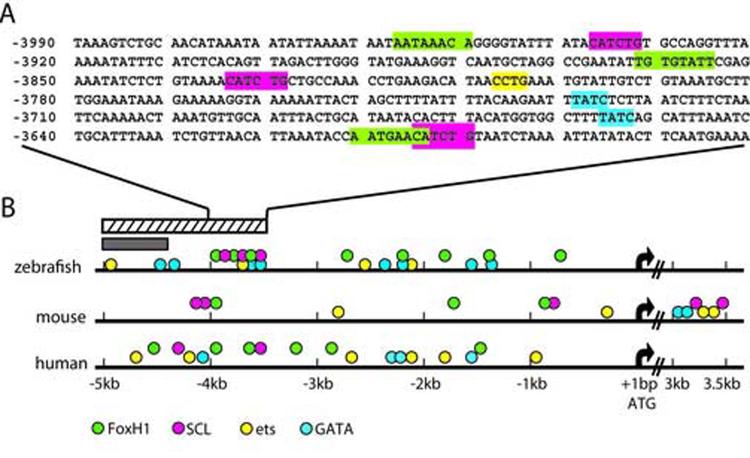
To further investigate regulatory mechanisms of flk1 gene expression, we searched for transcription factor binding sites in flk1 upstream sequences from −5045 to −3543. In the region that is critical for restricting flk1 expression to endothelial cells (−5043 to −4326), we noted an Ets and a GATA factor consensus binding site but did not identify binding sites for other transcription factors with known function in endothelial gene expression, including FoxH1 (see below) (Fig.4B). However, multiple consensus binding sites of transcription repressors, including five engrailed, three pbx and two brn-3 binding sites, were found in this region. Interestingly, all these genes are expressed in the brain and the eyes (DeCarvalho et al., 2004; Erickson et al., 2006), raising a possibility that they are part of the machinery restricting flk1 expression to endothelial cells. Further studies on molecular mechanisms by which −5045 to −4326 influences flk1 gene expression, including the regulatory effects of engrailed, pbx and brn-3 on flk1, should provide more insights into flk1 gene regulation.
Fig.4.
Cross-species comparison of flk1 5'-flanking sequences. (A) Sequence of flk1 enhancer from −3990 to −3571. (B) Schematic diagrams of 5'-flanking regions of zebrafish, mouse, and human flk1 genes. Consensus sequences of FoxH1, scl, ets and GATA factor binding sites are highlighted in green, red, yellow and blue, respectively in (A) and represented with dots with the same color scheme in (B). Translation initiation sites are marked by arrows. The hatched bar indicates the zebrafish flk1 upstream sequence from −5.0kb to −3.5kb and the grey bar indicates the region from −5.0kb to −4.3kb.
The flk1 upstream segment from −4353 to −3543 is critical for driving GFP expression in endothelial cells. In this region, we found multiple binding sites of scl, Ets and GATA factors. Interestingly, these transcription binding sites are also present in the mouse flk1 minimal enhancer and human flk1/KDR promoter (Patterson et al., 1995; Kappel et al., 2000) (Fig.4), suggesting that the regulatory machinery of flk1 gene expression is conserved from fish to mammals.
In addition to scl, ets and GATA binding sites, there are three FoxH1/Fast1 binding sites in the flk1 minimal regulatory element (−4353 to −3543) (Fig.4). While the role of FoxH1 in endothelial cells has not been investigated, the effects of TGFβ signaling on the proliferation, differentiation, migration and permeability of endothelial cells have been well documented in animal models and in cultured cells (for review, see (Goumans and Mummery, 2000; Goumans et al., 2002), raising a possibility that flk1 is a target gene of FoxH1-mediated TGFβ signaling. We therefore investigated whether FoxH1 binding sites are also present in the regulatory regions of mouse and human flk1/KDR genes. As illustrated in Fig.4B, multiple FoxH1 binding sites are present within a 5kb region upstream of the human and mouse flk1 genes, supporting the notion that FoxH1 is part of flk1 regulatory machinery that is conserved from fish to mammals.
FoxH1 binds to the flk1 minimal endothelial enhancer in vitro
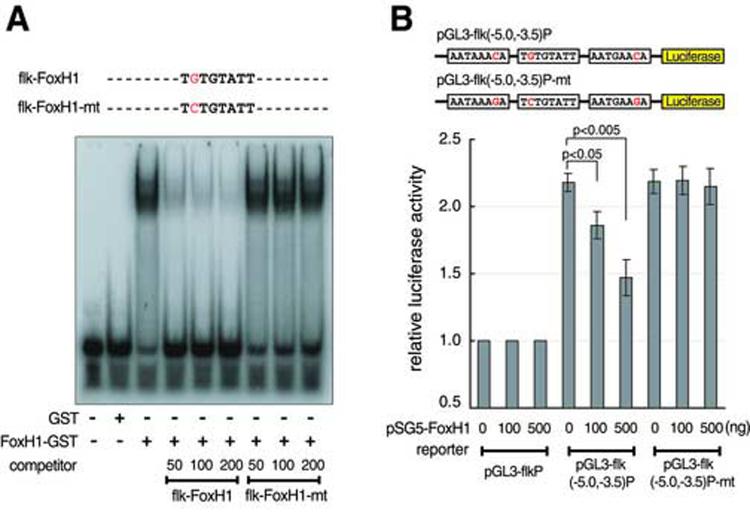
To explore the possibility that FoxH1 regulates flk1 gene expression directly, we tested whether FoxH1 could bind to the consensus FoxH1 binding sites in the flk1 minimal regulatory element. We performed a gel shift assay using a synthetic oligonucleotide, flk-FoxH1, corresponding to flk1 upstream sequences from −3870 to −3844, consisting of one FoxH1 binding site, as a probe. As shown in Fig.5A, FoxH1 binds to flk-FoxH1 in vitro and this binding is competed away by unlabeled probes in a dosage-dependent manner (Fig.5A). It was previously shown that a G to C substitution in the FoxH1 binding site consensus sequence abolishes its interaction with FoxH1 (Chen et al., 1996). We found that flk-FoxH1-mt, synthetic oligonucleotides harboring the same G to C mutation in the consensus sequences of the FoxH1 binding site did not compete out FoxH1 interaction with the labeled probes (Fig.5A), indicating that the binding of FoxH1 to the flk1 minimal endothelial enhancer is sequence-specific.
Fig.5.
FoxH1 regulates flk1 expression at the level of transcription. (A) FoxH1-GST fusion protein were purified and used in a gel shift assay with a radiolabeled probe (flk-FoxH1) corresponding to the flk1 enhancer. Unlabeled probe and unlabeled flk-FoxH1-mt, oligonucletides carrying a G to C substitution in the consensus sequences of the FoxH1 binding site were used as competitors in the gel shift assay at 50-, 100- or 200-fold molar excess. A representative experiment is shown. All assays were performed at least three times with comparable results. (B) HEK293T cells were transiently transfected with the indicated luciferase reporters and an expression vector encoding FoxH1. Values are relative to the luciferase activity of cells transfected with pGL3-flkP alone. Results of an average of four independent experiments are shown.
FoxH1 suppresses flk1 gene expression in HEK293T cells
Previous studies show that FoxH1 can function as an activator as well as a repressor (Labbe et al., 1998; Watanabe and Whitman, 1999; Osada et al., 2000; Saijoh et al., 2000; Kofron et al., 2004; Chen et al., 2005). We therefore explored the regulatory relationship between FoxH1 and flk1 in HEK293T cells. pGL3-flk(−5.0,−3.5)P is a construct that uses the flk1 upstream sequences from −5045 to −3543 and a 162bp flk1 basal promoter to drive luciferase expression. Co-transfecting pSG5-FoxH1 with pGL3-flk(−5.0,−3.5)P represses the production of luciferase in a dosage-dependent manner (p<0.05) (Fig.5B), demonstrating that FoxH1 functions as a repressor for flk1. To investigate if the repressive activity of FoxH1 is DNA-binding dependent, we created the pGL3-flk(−5.0,−3.5)P-mt construct where the Gs in all three FoxH1 binding sites on the minimal flk1 regulatory element were substituted with Cs (Chen et al., 1996). HEK293T cells transfected with pGL3-flk(−5.0,−3.5)P-mt have similar luciferase activity as those cells transfected with pGL3-flk(−5.0,−3.5)P, indicating that loss of FoxH1 binding does not affect the activity of flk1 minimal enhancer. Furthermore, no significant difference in luciferase activity was observed when FoxH1 expression plasmid was cotransfected with pGL3-flk(−5.0,−3.5)P-mt (Fig.5B), suggesting that the repressive effect of FoxH1 on flk1 expression is DNA-binding dependent.
FoxH1 negatively regulates zebrafish vascular formation
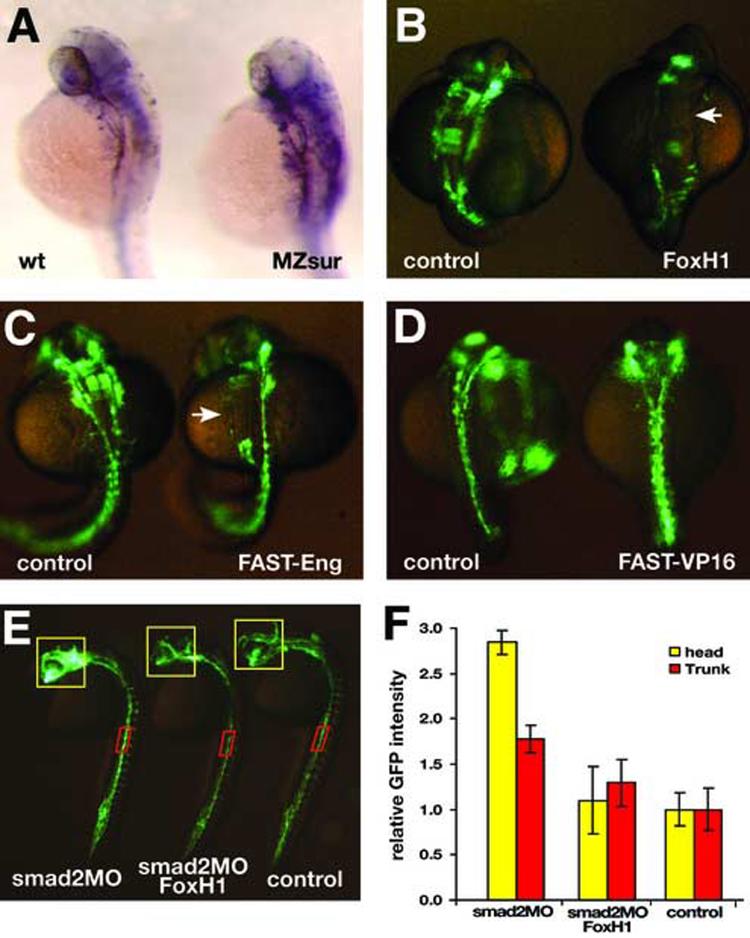
We further explored the regulatory effect of FoxH1 on the formation of the embryonic vascular system in zebrafish. Our data show that FoxH1 represses the transcription activity of the flk1 minimal endothelial enhancer in cultured cells. If the same regulatory relationships were relevant during embryonic vascular formation, one would expect flk1 expression levels to be increased in zebrafish embryos lacking FoxH1 function. The zebrafish schmalspur (sur) locus encodes FoxH1/Fast1 (Pogoda et al., 2000; Sirotkin et al., 2000). We analyzed flk1 expression levels in the maternal-zygotic sur homozygotes (MZsur) derived from the surty68b allele which carries a point mutation in the conserved FKH domain (Pogoda et al., 2000). As shown in Fig.6A, the flk1 expression levels are indeed moderately elevated in MZsur embryos, supporting the notion that FoxH1 is a repressor of flk1. We further analyzed the forced expression effects of FoxH1 on vascular formation in vivo. Data collected from three independent experiments showed that 44 out of 73 TG(flk1::GFP)la116 embryos injected 50pg of FoxH1 mRNA (60%) and 265 out of 282 TG(flk1::GFP)la116 embryos injected with 100pg of FoxH1 mRNA (94%) have missing patches of endothelial cells (Fig.6B), suggesting that FoxH1 negatively regulates vascular formation in zebrafish and that this negative effect of FoxH1 on zebrafish vascular formation is dose-dependent. Furthermore, we measured relative flk1 gene expression level in control and FoxH1 mRNA injected embryos using quantitative RT-PCR. From two independent experiments performed in triplicate, we consistently observed that the levels of flk1 transcripts in those embryos injected with 100 pg of FoxH1 mRNA were 48% of the levels in un-injected embryos (standard deviation = 4.2%, p<0.001), supporting the notion that FoxH1 represses flk1 gene expression.
Fig.6.
FoxH1 modulates vascular formation in zebrafish. (A) flk1 is expressed in the developing vasculature of embryos at 32hpf (left). The expression level of flk1 is elevated in MZsur embryos (right). (B) Injection of FoxH1 mRNA disrupts zebrafish vascular formation. Embryos were analyzed at the 18-somite stage. (C) Injection of mRNA encoding the FAST-Eng chimeric protein disrupts vascular formation (right), resembling the phenotype of FoxH1 overexpression. Embryos were analyzed at the 23-somite stage. (D) Injection of mRNA encoding FAST-VP16 chimeric protein does not disrupt the GFP expression pattern in TG(flk1:GFP)la116 embryos. Embryos were analyzed at the 18-somite stage. Arrows point to patches of endothelial cells missing in FoxH1 or FAST-Eng mRNA injected embryo. (E) Lateral view of the un-injected (right), smad2MO and FoxH1 mRNA co-injected (middle) and smad2MO injected (left) TG(flk1:GFP)la116 embryos at 24hpf. (F) Graph represents relative GFP intensities within the region of interests (indicated by the yellow and red boxes in panel E).
To further confirm that FoxH1 does indeed have a negative effect on vascular formation, we injected FAST-Eng and FAST-VP16 mRNA into TG(flk1::GFP)la116 embryos at the 1-cell stage. FAST-Eng and FAST-VP16 encode proteins that have the wild type FoxH1 forkhead DNA-binding domain fused to the transcriptional repressor domain of Engrailed (FAST-Eng) and the viral transcriptional activator domain of VP16 (FAST-VP16), respectively (Watanabe and Whitman, 1999). Embryos injected with 10pg of FAST-VP16 mRNA died before vascular formation. We, therefore, focused our analysis on lower levels of FAST-VP16 injection. None of the 130 embryos that received 4pg of FAST-VP16 had notable vascular defects (Fig.6D). On the other hand, embryos injected with FAST-Eng mRNA displayed a phenotype resembling that observed in FoxH1 mRNA injected embryos and this effect was dosage-dependent. While 43 out of 55 TG(flk1::GFP)la116 embryos injected with 10pg of mRNA encoding FAST-Eng had missing patches of endothelial cells, all 36 TG(flk1::GFP)la116 embryos injected with 25pg of FAST-Eng mRNA had missing patches of endothelial cells (Fig.6C). These data demonstrate that FoxH1 functions as a repressor and has a negative effect on zebrafish vascular development.
Studies in Xenopus have demonstrated both smad2-dependent and smad2-independent effects of FoxH1 in early embryogenesis (Kofron et al., 2004). We therefore evaluated whether the negative effects of FoxH1 on flk1 gene expression are smad2-dependent. If indeed the negative effects of FoxH1 on flk1 expression and zebrafish vascular formation were smad2-dependent, one would expect that knocking down smad2 activity could counteract the effects of FoxH1 overexpression. We found that while TG(flk1::GFP)la116 embryos injected with 0.8 ng of Smad2MO developed normally, the GFP signals in endothelial cells were brighter than their un-injected siblings (86%, n=112) and that co-injecting 50pg of FoxH1 mRNA with 0.8ng of smad2MO could suppress the smad2MO induced elevation of GFP signals (100%, n=102) (Fig.6E,F). We then quantified the GFP intensity at selected regions of interest in the brain and in the trunk using the NIH ImageJ software. As shown in Fig.6E and F, while there was no significant differences in the GFP signals between wild type embryos and embryos co-injected with smad2MO and FoxH1 mRNA (25 embryos analyzed from each group, p>0.05), the GFP signals were increased by 2.84-fold in head blood vessels and 1.78-fold in the dorsal aorta and cardinal vein of smad2 morphants when compared to wild type embryos (n=25 for each group, p<0.001). These data support the notion that the effects of FoxH1 on the developing zebrafish vasculature are smad2-dependent and indicate that TGFβ signaling plays a role in flk1 gene expression.
The role of FoxH1 in early embryogenesis has been studied extensively, but its relevance in vascular development has not been explored. In this report, we provide evidence that FoxH1 binds directly to the flk1 endothelial enhancer and modulates flk1 expression both in vivo and in cultured cells. We also show that FoxH1 has a negative effect on blood vessel formation in developing zebrafish embryos, offering the first evidence of the role of FoxH1 in patterning embryonic vasculature. It is interesting to note that TGFβ has both stimulating and inhibitory effects on endothelial cells (Pepper, 1997; Piek et al., 1999). Recent studies indicate that cellular effects of TGFβ signaling on endothelial cells are determined by the selection of downstream pathways (Goumans et al., 2002). In this model, TGFβ signaling via Alk1/smad1,5,8 pathway induces cell proliferation, differentiation and survival, whereas the Alk5/smad2,3 pathway which involves FoxH1, inhibits endothelial cell proliferation and differentiation, and induces cell death (for review, see (Goumans et al., 2003). Our finding that FoxH1 negatively regulates flk1 gene expression in a moderate and smad2-dependent manner is consistent with the negative effect of Alk5-mediated TGFβ signaling on endothelial cells and raises the possibility that the VEGF/flk1 pathway is a direct target of TGFβ signaling. Detailed investigation of the regulation of the VEGF pathway by TGFβ/FoxH1 will deepen our understanding of the interplay of these pathways in endothelial cells.
Acknowledgements
We thank A. Langenbacher for discussion and critical comments on the manuscript, K. Lyons for access to the tissue culture facility and D. Meyer for providing MZsur embryos. This work is supported by grants from NIH (HD41367), Margaret E. Early Foundation, and the Laubisch Fund to J.-N.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Attisano L, Silvestri C, Izzi L, Labbe E. The transcriptional role of Smads and FAST (FoxH1) in TGFbeta and activin signalling. Mol Cell Endocrinol. 2001;180:3–11. doi: 10.1016/s0303-7207(01)00524-x. [DOI] [PubMed] [Google Scholar]
- Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- Chen G, Nomura M, Morinaga H, Matsubara E, Okabe T, Goto K, Yanase T, Zheng H, Lu J, Nawata H. Modulation of androgen receptor transactivation by FoxH1. A newly identified androgen receptor corepressor. J Biol Chem. 2005;280:36355–63. doi: 10.1074/jbc.M506147200. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature. 1996;383:691–6. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Covassin LD, Villefranc JA, Kacergis MC, Weinstein BM, Lawson ND. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc Natl Acad Sci U S A. 2006;103:6554–9. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–2. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- DeCarvalho AC, Cappendijk SL, Fadool JM. Developmental expression of the POU domain transcription factor Brn-3b (Pou4f2) in the lateral line and visual system of zebrafish. Dev Dyn. 2004;229:869–76. doi: 10.1002/dvdy.10475. [DOI] [PubMed] [Google Scholar]
- Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, Rauter M, Plate K, Sieweke M, Breier G, Flamme I. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2003;278:7520–30. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- Erickson T, Scholpp S, Brand M, Moens CB, Jan Waskiewicz A. Pbx proteins cooperate with Engrailed to pattern the midbrain-hindbrain and diencephalic-mesencephalic boundaries. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet B, Weinstein BM, Serluca FC, Fishman MC. Vessel patterning in the embryo of the zebrafish: guidance by notochord. Dev Biol. 1997;183:37–48. doi: 10.1006/dbio.1996.8495. [DOI] [PubMed] [Google Scholar]
- Goumans MJ, Lebrin F, Valdimarsdottir G. Controlling the angiogenic switch: a balance between two distinct TGF-b receptor signaling pathways. Trends Cardiovasc Med. 2003;13:301–7. doi: 10.1016/s1050-1738(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Goumans MJ, Mummery C. Functional analysis of the TGFbeta receptor/Smad pathway through gene ablation in mice. Int J Dev Biol. 2000;44:253–65. [PubMed] [Google Scholar]
- Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. Embo J. 2002;21:1743–53. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S. Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol. 2002;12:1405–12. doi: 10.1016/s0960-9822(02)01044-8. [DOI] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbe E, Attisano L, Rossant J, Wrana JL. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes Dev. 2001;15:1257–71. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Kappel A, Schlaeger TM, Flamme I, Orkin SH, Risau W, Breier G. Role of SCL/Tal-1, GATA, and ets transcription factor binding sites for the regulation of flk-1 expression during murine vascular development. Blood. 2000;96:3078–85. [PubMed] [Google Scholar]
- Kofron M, Puck H, Standley H, Wylie C, Old R, Whitman M, Heasman J. New roles for FoxH1 in patterning the early embryo. Development. 2004;131:5065–78. doi: 10.1242/dev.01396. [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–20. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–18. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–6. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, Peters K, Grunwald DJ, Stainier DY. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–9. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development. 2000;127:4303–13. doi: 10.1242/dev.127.20.4303. [DOI] [PubMed] [Google Scholar]
- Osada SI, Saijoh Y, Frisch A, Yeo CY, Adachi H, Watanabe M, Whitman M, Hamada H, Wright CV. Activin/nodal responsiveness and asymmetric expression of a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development. 2000;127:2503–14. doi: 10.1242/dev.127.11.2503. [DOI] [PubMed] [Google Scholar]
- Patterson C, Perrella MA, Hsieh CM, Yoshizumi M, Lee ME, Haber E. Cloning and functional analysis of the promoter for KDR/flk-1, a receptor for vascular endothelial growth factor. J Biol Chem. 1995;270:23111–8. doi: 10.1074/jbc.270.39.23111. [DOI] [PubMed] [Google Scholar]
- Pepper MS. Transforming growth factor-beta: vasculogenesis, angiogenesis, and vessel wall integrity. Cytokine Growth Factor Rev. 1997;8:21–43. doi: 10.1016/s1359-6101(96)00048-2. [DOI] [PubMed] [Google Scholar]
- Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, Lo B, Weinstein BM. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek E, Heldin CH, Ten Dijke P. Specificity, diversity, and regulation in TGF-beta superfamily signaling. Faseb J. 1999;13:2105–24. [PubMed] [Google Scholar]
- Pogoda HM, Solnica-Krezel L, Driever W, Meyer D. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr Biol. 2000;10:1041–9. doi: 10.1016/s0960-9822(00)00669-2. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Stringer SE. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J Cell Sci. 2001;114:853–65. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- Roman BL, Weinstein BM. Building the vertebrate vasculature: research is going swimmingly. Bioessays. 2000;22:882–93. doi: 10.1002/1521-1878(200010)22:10<882::AID-BIES3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, Miyazono K, Whitman M, Hamada H. Left-right asymmetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–6. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Sirotkin HI, Gates MA, Kelly PD, Schier AF, Talbot WS. Fast1 is required for the development of dorsal axial structures in zebrafish. Curr Biol. 2000;10:1051–4. doi: 10.1016/s0960-9822(00)00679-5. [DOI] [PubMed] [Google Scholar]
- Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–45. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development. 1999;126:5621–34. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. University of Oregon; 2000. [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes Dev. 2001;15:1242–56. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]