Abstract
Introduction
The human multi-drug resistance gene (MDR1, ABCB1) codes for P-glycoprotein (P-gp), an important membrane-bound efflux transporter known to confer anti-cancer drug resistance as well as affect the pharmacokinetics of many drugs and xenobiotics. A number of single nucleotide polymorphisms (SNPs) have been identified throughout the ABCB1 gene which may have an effect on P-gp expression levels and function. Haplotype as well as genotype analysis of SNPs is becoming increasingly important in identifying genetic variants underlying susceptibility to human disease. Three SNPs, 1236C>T, 2677G>T, and 3435C>T have been repeatedly shown to predict changes in the function of P-gp. The frequencies with which these polymorphisms exist in a population have also been shown to be ethnically related.
Methods
In this study, 95 individuals representative of the entire ethnic make-up of the United States were compared to 101 individuals from an Ashkenazi Jewish population. These individuals were analyzed by genomic sequencing and PCR-RFLP to calculate their genotype frequencies.
Results
Twenty-five SNPs were located in the exons of the ABCB1 gene. All of the polymorphisms identified were in parts of the ABCB1 gene product predicted to be intracellular, and 16 appear to be novel as compared to those listed by NCBI. Frequencies of the 1236C>T and 2677G>T/A/C SNPs were similar for the American and Ashkenazi populations (64.2% and 60.4% respectively for 1236C>T – χ2 is 0.30 p≤1; 55.8% and 64.4% for 2677G>T/A/C χ2 is 1.49 p≤1), but were different for 3435C>T (24.2% for the American population and 69.3% for the Ashkenazi population χ2 is 39.927 p<0.001). The 1236T/2677T/3435T haplotype occurred in 23.6% (SE 0.013) of the Ashkenazi population.
Conclusion
The SNP at location 3435C>T plays a significant role in the ABCB1 gene. The haplotype and genotype analysis from these data may be used as a basis for studies on the relationship between ABCB1 genotypes and drug efficacy, drug toxicity, disease susceptibility or other phenotypes.
Keywords: P-glycoprotein, ABCB1, ethnicity, haplotypes, SNPs
Introduction
The efficiency of chemotherapy in the treatment of cancer is limited by the development of resistant cell variants. A common mechanism of multidrug resistance (MDR) in cancer cells is the expression of higher than normal levels of a transmembrane protein which serves as an energy dependent efflux pump, thus causing a reduction in the amount of drug that accumulates within the cancer cell [1, 2]. This appears to be a primary reason for failure of chemotherapeutic treatment despite the application of a combination of pharmacological agents.
The human MDR1 gene (ABCB1) encodes a 170-kDa plasma membrane glycoprotein [P-glycoprotein (P-gp)], which is a member of the ATP-dependent ABC transporters superfamily. P-gp has an effect on the pharmacokinetics of a variety of anticancer drugs and other drugs including digoxin [3], HIV protease inhibitors [4], statins [5], antihistamines [6] and numerous other drugs and xenobiotics.
ABCB1 is composed of 28 exons ranging in size from 49 to 209 base pairs (bps) encoding an mRNA of 4.5 kb. When the human genome was first sequenced, researchers estimated that on average, one single nucleotide polymorphism (SNP) occurs per 1,250 bps in the human genome [7]. Since then however, this estimate has been revised to about one in 300 nucleotides, resulting in approximately 11 million SNPs [8]. Notably, this latter estimate depends in turn on the definition of an SNP: a polymorphism present in at least 1% of the human population. Lowering this 1% threshold would result in a further increase in the total number of SNPs in the human genome. This number of SNPs along the ABCB1 cDNA (4.5 kb) is consistent with the average in the genome. Initially, only 15 different SNPs were found in the exons of ABCB1 in a Caucasian population [9]. Other researchers detected a total of 50 SNPs and insertion/deletion polymorphisms in the cDNA [10–13]. Many of these SNPs are silent (synonymous), and do not produce a change in amino acid sequence.
Among various population groups, the most common polymorphisms found in ABCB1 are 1236C>T, 2677G>T/A/C, and 3435C>T. Studies of polymorphisms occurring in two ethnic groups of the U.S. (European Americans and African Americans), polymorphic analysis and direct sequencing of exonic ABCB1 in these groups, identified 10 SNPs, of which 6 were non-synonymous variants. Of the synonymous SNPs, two (1236C>T and 3435C>T) were linked to a non-synonymous SNP (2677G>T, Ala893Ser) and occurred in 62% of the European Americans tested, but appeared in only 13% of the African Americans [14] (NCBI SNP website). Researchers searching for a correlation between the silent polymorphism 3435C>T, the non-synonymous polymorphism 2677G>A, and the expression of P-gp in human liver cells, reported a great variation in P-gp expression, but concluded it was not due to polymorphic alleles [15].
The Ashkenazi Jewish population, the largest Jewish ethnic group globally, representing the majority of North-American Jewry, consists of the descendants of central and eastern European Jews. It is considered relatively homogenous, due to a bottleneck which occurred fewer than 100 generations ago that was followed by very rapid expansion during the 17th to 19th centuries and is therefore a suitable ethnic group for certain types of genetic research [16, 17]. Various studies on this population have been performed, and a predisposition to different diseases has been demonstrated, including Tay-Sachs [18], familial Mediterranean fever [19], hereditary breast and ovarian cancers [20], Gaucher’s and Parkinson’s diseases [21], and Crohn’s disease [22]. However, only a single study has been conducted regarding ABCB1 in the Ashkenazi Jewish population and it only addressed the SNP 3435C>T variant [23].
Haplotype analysis, in addition to the customary analysis of SNPs, may play an important role in the identification of genetic variations between ethnic groups. In addition, the functional effects of P-gp activity may also be related to haplotypes in the ABCB1 gene. Kroetz and colleagues performed an in-depth haplotype analysis of several ethnic groups [24] and analyzed some of these haplotypes in vitro for P-gp activity. It has been shown that several haplotypes in the ABCB1 gene have clinical relevance. For example, the 2677G/3435T haplotype is associated with significantly higher bioavailability after oral digoxin administration, while 2677G/3435C was correlated with lower digoxin bioavailability [25]. In another study, haplotype 1236T/2677G/3435T carriers demonstrated higher P-gp activity when compared to non-carriers [14]. The 1236T/2677T/3435T haplotype was also shown to be associated with a slightly greater risk of developing refractory Crohn’s disease (P=0.044) [26]. The same study showed that when each SNP occurred separately, there was no significant correlation with the development of the disease. Therefore, haplotype analysis may provide an advantage in the diagnosis of these complex diseases. Researchers also believe that better understanding of haplotypes, rather than single SNP analysis, will lead to better identification of genetic variants underlying complex traits [27].
Our current study analyzes existing ABCB1 polymorphisms in two populations, the ethnically homogenous Ashkenazi Jews and the ethnically diverse US population. Within the American population (with a full representation of different ethnicities in the United States) we performed an in depth analysis of exonic polymorphic changes and their locations along the gene. Within the population of Ashkenazi Jews, we examined the three most frequently occurring SNPs and their haplotypes (1236C>T, 2677G>T/A/C and 3435C>T). We report here our findings and discuss their plausible significance for pharmacogenomic applications.
Materials and Methods
Study groups
Ninety individual DNA samples were purchased from the Coriell Repository, with 5 additional samples kindly provided by Tito Fojo (NCI, NIH) after obtaining written informed consent for genetic analysis from all individuals. The Coriell sample is a proportional sample of all major ethnic groups within the entire U.S. population; however, the ethnicity of each individual sample was not identified to the researchers. One hundred and one DNA samples from unrelated adult individuals representing the Ashkenazi Jewish population in Israel were obtained from the National Laboratory for the Genetics of Israeli Populations at Tel-Aviv University, Israel (http://nlgip.tau.ac.il) after receiving informed consent for genetic analysis from those individuals.
Genotype and haplotype analysis
For the samples supplied by the Coriell Repository and those given by Dr. Fojo, primer design, PCR and sequencing were performed on each sample by the NCI Sequencing Facility. Initially, sequencing was performed from 5′ to 3′ with fragments of 600 base pairs and a 50 to 100 base pair overlap between each of the fragments. The sequencing took place over the entire exonic region of the gene, excluding the 5′ and 3′ untranslated regions. Numerous samples were also sequenced from 3′ to 5′. Additional sequence tests were deemed necessary for a number of the samples and the sequencing was repeated several times (up to six repetitions). The sequences obtained were compared with the wild-type sequence of the ABCB1 gene (NM_000927.3) to search for possible polymorphisms throughout the population studied. The inclusion criterion for the sequencing results was a base quality greater than 10, based on the NCI sequencing facility criteria. Sequencing results that were below this criterion were excluded. Polymorphic sequences were grouped according to the reproducibility of the sequencing results. Polymorphisms appearing with reproducibility lower than 50% were excluded from the study. By this method, individuals were not identified as heterozygous or homozygous for the polymorphic allele, but rather as polymorphic or wild-type. Final analysis was performed on the data obtained from the remaining samples.
The three regions of the ABCB1 gene including SNPs 1236C>T, 2677G>T/A/C, and 3435C>T in the Ashkenazi Jewish Population were amplified using the High Fidelity 2 PCR kit (Clontech, Mountain View CA). We found that the Hot Start procedure was the optimal procedure for the PCR products. The primers, PCR annealing conditions and restriction enzymes are summarized in Table 1.
Table 1.
PCR and digestion to detect common MDR1 polymorphisms
| Polymorphism (location) | Forward primer | Reverse primer | Product length (bp) | PCR annealing temperature | Restriction enzyme |
|---|---|---|---|---|---|
| GGC>GGT | 5’AAGGGAAATTTG
GAATTCAGAAAT3’ |
5’TTGTGCTCTTCCCA
CAGCCACTGTTT3’ |
92 | 60°C | Pho I |
| (1236C>T) | |||||
| GCT>TCT | 5’ATTGCAATAGCA
GGAGTTGTTGAAA |
5’TAATCAATCATATT
TAGTTTGACTCAC3’ |
57 | 55°C | Bse YI |
| (2677G>T/A/C) | T3’ | ||||
| ATC>ATT | 5’AACATTGCCTAT
GGAGACAACA3’ |
5’AGTGACTCGATGA
AGGCATGTAT3’ |
50 | 60°C | Bfu CI |
| (3435C>T) |
The amplified DNA resulting from the PCR was then verified by gel electrophoresis analysis. Once the product size was confirmed, the remaining product was purified using the QIAquick® PCR Purification Kit (Qiagen Inc., Chatsworth, CA). Following purification, the purified PCR product was digested with a restriction enzyme that could distinguish between genotypes and was chosen using the website http://tools.neb.com/NEBcutter2/index.php (PCR-restriction fragment length polymorphism – PCR-RFLP).
To verify results obtained from gel electrophoresis analysis of restriction enzymes, samples of each genotype (homozygous wild-type, heterozygous, and homozygous polymorphism; total of 15 samples) were sequenced to confirm their genotypes, which permit us to use the common nomenclature 1236C>T, 2677G>T/A/C, and 3435C>T for these sites.
The haplotypes of the Ashkenazi population in this study were statistically inferred by the use of PHASE software (version 2.1), which utilizes an algorithm based on Bayeian interference [28, 29]. Haplotypes were deduced after running PHASE a total of 10 times, in which each time the program returned the same combinations of the most likely haplotypes. The standard error in calculation of haplotypes was consistently less than 1.5%.
Statistical analysis in this study was performed with the Chi-square test for comparison of genotype frequencies between populations. Statistical significance between the observed and expected Hardy-Weinberg genotype frequencies was determined. A P value of less than 0.05 was considered to be statistically significant.
Results
Common characteristics of exonic polymorphisms along the ABCB1 gene
Sequencing results revealed a total of 25 SNPs in the exonic regions of the ABCB1 gene, but these SNPs are not evenly distributed along the gene. Most of them are located in the region between bp 150,000 and 200,000 (Figure 1). Ten of the 25 exonic polymorphisms detected are synonymous polymorphisms and 15 are non-synonymous. Of these 25 polymorphisms, 16 are newly identified SNPs as compared to those listed by NCBI (Table 2), however some appear only in one individual. Fifty-six of the ninety-five American samples had the non-synonymous 2677G>T/A/C polymorphism, resulting in amino acids Serine, Threonine and Proline, respectively. Five individuals had a non-synonymous SNP at site 2959, resulting in an Alanine to Proline alteration (the latter usually encodes a helix termination) (Table 2). Structure analysis reveals that this change appears adjacent to the ATP binding site of the P-gp protein (Figure 1). Analysis of the locations of the SNPs demonstrates that all non-synonymous SNPs are intracellular, none of which appear at ATP binding sites.
Figure 1. Hypothetical structure of P-gp and corresponding SNP locations within the ABCB1 exonic region of the American population.
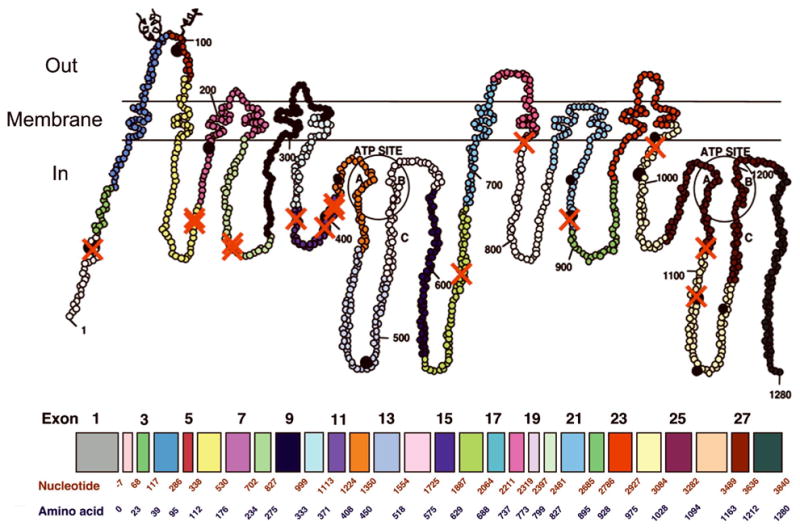
This figure illustrates the approximate locations of amino acid changes along the P-gp protein resulting from non-synonymous polymorphisms. A red “X” marks the approximate location of each polymorphism identified in this study. Black dots represent the approximate locations of common amino acid changes previously reported in the P-gp protein. Exons 1–28 are labeled by color and nucleotide length.
Table 2.
ABCB1 polymorphisms in the American population (n=95)
| Exon | Polymorphism | Nucleotide change | Amino acid change | Amino acid position | Frequency (%) |
|---|---|---|---|---|---|
| 2 | 60* | T to C | Synonymous | 20 | 2.1 |
| 2 | 61 | A to G | Asn to Asp | 21 | 4.2 |
| 6 | 502* | G to A | Val to Ile | 168 | 1.1 |
| 6 | 508* | G to A | Glu to Lys | 170 | 1.1 |
| 8 | 729 | A to G | Synonymous | 243 | 1.1 |
| 8 | 781 | A to G | Ile to Val | 261 | 1.1 |
| 8 | 785* | G to A | Arg to Lys | 262 | 1.1 |
| 11 | 1131* | C to G | Ser to Arg | 377 | 1.1 |
| 11 | 1170* | A to G | Synonymous | 390 | 1.1 |
| 11 | 1189* | G to T | Val to Phe | 397 | 1.1 |
| 11 | 1199 | G to A | Ser to Asn | 400 | 1.1 |
| 11 | 1201* | T to A | Tyr to Asn | 401 | 1.1 |
| 12 | 1236 | C to T | Synonymous | 412 | 64.2 |
| 14 | 1659 | G to C | Synonymous | 553 | 2.1 |
| 16 | 1991* | G to T | Arg to Ile | 664 | 1.1 |
| 18 | 2315* | T to C | Leu to Pro | 772 | 1.1 |
| 21 | 2650 | C to T | Synonymous | 884 | 1.1 |
| 21 | 2676* | T to G | Synonymous | 892 | 2.1 |
| 21 | 2677 | G to T/A/C | Ala to Ser/Thr/Pro | 893 | 55.8 |
| 22 | 2712* | C to T | Synonymous | 904 | 1.1 |
| 24 | 2959* | G to C | Ala to Pro | 987 | 5.3 |
| 24 | 3048* | T to C | Synonymous | 1016 | 3.2 |
| 25 | 3262* | G to T | Asp to Tyr | 1088 | 1.1 |
| 26 | 3346* | G to A | Val to Met | 1116 | 1.1 |
| 26 | 3435 | C to T | Synonymous | 1145 | 24.2 |
Newly-identified polymorphism, as compared to the NCBI database.
Combination of SNPs in the American population
The most common combination of SNPs, appearing in 47.4% (n=45) of the individuals in the American population, was 1236C>T, 2677G>T/A/C. Furthermore, in 14.7% (n=14) of the population the SNPs 1236C>T, 2677G>T/A/C, and 3435C>T were found together. The SNP 1236C>T, appearing is a sole SNP or accompanied by other SNPs, appeared in nearly two thirds of all individuals (Table 3).
Table 3.
Combination of SNPs in the American population
| SNPs | Number of individuals out of 95 | Percent of total |
|---|---|---|
| 1236C>T, 3048T>C | 2 | 2.1 |
| 1236C>T, 60T>C | 2 | 2.1 |
| 2677G>T/A/C, 60T>C | 2 | 2.1 |
| 1236C>T, 61A>G | 3 | 3.2 |
| 2677G>T/A/C, 2959G>C | 3 | 3.2 |
| 1236C>T, 2959G>C | 3 | 3.2 |
| 2677G>T/A/C, 61A>G | 4 | 4.2 |
| 2677G>T/A/C, 3435C>T | 16 | 16.8 |
| 1236C>T, 3435C>T | 17 | 17.9 |
| 1236C>T, 2677G>T/A/C | 45 | 47.4 |
| 1236C>T, 2677G>T/A/C, 2959G>C | 2 | 2.1 |
| 1236C>T, 2677G>T/A/C, 60T>C | 2 | 2.1 |
| 1236C>T, 2677G>T/A/C, 61A>G | 3 | 3.2 |
| 1236C>T, 2677G>T/A/C, 3435C>T | 14 | 14.7 |
Bold numbers indicate high relative percentage
Frequency of the common genotypes and haplotypes in the Ashkenazi Jewish population
PCR-RFLP results were confirmed by sequencing analysis (Figure 2). All of the 101 individuals in the Ashkenazi population were analyzed for the three common exonic SNP locations (1236C>T, 2677G>T/A/C, and 3435C>T). Sequencing was used to confirm the restriction enzyme method to determine individual genotypes. Of the samples studied, 39.6% (n=40) were homozygous for the 1236C allele, 23.8% (n=24) for the 1236T allele, and 36.6% (n=37) carried the 1236CT heterozygote. For the ABCB1 2677 genotype, 35.6% (n=36) were homozygous for the 2677G allele, 16.8% (n=17) for the 2677T polymorphic allele, and 47.5% (n=48) contained 2677GT heterozygotes. For location 3435, 30.7% (n=31) were homozygous for the 3435C allele, 30.7% (n=31) for the 3435T allele, and 38.6% (n=39) were 3435CT heterozygotes.
Figure 2. Sequencing and electrophoresis results confirm individual genotype status.
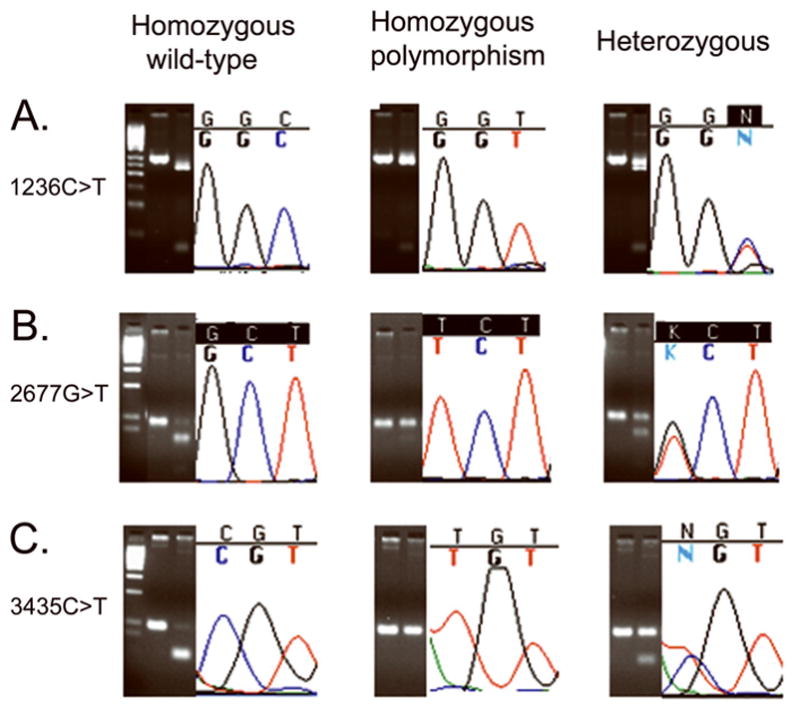
DNA sequencing was used to ensure that results obtained by PCR-RFLP were accurate. A. SNP 1236C>T occurs in the third base-pair of the codon B. SNP 2677G>T occurs in the first base-pair of the codon, C. SNP 3435C>T occurs in the first base-pair of the codon. Electrophoresis was performed on 4% TBE agarose gels with a 50 bp Low Range Ladder.
The most common haplotypes of the three polymorphisms, calculated by PHASE software, are summarized in Table 5. The occurrence of haplotype 1236T/2677T/3435T was 23.6% (SE 0.013). A comparison of the SNP frequencies in the American Population and the Ashkenazi population revealed that the Ashkenazi population had a significantly higher percentage of the 3435C>T polymorphism (69.3%, n=70); only 24.2% (n=23) of the American population carried this SNP (χ2 =39.927 p<0.001). However, similar frequencies were found for 1236C>T (64.2% in the American and 60.4% in the Ashkenazi group) and for 2677G>T (55.8% in the American and 64.4% in the Ashkenazi group). A detailed comparison of allele and genotype frequencies among Ashkenazi Jews (this study) and other groups such as African Americans and Chinese is provided in Table 4. For the 1236C>T SNP, two sets of data are listed for Caucasians, as there were considerable differences.
Table 5.
Haplotypes in the Ashkenazi population
| Position
|
Estimated frequency (%) | S.E. | ||
|---|---|---|---|---|
| 1236 | 2677 | 3435 | ||
| C | G | C | 31.7 | 0.014 |
| T | T | T | 23.6 | 0.013 |
| C | G | T | 14.4 | 0.013 |
| T | G | C | 6.9 | 0.009 |
| T | G | T | 6.4 | 0.009 |
| C | T | C | 6.2 | 0.011 |
| C | T | T | 5.6 | 0.012 |
| T | T | C | 5.2 | 0.01 |
Table 4.
Allele and genotype frequency comparisons between ethnicities
| SNP | Population | N | Allele frequency | Genotype frequency | Reference | |||
|---|---|---|---|---|---|---|---|---|
| C | T | CC | CT | TT | ||||
|
| ||||||||
| 1236C>T | Ashkenazi | 101 | 0.58 | 0.42 | 0.40 | 0.37 | 0.24 | This study |
| Caucasian | 188 | 0.62 | 0.38 | 0.38 | 0.49 | 0.13 | [9] | |
| Caucasian | 31 | 0.53 | 0.47 | 0.32 | 0.42 | 0.26 | NCBI | |
| Chinese | 45 | 0.31 | 0.69 | 0.11 | 0.40 | 0.49 | NCBI | |
| African American | 24 | 0.81 | 0.19 | 0.63 | 0.37 | 0.00 | NCBI | |
| G | T | GG | GT | TT | ||||
|
| ||||||||
| 2677G>T | Ashkenazi | 101 | 0.59 | 0.41 | 0.36 | 0.48 | 0.17 | This study |
| Caucasian | 31 | 0.57 | 0.43 | 0.32 | 0.48 | 0.19 | NCBI | |
| African American | 24 | 0.85 | 0.11 | 0.75 | 0.21 | 0.00 | NCBI | |
| Asian | 44 | 0.50 | 0.50 | 0.32 | 0.36 | 0.32 | NCBI | |
| C | T | CC | CT | TT | ||||
|
| ||||||||
| 3435C>T | Ashkenazi | 101 | 0.50 | 0.50 | 0.31 | 0.39 | 0.31 | This study |
| Caucasian | 188 | 0.52 | 0.48 | 0.28 | 0.48 | 0.24 | [9] | |
| African American | 88 | 0.84 | 0.16 | 0.68 | 0.31 | 0.01 | [55] | |
| Chinese | 265 | 0.56 | 0.44 | 0.32 | 0.48 | 0.20 | [56] | |
Discussion
Twenty-five exonic SNPs were revealed in the American population, 16 of which are newly-identified as compared to the NCBI database. In the Ashkenazi Jewish population, the SNP in position 3435T was significantly more common than in the American population (χ2 is 39.927 p<0.001). The estimated frequency of the most common haplotype, 1236T/2677T/3435T, is 23.6%.
Analysis of the locations of the SNPs demonstrates that all non-synonymous SNPs occur in coding regions predicted to be intracellular, and moreover, none of them appear at the ATP binding sites. Nonetheless, several non-synonymous SNP sites could potentially affect either the nearby ATP sites, or the function of the ABCB1 protein as a whole. The intracellular loops of the ABCB1 sequence are less conserved (20%) than the extracellular loops (40%) among various species, which is consistent with the higher probability of SNPs in the intracellular loops (http://genome.ucsc.edu for ABCB1).
To date, the most commonly reported polymorphism linked to different responses of patients to various ABCB1 substrates is located at exon 26, 3435C>T, and does not result in an amino acid change [30]. The 1236C>T and 2677G>T/A/C polymorphisms have also been linked to several diseases such as pharmacoresistant epilepsy and Parkinsons disease [31–45]. Studies on these polymorphisms have yielded contradictory results, possibly due to small sample sizes and the isolated nature of the study populations. Also, some studies did not include a control group for comparison with cancer patients. Several researchers performed an in-depth analysis of the polymorphisms along the ABCB1 gene, while others researched specific SNPs without investigating the existence of other polymorphic locations. The contradictory results may indicate that SNPs of the ABCB1 gene should be analyzed according to complete haplotypes instead of individually. Several different studies have found the 3435C>T polymorphism to be associated with a change in the expression of P-gp [30, 46], although in transfection studies of cells, no differences in either mRNA or protein levels are observed [47].
Ostrovsky and colleagues found a significantly different frequency of the T allele at site 3435 of the ABCB1 gene among Near Eastern Jews (from Iraq, Iran, and Buchara) when compared with other Jewish populations (Ashkenazi, Yemenite and North-African) [23]. Our results for the homozygous polymorphism at this site (38.6%) are more comparable to those found in Near Eastern Jews (31%) than the Ashkenazi Jews (12%) in that study. A comparison between the allele and genotype frequencies of the Ashkenazi population and other ethnic groups from previous studies is summarized in Table 4. In each of the three SNP locations, the Ashkenazi population is comparable in its allele and genotype frequencies to that of Caucasian populations. According to studies submitted to the National Center of Bioinformatics, the incidence of the 3435T allele in the studied American population is similar to African American populations (24.2% and 16% respectively). Studies on the 3435C>T SNP show a correlation between allele frequency and risk of cancer development, as well as various responses to drug treatments. While no association has been observed between the TT genotype and the lung cancer phenotype [9, 48], homozygous and heterozygous carriers of the T allele have been linked to a greater risk of developing nonclear cell renal cell carcinoma than individuals carrying the C allele [49]. Carriers of the TT genotype are more at risk of developing Acute Lymphoblastic Leukemia (ALL) than other individuals, whereas the CC genotype carriers exhibit a different prognosis [50].
The American and Ashkenazi populations both expressed similar percentages of the 1236C>T and 2677G>T/A/C SNPs in the ABCB1 gene. However, the percentage of individuals expressing the 3435C>T polymorphism in the Ashkenazi population was significantly higher when compared to the American population (χ2 is 39.927 p<0.001). The high prevalence of this SNP in the Ashkenazi population may reflect a founder effect that preceded the large expansion of the Ashkenazi Jews between the 16th to the 19th centuries. Later on, the polymorphism was fixed in this population due to its rapid expansion and not necessarily because of a selective advantage of carrying this SNP. Of note, the high prevalence of BRCA1 185delAG and APC 11307K in Ashkenazi Jews was similarly explained by a founder effect occurring between 947 and 195 BC rather than by selective advantage [51].
An increased risk for the development of colorectal cancer has also been found in carriers of the 3435T allele under the age of fifty. Carriers of the ABCB1 3435TT genotype had a 2.7-fold greater risk of the development of colorectal cancer [32]. This SNP may therefore be linked to a high prevalence of colon cancer in the Ashkenazi population. Similarly, refractory Crohn’s disease has also been found to be associated with the 2677G>T/A/C, 3435C>T, and 1236C>T SNPs [31]. This could also be linked to a high prevalence of the disease in the Ashkenazi Jewish population, as noted by the high prevalence of the 3435T allele discovered in our Ashkenazi population. The mutation ΔF508 in Cystic fibrosis also has a high incidence in the Ashkenazi population [52]. This mutation in another ATP transporter was associated with protection against cholera and typhoid fever [53, 54]. However, it has yet to be determined if the high prevalence of the 3435C>T SNP has a heterozygote advantage in these diseases.
In summary, this study has investigated differences in the number and location of single nucleotide polymorphisms along the ABCB1 gene between an ethnically diverse American population and an ethnically homogeneous Ashkenazi Jewish population. Using sequencing methods as well as PCR-RFLP, polymorphisms were located at three primary locations along the ABCB1 gene (1236C>T, 2677G>T/A/C, and 3435C>T). The PCR-RFLP method proved to be a very accurate and reliable method for identifying specific SNPs at a lower cost then genomic sequencing. Results of this analysis showed that the 3435C>T polymorphism plays a significant role in the determination of haplotypes of the ABCB1 gene. However, due to the prevalence of the 1236T/2677T/3435T haplotype in the Ashkenazi population in individuals that possess the 3435C>T SNP, it may be beneficial to also search for this particular haplotype.
The overall frequency of SNPs is consistent with current understanding of the prevalence of SNPs in the genome. Notably, a comparative analysis of DNA sequences of 132 introns and 140 exons from 42 pairs of orthologous mouse and rat genes has shown that on average, non-synonymous exonic changes between these two species were 3-fold less common compared with intronic inter-species changes [28]. Thus, we can conclude that the higher degree of conservation of the ABCB1 gene may indeed reflect the function of the ABCB1 protein as a poly-specific detoxifying transporter of foreign compounds and its need to adapt to new challenges.
Outlook
Detecting the exact haplotype of an individual can be used as a basis for further studies of the effect of pharmacogenomic analysis of this haplotype on drug safety and efficacy, as well as disease susceptibility. The results reported here may be used to compare the effects of individual drug response in ethnic groups as well as for the Ashkenazi Jewish population. The higher incidence of the 1236T/2677T/3435T haplotype in the Ashkenazi Jewish population may predict altered sensitivity, or possibly higher toxicity from many drugs that are primarily P-gp substrates such as digoxin and fexofenidine. Furthermore, the relative similarity between allele and genotype frequencies of the Ashkenazi population, as compared to Caucasian populations, may help to predict drug dose requirements in Ashkenazi Jews, which would be very similar to Caucasians. Further studies are needed to determine the effects of these ABCB1 gene haplotypes on drug pharmacokinetics.
Highlights
Twenty-five SNPs, 16 of which have not previously been reported, were located in exons of the ABCB1 gene, all of which were in parts of the ABCB1 gene product predicted to be intracellular.
A total of 101 individuals from an Ashkenazi-Jewish population were tested for the 3 most common SNPs and the estimated frequency of the 1236T/2677T/3435T haplotype is 23.6%.
The SNP at location 3435C>T plays a significant role in prediction of haplotypes in the ABCB1 gene.
Allele and genotype frequencies of the Ashkenazi population are very similar to those of Caucasian populations.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. We would like to thank Ken Buetow, Eric Green, Michael Edmonson, and Jenny Kelley (National Institutes of Health) for all their contributions in DNA sequencing for this experiment. Also, a special thanks to George Leiman for insightful editorial assistance.
Bibliography
- 1.Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–92. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–27. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 3.Mayer U, Wagenaar E, Beijnen JH, et al. Substantial excretion of digoxin via the intestinal mucosa and prevention of long-term digoxin accumulation in the brain by the mdr 1a P-glycoprotein. Br J Pharmacol. 1996;119(5):1038–44. doi: 10.1111/j.1476-5381.1996.tb15775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee CG, Gottesman MM, Cardarelli CO, et al. HIV-1 protease inhibitors are substrates for the MDR1 multidrug transporter. Biochemistry. 1998;37(11):3594–601. doi: 10.1021/bi972709x. [DOI] [PubMed] [Google Scholar]
- 5.Bogman K, Peyer AK, Torok M, Kusters E, Drewe J. HMG-CoA reductase inhibitors and P-glycoprotein modulation. Br J Pharmacol. 2001;132(6):1183–92. doi: 10.1038/sj.bjp.0703920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiou WL, Chung SM, Wu TC, Ma C. A comprehensive account on the role of efflux transporters in the gastrointestinal absorption of 13 commonly used substrate drugs in humans. Int J Clin Pharmacol Ther. 2001;39(3):93–101. doi: 10.5414/cpp39093. [DOI] [PubMed] [Google Scholar]
- 7.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 8.Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim Biophys Acta. 2006;1762(1):17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97(7):3473–8. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkmann U, Roots I, Eichelbaum M. Pharmacogenetics of the human drug-transporter gene MDR1: impact of polymorphisms on pharmacotherapy. Drug Discov Today. 2001;6(16):835–839. doi: 10.1016/s1359-6446(01)01892-x. [DOI] [PubMed] [Google Scholar]
- 11.Saito S, Iida A, Sekine A, et al. Three hundred twenty-six genetic variations in genes encoding nine members of ATP-binding cassette, subfamily B (ABCB/MDR/TAP), in the Japanese population. J Hum Genet. 2002;47(1):38–50. doi: 10.1007/s10038-002-8653-6. [DOI] [PubMed] [Google Scholar]
- 12.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–49. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 13.Schwab M, Eichelbaum M, Fromm MF. Genetic polymorphisms of the human MDR1 drug transporter. Annu Rev Pharmacol Toxicol. 2003;43:285–307. doi: 10.1146/annurev.pharmtox.43.100901.140233. [DOI] [PubMed] [Google Scholar]
- 14.Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70(2):189–99. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 15.Owen A, Goldring C, Morgan P, Chadwick D, Park BK, Pirmohamed M. Relationship between the C3435T and G2677T(A) polymorphisms in the ABCB1 gene and P-glycoprotein expression in human liver. Br J Clin Pharmacol. 2005;59(3):365–70. doi: 10.1111/j.1365-2125.2005.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behar DM, Hammer MF, Garrigan D, et al. MtDNA evidence for a genetic bottleneck in the early history of the Ashkenazi Jewish population. Eur J Hum Genet. 2004;12(5):355–64. doi: 10.1038/sj.ejhg.5201156. [DOI] [PubMed] [Google Scholar]
- 17.Behar DM, Metspalu E, Kivisild T, et al. The matrilineal ancestry of Ashkenazi Jewry: portrait of a recent founder event. Am J Hum Genet. 2006;78(3):487–97. doi: 10.1086/500307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slatkin M. A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am J Hum Genet. 2004;75(2):282–93. doi: 10.1086/423146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershoni-Baruch R, Shinawi M, Leah K, Badarnah K, Brik R. Familial Mediterranean fever: prevalence, penetrance and genetic drift. Eur J Hum Genet. 2001;9(8):634–7. doi: 10.1038/sj.ejhg.5200672. [DOI] [PubMed] [Google Scholar]
- 20.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 21.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351(19):1972–7. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 22.Tukel T, Shalata A, Present D, et al. Crohn disease: frequency and nature of CARD15 mutations in Ashkenazi and Sephardi/Oriental Jewish families. Am J Hum Genet. 2004;74(4):623–36. doi: 10.1086/382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrovsky O, Nagler A, Korostishevsky M, Gazit E, Galski H. Genotype and allele frequencies of C3435T polymorphism of the MDR1 gene in various Jewish populations of Israel. Ther Drug Monit. 2004;26(6):679–84. doi: 10.1097/00007691-200412000-00015. [DOI] [PubMed] [Google Scholar]
- 24.Kroetz DL, Pauli-Magnus C, Hodges LM, et al. Sequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) gene. Pharmacogenetics. 2003;13(8):481–94. doi: 10.1097/00008571-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Johne A, Kopke K, Gerloff T, et al. Modulation of steady-state kinetics of digoxin by haplotypes of the P-glycoprotein MDR1 gene. Clin Pharmacol Ther. 2002;72(5):584–94. doi: 10.1067/mcp.2002.129196. [DOI] [PubMed] [Google Scholar]
- 26.Potocnik U, Ferkolj I, Glavac D, Dean M. Polymorphisms in multidrug resistance 1 (MDR1) gene are associated with refractory Crohn disease and ulcerative colitis. Genes Immun. 2004;5(7):530–9. doi: 10.1038/sj.gene.6364123. [DOI] [PubMed] [Google Scholar]
- 27.Zhao H, Pfeiffer R, Gail MH. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003;4(2):171–8. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
- 28.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(5):1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22(47):7468–85. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- 31.Asano T, Takahashi KA, Fujioka M, et al. ABCB1 C3435T and G2677T/A polymorphism decreased the risk for steroid-induced osteonecrosis of the femoral head after kidney transplantation. Pharmacogenetics. 2003;13(11):675–82. doi: 10.1097/00008571-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Kurzawski M, Drozdzik M, Suchy J, et al. Polymorphism in the P-glycoprotein drug transporter MDR1 gene in colon cancer patients. Eur J Clin Pharmacol. 2005;61(5–6):389–94. doi: 10.1007/s00228-005-0926-5. [DOI] [PubMed] [Google Scholar]
- 33.Tan EK, Chan DK, Ng PW, et al. Effect of MDR1 haplotype on risk of Parkinson disease. Arch Neurol. 2005;62(3):460–4. doi: 10.1001/archneur.62.3.460. [DOI] [PubMed] [Google Scholar]
- 34.Goreva OB, Grishanova AY, Mukhin OV, Domnikova NP, Lyakhovich VV. Possible prediction of the efficiency of chemotherapy in patients with lymphoproliferative diseases based on MDR1 gene G2677T and C3435T polymorphisms. Bull Exp Biol Med. 2003;136(2):183–5. doi: 10.1023/a:1026331326648. [DOI] [PubMed] [Google Scholar]
- 35.Zhu D, Taguchi-Nakamura H, Goto M, et al. Influence of single-nucleotide polymorphisms in the multidrug resistance-1 gene on the cellular export of nelfinavir and its clinical implication for highly active antiretroviral therapy. Antivir Ther. 2004;9(6):929–35. [PubMed] [Google Scholar]
- 36.Sills GJ, Mohanraj R, Butler E, et al. Lack of association between the C3435T polymorphism in the human multidrug resistance (MDR1) gene and response to antiepileptic drug treatment. Epilepsia. 2005;46(5):643–7. doi: 10.1111/j.1528-1167.2005.46304.x. [DOI] [PubMed] [Google Scholar]
- 37.Yamauchi A, Ieiri I, Kataoka Y, et al. Neurotoxicity induced by tacrolimus after liver transplantation: relation to genetic polymorphisms of the ABCB1 (MDR1) gene. Transplantation. 2002;74(4):571–2. doi: 10.1097/00007890-200208270-00024. [DOI] [PubMed] [Google Scholar]
- 38.van den Heuvel-Eibrink MM, Wiemer EA, de Boevere MJ, et al. MDR1 gene-related clonal selection and P-glycoprotein function and expression in relapsed or refractory acute myeloid leukemia. Blood. 2001;97(11):3605–11. doi: 10.1182/blood.v97.11.3605. [DOI] [PubMed] [Google Scholar]
- 39.Meissner K, Jedlitschky G, Meyer zu Schwabedissen H, et al. Modulation of multidrug resistance P-glycoprotein 1 (ABCB1) expression in human heart by hereditary polymorphisms. Pharmacogenetics. 2004;14(6):381–5. doi: 10.1097/00008571-200406000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Kurata Y, Ieiri I, Kimura M, et al. Role of human MDR1 gene polymorphism in bioavailability and interaction of digoxin, a substrate of P-glycoprotein. Clin Pharmacol Ther. 2002;72(2):209–19. doi: 10.1067/mcp.2002.126177. [DOI] [PubMed] [Google Scholar]
- 41.Horinouchi M, Sakaeda T, Nakamura T, et al. Significant genetic linkage of MDR1 polymorphisms at positions 3435 and 2677: functional relevance to pharmacokinetics of digoxin. Pharm Res. 2002;19(10):1581–5. doi: 10.1023/a:1020433422259. [DOI] [PubMed] [Google Scholar]
- 42.Moriya Y, Nakamura T, Horinouchi M, et al. Effects of polymorphisms of MDR1, MRP1, and MRP2 genes on their mRNA expression levels in duodenal enterocytes of healthy Japanese subjects. Biol Pharm Bull. 2002;25(10):1356–9. doi: 10.1248/bpb.25.1356. [DOI] [PubMed] [Google Scholar]
- 43.Tang K, Ngoi SM, Gwee PC, et al. Distinct haplotype profiles and strong linkage disequilibrium at the MDR1 multidrug transporter gene locus in three ethnic Asian populations. Pharmacogenetics. 2002;12(6):437–50. doi: 10.1097/00008571-200208000-00004. [DOI] [PubMed] [Google Scholar]
- 44.Tanabe M, Ieiri I, Nagata N, et al. Expression of P-glycoprotein in human placenta: relation to genetic polymorphism of the multidrug resistance (MDR)-1 gene. J Pharmacol Exp Ther. 2001;297(3):1137–43. [PubMed] [Google Scholar]
- 45.Furuno T, Landi MT, Ceroni M, et al. Expression polymorphism of the blood-brain barrier component P-glycoprotein (MDR1) in relation to Parkinson’s disease. Pharmacogenetics. 2002;12(7):529–34. doi: 10.1097/00008571-200210000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Markova S, Nakamura T, Sakaeda T, et al. Genotype-dependent down-regulation of gene expression and function of MDR1 in human peripheral blood mononuclear cells under acute inflammation. Drug Metab Pharmacokinet. 2006;21(3):194–200. doi: 10.2133/dmpk.21.194. [DOI] [PubMed] [Google Scholar]
- 47.Kimchi-Sarfaty C, Oh J-M, Kim I-W, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 48.Sinues B, Fanlo A, Bernal ML, et al. MDR-1 C3435T genetic polymorphism and tobacco-related lung cancer. Oncology. 2003;64(2):183–5. doi: 10.1159/000067770. [DOI] [PubMed] [Google Scholar]
- 49.Siegsmund M, Brinkmann U, Schaffeler E, et al. Association of the P-glycoprotein transporter MDR1(C3435T) polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;13(7):1847–54. doi: 10.1097/01.asn.0000019412.87412.bc. [DOI] [PubMed] [Google Scholar]
- 50.Jamroziak K, Mlynarski W, Balcerczak E, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72(5):314–21. doi: 10.1111/j.1600-0609.2004.00228.x. [DOI] [PubMed] [Google Scholar]
- 51.Niell BL, Long JC, Rennert G, Gruber SB. Genetic anthropology of the colorectal cancer-susceptibility allele APC I1307K: evidence of genetic drift within the Ashkenazim. Am J Hum Genet. 2003;73(6):1250–60. doi: 10.1086/379926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronn D, Jansen V, Ostrer H. Carrier screening for cystic fibrosis, Gaucher disease, and Tay-Sachs disease in the Ashkenazi Jewish population: the first 1000 cases at New York University Medical Center, New York, NY. Arch Intern Med. 1998;158(7):777–81. doi: 10.1001/archinte.158.7.777. [DOI] [PubMed] [Google Scholar]
- 53.Pier GB, Grout M, Zaidi T, et al. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;393(6680):79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 54.Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ. Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science. 1994;266(5182):107–9. doi: 10.1126/science.7524148. [DOI] [PubMed] [Google Scholar]
- 55.Ameyaw MM, Regateiro F, Li T, et al. MDR1 pharmacogenetics: frequency of the C3435T mutation in exon 26 is significantly influenced by ethnicity. Pharmacogenetics. 2001;11(3):217–21. doi: 10.1097/00008571-200104000-00005. [DOI] [PubMed] [Google Scholar]
- 56.Li Y, Wang Y, Sun J, Li Y, Yang L. Distribution of the functional MDR1 C3435T polymorphism in the Han population of China. Swiss Med Wkly. 2006;136(23–24):377–82. doi: 10.4414/smw.2006.11113. [DOI] [PubMed] [Google Scholar]


