Abstract
The C-terminal binding protein (CtBP) is an evolutionarily conserved transcriptional corepressor found in multicellular eukaryotes. Multiple forms of the protein are typically found in animal cells, produced from separate genes and by alternative splicing. CtBP isoforms have also been implicated in cytoplasmic functions, including Golgi fission and vesicular trafficking. All forms of CtBP contain a conserved core domain that is homologous to α-hydroxyacid dehydrogenases, and a subset of isoforms (CtBPL) contain extensions at the C-terminus. Despite distinct developmental profiles and knockout phenotypes in the mouse, the properties of different isoforms of the protein are found to be similar in many transcriptional assays. We have investigated the expression and conservation of distinct isoforms of the CtBP protein in insects, and find that the expression of multiple, developmentally regulated isoforms is widely conserved. In a variety of Drosophila species, the relative abundance of CtBPL to CtBPS drops sharply after embryogenesis, revealing a conserved developmental shift. Despite the overall lower levels of this isoform, bioinformatic analysis reveals that exons encoding the C-terminal extension in CtBPL are conserved from Diptera to Coleoptera, suggesting that the CtBPL isoform contributes an important, evolutionarily conserved function.
Keywords: C-terminal binding protein, CtBP, transcription, corepressor
Introduction
The C-terminal binding protein (CtBP) is an evolutionarily conserved factor that has been implicated in a variety of cellular processes, including transcriptional repression, Golgi function, and vertebrate retinal synapse activity (Chinnadurai, 2005). Originally identified by its ability to interact with the C-terminus of the adenovirus E1A protein, CtBP has been shown to directly bind to a variety of transcription factors in vertebrate cells and in Drosophila, and recruit chromatin-modifying factors including histone deacetylases and histone demethylases (reviewed in Turner and Crossley, 2001; Chinnadurai, 2003). CtBP proteins share a high degree of similarity within a central domain comprised of an NAD-binding domain and a substrate-binding fold. The proteins form dimers, and demonstrate extensive structural similarity to NAD-dependent dehydrogenases (Kumar et al., 2002; Nardini et al., 2003). CtBP proteins also possess C-terminal sequences of variable lengths that are likely to be unstructured (Nardini et al., 2006).
CtBP proteins exhibit a weak NAD-dependent catalytic activity in vitro, however the physiological relevance of this activity is unknown (Kumar et al., 2002; Balasubramanian et al., 2003; Barnes et al., 2003). NAD binding has also been suggested to regulate CtBP allostery, permitting the interaction of the protein with binding partners. In vitro, association of CtBP with E1A proteins is stimulated by NAD and NADH, suggesting a possible molecular switch that might regulate CtBP activity (Kumar et al., 1987; Barnes et al., 2003). Differential affinity of the protein for NADH relative to NAD has been suggested to endow CtBP with the potential to respond to differing intracellular levels of these cofactors, possibly linking gene regulation to cellular redox states (Zhang et al., 2002). A possible lysophosphatidic acid acyl transferase activity relevant to membrane trafficking has also been ascribed to one form of CtBP (CtBP3/CtBP1-S/BARS), however this result has been disputed (Weigert et al., 1999; Gallop et al., 2005).
Distinct CtBP isoforms are expressed as a result of alternative splicing, alternative promoter usage, and different genes. In vertebrates, the ctbp1 and ctbp2 genes are expressed in overlapping patterns during development and exhibit distinct functions. ctbp1 knockout mice are viable, but are smaller and show increased postnatal mortality, while the ctbp2 mutation is embryonic lethal (Hildebrand and Soriano, 2002; Van Hateren et al., 2006). The rat CtBP1 isoform termed CtBP1-S/CtBP3/BARS lacks a short region at the N terminus; this protein has been implicated in membrane fission events that are required for Golgi trafficking and Golgi fragmentation during mitosis (reviewed in Corda et al., 2006). In vertebrates, the RIBEYE splice form of CtBP2 produces a protein containing CtBP residues fused to a unique N terminus; this protein is localized to synaptic vesicles of the retina (Schmitz et al., 2000).
Posttranslational modifications and association with other binding proteins have been shown to regulate the stability, activity and localization of CtBP proteins in vertebrates. Some of these modifications target the central conserved region of the protein; the Pak1 kinase phosphorylates CtBP1 at Ser158, stimulating nuclear to cytoplasmic translocation and downregulating CtBP repression activity (Barnes et al., 2003). Other modifications are targeted to the C-terminal nonconserved portion of the protein; phosphorylation of CtBP1 ser422 by the HIPK2 kinase promotes degradation of the protein, whereas SUMOylation of the C-terminus is required for nuclear localization of CtBP1 ()(Kagey et al., 2003; Lin et al., 2003; Zhang et al., 2005). In addition to being covalently modified, the C-terminus can also serve as the binding target for a PDZ-domain containing protein, neuronal nitric oxide synthase, that drives cytoplasmic localization of the CtBP1 (Riefler and Firestein, 2001).
In contrast to vertebrates, distinct Drosophila CtBP proteins are produced from a single gene. Two major isoforms, termed CtBPL and CtBPS, differ by the presence or absence of a ~90 amino acid extension at the C-terminus, which, although of similar size and amino acid composition, is not homologous to C-terminal extensions found in vertebrate CtBP proteins (Poortinga et al., 1998; Nibu et al., 1998a). In light of the fact that the unstructured C-terminus can play a regulatory role in vertebrates, it seems possible that Drosophila CtBPL may be subject to similar covalent modifications as those found in vertebrates, but currently there is little understanding of the biological importance of the different isoforms. In vitro, both CtBPL and CtBPS are able to bind to short-range transcriptional repressors such as Knirps and Krüppel, and in transcriptional assays, both isoforms exhibit similar activities (Sutrias-Grau and Arnosti, 2004; Fang et al., 2006). Therefore, we have utilized biochemical and phylogenetic analysis to study expression of the protein in disparate orders to gain more insight into the significance of distinct isoforms of this widely conserved protein. Biochemical and phylogenetic evidence indicates that the alternatively spliced CtBPL isoform represents a conserved, developmentally regulated form of the protein, suggesting a specific functional role for this protein.
Materials and Methods
Insect stocks and lysate preparation
The fly stocks used in this study were: D. melanogaster yw67 (Arnosti lab), D. sechellia, D. mojavensis (Tucson Drosophila Stock Center), D. virilis (Dr. Scott Pitnick). Tribolium castaneum was a gift from Dr. Susan Brown, Anopheles gambiae from Dr. Ned Walker and Apis mellifera from Dr. Zachary Huang. All flies were maintained on standard cornmeal/molasses food and embryos collected at 25°C on apple juice/agar. For developmental expression analysis, staged embryos were collected, dechorionated and resuspended in lysis buffer (150mM NaCl, 50mM Hepes, pH 7.9, 10% glycerol, 0.1mM EDTA with Complete mini-EDTA free protease inhibitor cocktail tablet, Roche) and sonicated using a Branson-250 sonifier. Larvae, pupae and adults were homogenized in lysis buffer with a steel pestle and then sonicated under the same conditions. Lysates were cleared by centrifugation and total protein concentration of the supernatant was measured by a Bradford assay with BSA as the standard. The supernatant was mixed with Laemmli sample buffer for SDS-PAGE analysis. For identification of CtBP isoforms in bee, beetle, and mosquito and flies, whole adult animals were homogenized in Laemmli sample buffer for SDS-PAGE analysis.
Western Blot Analysis
Immunoblotting was performed using 10% SDS-PAGE gels in a tank transfer system (Bio-Rad Mini Trans-Blot® Cell) and proteins were transferred to Immuno-BlotTM PVDF membrane (Bio-Rad). Antibody incubation was performed in TBST (20mM Tris-HCl, pH 7.5, 120mM NaCl, 0.1% Tween-20) supplemented with 5% non fat dry milk as a blocking agent. Rabbit polyclonal antibodies used to detect CtBP (1:10,000) and monoclonal mouse antibody for tubulin (1:6000, Iowa Hybridoma Bank) were visualized using HRP-conjugated secondary antibodies (Pierce) and SuperSignal® West Pico chemiluminiscent substrate (Pierce). Western blots shown are representative of at least three biological replicates for each experiment.
Antibodies and recombinant CtBP proteins
Polyclonal anti-CtBP antibodies were generated as described in Struffi et al. (2005). For the production of recombinant proteins, the cDNAs for CtBPL and CtBPs bearing two Flag epitope tags at the C-terminal end was cloned into the pET15b expression vector and used to transform E.coli BL-21 cells. Expression of bacterial proteins was induced by treating log-phase cultures with 0.4mM IPTG. The cells were then sonicated in lysis buffer, centrifuged and supernatant was dissolved in Laemmli sample buffer for western analysis.
RT-PCR measurements of splice form abundances in embryos and adults
Total RNA of D. melanogaster embryos (stage 0–12) and adults was isolated by tissue homogenization in Trizol reagent (Sigma) according to the manufacturer's protocol. RNA was treated with Rnase-free DNase (RQ1, Promega) to remove contaminating genomic DNA. RT-PCR was performed using AccessQuickTM RT-PCR System from Promega. Transcripts for CtBPL were amplified using primer pairs DA1147 – 5’ CCCCACAGTACAACCAACCT 3’ and DA1148 – 5’ TCCGTTTTTATGCTCGATGA 3’, CtBPs using primer pairs DA 1146 – 5’ CTCAACGAGCACAACCATCATTTAATC 3’ and DA 1150 – 5’ CTCTACTTTTCTTGATTTGATATCATTTGTAG 3’ and total CtBP was amplified using primer pairs DA 1146 - 5’ CTCAACGAGCACAACCATCATTTAATC 3’ and DA 1151 –5’GCACGTCTGGAATATTGCCGAC 3’. All primer pairs spanned an intron such that amplification of contaminating genomic DNA could be distinguished from the RT-PCR amplified products. The RT step was performed at 45°C for 45 mins followed by 30 cycles of PCR in a 25 μl reaction mix for 94°C for 1min, 56°C for 1 min and 72°C for 1 min. PCR products were visualized by agarose gel electrophoresis and quantitated using BioRad Quantity One software Version 4.4.1. The data shown in Figure 1C is a representative result of RT-PCR analysis of biological triplicates that were each analyzed at least two times.
Fig. 1. Developmental expression profile of CtBP isoforms in Drosophila melanogaster.
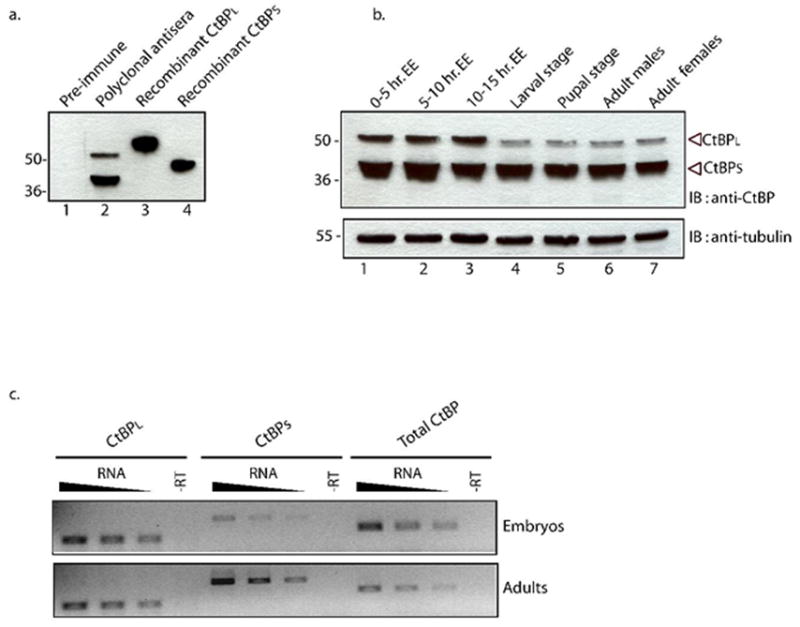
A.Specificity of α-CtBP antibody tested in Western blot with Drosophila melanogaster embryonic extract (lanes 1,2) or bacterial extracts containing recombinant CtBPL (lane 3) or CtBPS (lane 4). Preimmune serum did not cross react with any proteins in embryo extract, while α-CtBP recognized two isoforms of approximately 42 and 50 kDa in embryonic extracts. Recombinant proteins migrate slower than endogenous counterparts due the presence of an N-terminal hexahistidine tag and a C-terminal Flag tag. Markers (kDa) are indicated to the left. B. Expression of CtBP isoforms in embryos, larvae, pupae, and adults. 50 μg of total soluble protein was loaded on 10% SDS-PAGE and analyzed by immunoblotting with anti-CtBP. Relative CtBPL and CtBPS levels were unchanged during embryogenesis. A marked reduction in the relative level of CtBPL was observed from the larval through adult stages. CtBPs levels remained relatively unchanged throughout the developmental time course. The bottom panel shows β-tubulin as a loading control. C. Steady-state levels of CtBP mRNAs measured by RT-PCR analysis. Total mRNA from embryos and adults was reverse transcribed and PCR amplified using primers specific to CtBPL exons, CtBPS regions, or a region common to both isoforms as indicated. Reverse transcription reactions were primed with 60, 30, or 15 ng of total RNA, as indicated by triangular symbol. The –RT control reactions were primed with 60 ng of RNA. Based on quantitation of biological replicates, the ratio of CtBPL to CtBPS products was measured to be approximately 1:1 in adults compared to 4:1 in embryos.
Sequence alignments
To determine the conservation of CtBP exons in diverse insect genomes we searched the Flybase database (Release 4.2) using FLYBASE BLAST for the assembled genomes of Drosophila melanogaster, D. sechellia, D. persimilis, D. mojavensis, D. virilis, D. grimshawi, Anopheles gambiae, Aedes aegypti, Apis mellifera and Tribolium castaneum. Matches to conserved exons 1–4 of CtBP were obtained for D. sechellia ( AAKO01000254.1) D. persimilis (AAIZ01000471), D. mojavensis (contig_8705), D. virilis (contig_15233), D. grimshawi (contig_21987), A.gambiae (AAAB01008805), Aedes aegypti (supercontig_1.155), A.mellifera (AADG05006060) and T. castaneum (CM000284.1). These automated alignments generally did not identify exons 6 and 7, however, therefore sequences 3’ to the conserved exons were searched in all three reading frames for conserved coding information and aligned using Clustal W. Predicted gene sequences for A. mellifera (XM_392682) and T. castaneum (XP_972241) were included in these alignments.
Results
Expression of CtBP isoforms in Drosophila
Four major CtBP transcripts are detected ubiquitously during development and are predicted to produce proteins of 383, 386, 476 and 479 amino acids (Poortinga et al., 1998; Nibu et al., 1998b; Sutrias-Grau and Arnosti, 2004). To analyze endogenous CtBP proteins in Drosophila, we generated polyclonal rabbit antibodies against CtBPL protein (aa1-479) expressed in E.coli. The antibodies are specific and detect proteins of the expected sizes in embryonic extracts, approximately 42 (CtBPS) and 50 KDa (CtBPL ) (Fig. 1A). Immunostaining revealed that CtBP proteins are ubiquitously present in the nuclei of pre- and post-blastoderm embryos and imaginal discs from third instar larvae (data not shown).
To analyze the developmental expression profile of CtBP isoforms, we analyzed soluble extracts from different developmental stages of the fly (Fig. 1B). Both CtBPL and CtBPS isoforms are detected throughout the first 15 hours of embryogenesis, with relatively higher level of CtBPS than CtBPL (antibody recognition of CtBPL is expected to be equal or better than that of CtBPS because the two proteins are virtually identical in the central domain, and the antibody was raised against CtBPL). The relative levels of CtBPL to CtBPS drop further after embryogenesis, showing weak expression of CtBPL in the larva, pupa, and adult (Fig. 1B). The lower abundance of CtBPL in postembryonic stages is not simply due to sequestration of the protein in an insoluble form, because similar low levels of CtBPL were observed in whole animal extracts prepared in boiling SDS (discussed below).
We measured the relative levels of specific CtBP mRNA splice forms in embryonic and adult stages to determine if this developmental switch reflects a change in alternative mRNA isoform abundance. Primer pairs specific to the CtBPS, CtBPL, and to a region of the gene common to both isoforms were used in RT-PCR reactions. The absolute amounts of CtBPS and CtBPL RT-PCR products are not directly comparable because different primer sets were used, however the relative ratios in different stages of development are informative. The ratio of CtBPS to CtBPL transcripts undergo a marked shift between these two stages, with relative levels of CtBPS increasing approximately 4 fold with respect to CtBPL (Fig. 1C). This change suggests that the developmental protein profile may be largely determined by changes in the abundance of distinct splice forms of the mRNA. Additional post-transcriptional effects may also contribute to the decreased CtBPL protein levels observed.
Identification of conserved CtBPL-specific coding information
We examined genomic sequences of 10 different insects representing >300 million years of evolutionary divergence – the fruit flies D. melanogaster, D. sechellia, D. persimilis, D.mojavensis, D. grimshawi and D. virilis, the mosquitoes Anopheles gambiae and Aedes aegypti (Diptera), the honey bee Apis mellifera (Hymenoptera) and the red flour beetle Tribolium castaneum (Coleoptera) - to determine if these organisms might also express diverse isoforms of CtBP. Analysis of putative open reading frames 3’ of core conserved CtBP sequences identified regions homologous to D. melanogaster exons 6 and 7, which encode the C-terminal extension of CtBPL (Fig. 2A). In Drosophila species, the sequences of exon 6 appear to be separated from an upstream exon by a ~3 kbp intron, while the intron is of smaller size in mosquito and beetle. In the honey bee, this intron appears to have been entirely eliminated. The overall similarity among putative C-terminal coding regions is clearly lower than that observed for the core CtBP sequences, suggesting a lowered level of constraint. However, the similarities include several distinctive motifs involving less abundant amino acids, not simply tracts of repeating residues that would show similarities by chance. Splice signals following the terminal codons for exon 5 (YPEG), are conserved in all Drosophila, as well as lower Diptera and Tribolium, suggesting that the downstream coding information is likely to be incorporated into mRNAs (Fig. 2B). Splice acceptor sites are present immediately 5’ of the conserved LNGGYYT coding region of exon 6 in Drosophila species. A conserved splice acceptor sequence is not found directly 5’ of I/VNGGY coding sequences present in Tribolium and Anopheles, raising the possibility that acceptor sites in alternative locations may be used (Fig 2B). In the bee, the information for the tail extension seems to be fused to the core sequences, supporting the notion that these are indeed coding sequences. Similar to the case with vertebrate CtBP proteins, the predicted C-terminal extensions of these CtBP isoforms are probably unstructured in solution. The sequences are rich in disorder promoting amino acids (ala, gly, pro, ser) and are predicted to not assume a globular structure by the GlobPlot program (not shown) (Linding et al., 2003).
Fig. 2. Conservation of coding information for CtBPL-specific C-terminus.
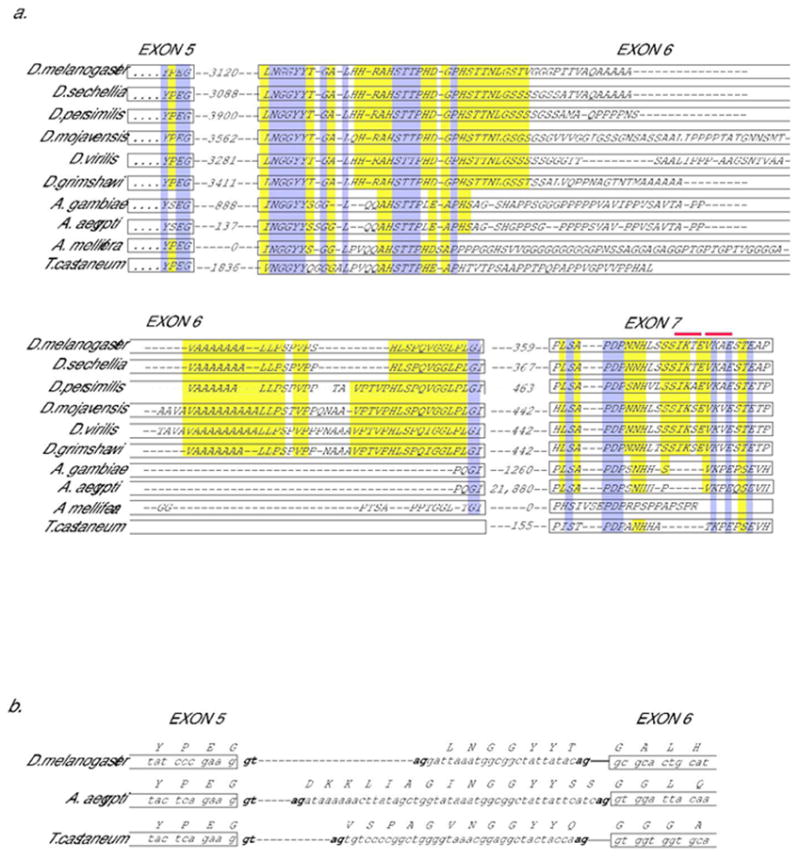
A. Peptide coding information present in dipterans, bee, and beetle genomic sequences homologous to alternatively spliced exon 6 and 7 in Drosophila melanogaster encoding CtBP “tail” region. Conceptual translations of genomic sequences are shown below sequence of CtBPL, in which YPEG represents the end of the exon 5 coding sequence for the CtBPL isoform. Predicted intron size in nucleotides is indicated between exons. The introns in Apis mellifera have apparently been eliminated. Dark gray (purple) shading indicates widely conserved sequences; light gray (yellow) shading partially conserved sequences. Possible sumoylation sites (I/V K X E) are indicated by gray (red) bars above exon 7 residues. An alternative splice acceptor site 5’ of the junction shown for exon 7 would produce an mRNA encoding an additional VSSQS motif at the beginning of exon 7; this sequence is not conserved outside of Drosophila, unlike the case shown in 2B. B. The cDNA sequences reported for D. melanogaster CtBPL contain alternative splice acceptor sites for the 5’ end of exon 6; a sequence isolated from adult head uses a downstream acceptor site (NP_001014617), while a different sequence isolated from embryo uses a more upstream acceptor ((Sutrias-Grau and Arnosti, 2004)) incorporating the residues LNGGYYT. This portion of the protein is evolutionarily conserved and contains appropriate splice acceptor sequences both 5’ and 3’ of region, thus alternative splicing may be a conserved feature here as well.
Developmental expression of alternative isoforms in D. mojavensis and D. virilis
The presence of the regions correlating to D. melanogaster exons 6 and 7 does not in itself reveal whether distinct CtBP isoforms are produced, therefore we measured CtBP protein levels in embryos, larvae, pupae and adults from D. mojavensis and D. virilis, which are estimated to have shared the last common ancestor with D. melanogaster about 40–60 million years ago. Western blot analysis revealed that two major bands of sizes similar to CtBPS and CtBPL were present in these species (Fig. 3). The relative abundance of the CtBPL isoform decreases in larval and pupal stages, staying low in D. mojavensis in the adult, but increasing again in adult D. virilis. While differing in details, these changes suggest that developmental changes in relative abundances of CtBP isoforms are a conserved feature in Drosophilids.
Fig. 3. Conserved developmental regulation of CtBP protein expression in D. mojavensis and D. virilis.
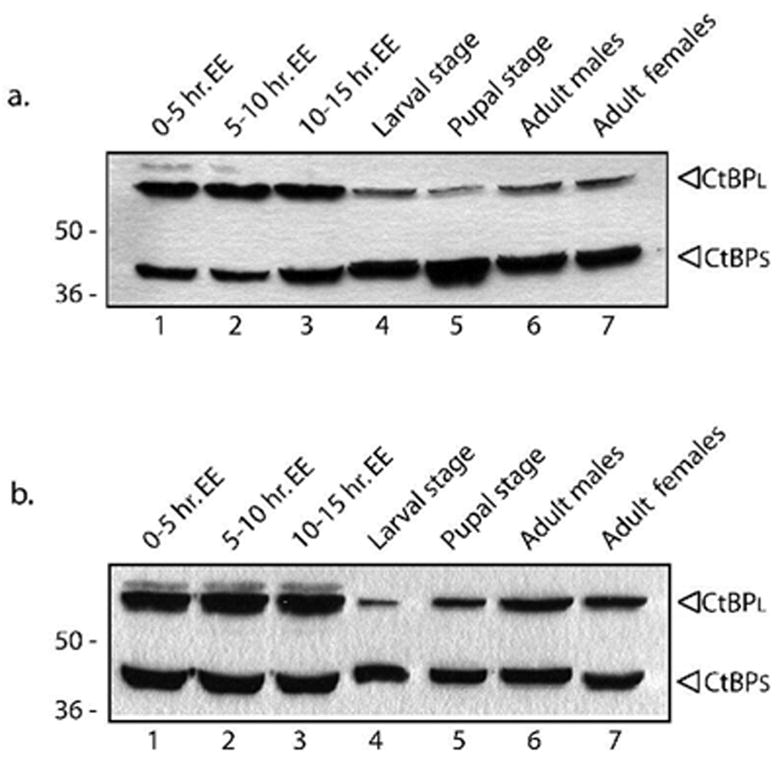
Expression of CtBP isoforms in embryos, larvae, pupae, and adults of D. mojavensis (A.) and D. virilis (B.). As in D. melanogaster, two predominant species were observed in both species, but the CtBPL isoform has a lower mobility (~60kDa vs. 50kDa in D. melanogaster). The relative levels of CtBPL to CtBPS in the embryo was greater in these species than in D. melanogaster, but just as in that species there is a pronounced decrease in relative levels of CtBPL in the larva and pupa. Adult levels of CtBPL remain low in D. mojavensis, but recover in D. virilis. 50 μg of total soluble protein was loaded on 10% SDS-PAGE and analyzed by immunoblotting with anti-CtBP.
Expression of CtBP isoforms in diverse orders
To determine whether expression of CtBPS and CtBPL-like isoforms is generally conserved in insects, we measured expression of CtBP proteins in organisms whose sequenced genomes had been examined for CtBPL-specific coding information (Fig. 2A). Crude extracts from adults were analyzed by Western blotting, including three Drosophila species of increasing phylogenetic distance from D. melanogaster, Anopheles gambiae (lower Dipteran), Apis mellifera (Hymenoptera), and Tribolium castaneum (Coleoptera) (Fig. 4). Relative to D. melanogaster, the closely related D. sechellia, (diverged ~3 Mya) had CtBPS and CtBPL isoforms of the same size. The extracts from the more distantly related species D. mojavensis and D. virilis (diverged 40–60 ~Mya) contained proteins of similar size to CtBPS (~42 kDa) and an additional, lower mobility form (60 kDa) that migrated slower than D. melanogaster CtBPL. Two proteins were also evident in the mosquito, both of somewhat faster mobility than the Drosophila counterparts. Three cross-reacting species were found in the honeybee, all of similar abundance, including one protein of ~25 kDa that migrates considerably faster than CtBPS, similar to a minor species noted in D. mojavensis extracts. Only one major isoform of ~50 kDa was detected in extracts from Tribolium, similar in mobility to Drosophila CtBPL, although upon overexposure, weak bands of faster mobility could be seen. In this figure, the relative levels of CtBPL and CtBPS in Drosophila appear to be similar, but this is only because the gel was exposed for a long time to bring out the weaker A. mellifera bands. A Western blot of the Drosophila extracts in which the exposure was shorter reveals that the ratio of CtBPL to CtBPS in adults was low in all Drosophila species except D. virilis (Fig. 4, lanes 8–11), which is consistent with our developmental profiles for D. virilis and D. mojavensis.
Fig. 4. Adult expression of CtBP proteins in four Drosophila species, Anopheles gambiae, Apis mellifera, and Tribolium casteneum.
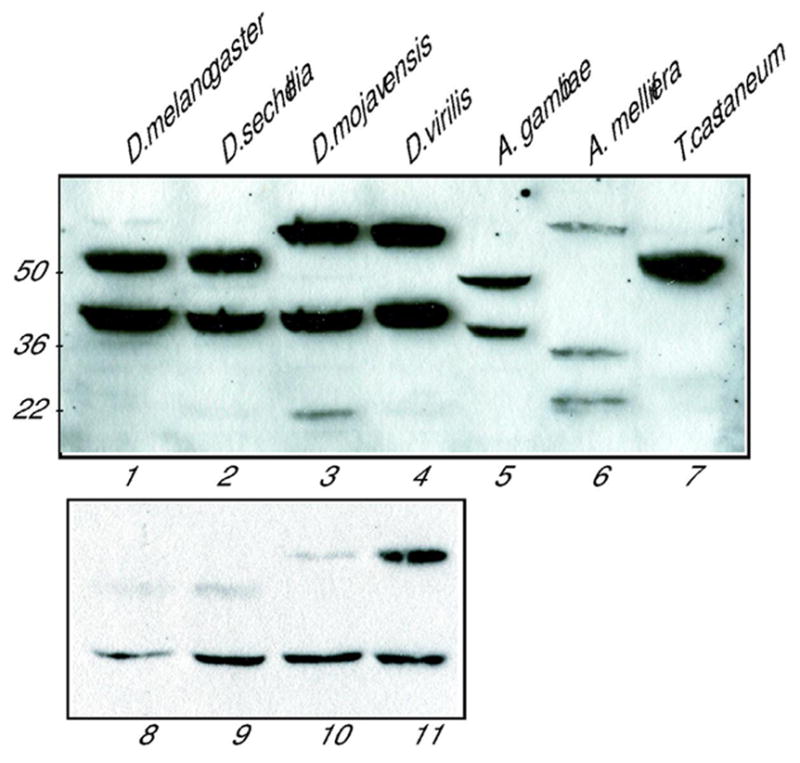
Soluble extracts from adults were analyzed by Western blotting using anti-CtBP. Cross-reacting species similar in size to CtBPS were noted in all Dipterans. Slower mobility proteins consistent with CtBPL-like species were present in all extracts; multiple bands were detected in extracts from all species except Tribolium. The relative abundance of CtBPL and CtBPS is masked by the long exposure of the gel; lower panel shows a separate Western blot (lanes 8-11) that was exposed for a shorter time to demonstrate the lower abundance of CtBPL to CtBPS in D. melanogaster, D. sechellia, and D. mojavensis adults.
Discussion
In the mouse, the CtBP1 and CtBP2 genes have been found to provide overlapping but functionally distinct activities in development (Hildebrand and Soriano, 2002). These different activities might be transcriptionally based, a situation in which homologous genes encode functionally interchangeable products, but the distinct timing and levels of transcriptional activity of the promoters are unique, as has been described for the Drosophila prd, gsb, and gsbn genes (Li & Noll 1994). However, this model cannot be applied to cover all vertebrate CtBP proteins, because the RIBEYE spliceform of CtBP2 and CtBP1-S/BARS splice variant of CtBP1 encode distinct polypeptides, and appear to have acquired unique roles in retinal function and membrane trafficking, respectively (Corda et al., 2006). With respect to the transcriptional regulatory forms of CtBP1 and 2, biochemical studies have identified molecular modifications that may distinguish the two isoforms functionally. CtBP1 is phosphorylated at serine158, a modification that induces nuclear to cytoplasmic translocation (Barnes et al., 2003). CtBP2 has a unique N-terminus that is acetylated, which facilitates nuclear retention of the protein (Zhao et al., 2006). Whether these differences play a role directly in transcription is unclear; both proteins may function similarly when recruited to promoters.
In Drosophila, less is known about distinctions among isoforms. Previous work from our and other laboratories has indicated that multiple CtBP isoforms are expressed in Drosophila, but no functional distinctions have been drawn between CtBPS and CtBPL isoforms until now. Our study provides evidence that the presence of these isoforms is not simply “noise”, for example, aberrant splicing that is tolerated by the system. The evolutionary conservation of multiple isoforms and developmental regulation strongly points to functional differentiation between these proteins. It is striking that all the organisms surveyed express proteins whose size corresponds to the D. melanogaster CtBPL isoform. In addition, all contain conserved coding sequences in their genomes for the unstructured C-terminal extension of the protein, which in the case of mammals is the subject of sumoylation, phosphorylation, and binding of regulatory proteins in mammals. Putative sumoylation signals are conserved in Dipteran sequences (Fig. 2A), suggesting that insect CtBP proteins may similarly be modified by SUMO. All vertebrate CtBP proteins possess some form of C-terminal extension, however the presence of CtBPS isoforms in insects may indicate that potential regulation by modification of the C-terminus may not be required, at least in some stages or roles. Additional biochemical and genetic studies will be required to identify possible functional distinctions between these isoforms.
Acknowledgments
We thank Scott Pitnick, Susan Brown, Zachary Huang, Ned Walker and Tucson Drosophila Stock Center for flies and insects used in this study and Dr. Casey Bergman for advice on alignments. We thank Paolo Struffi for generating the anti-CtBP antibody, as described in (Struffi et al., 2005), and Montserrat Sutrias-Grau for pointing out the potential SUMOylation sequences in CtBPL. This project was supported by NIH GM56976 to D.N.A.
References
- Balasubramanian P, Zhao LJ, Chinnadurai G. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 2003;537:157–160. doi: 10.1016/s0014-5793(03)00119-4. [DOI] [PubMed] [Google Scholar]
- Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 2003;10:622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP family proteins: more than transcriptional corepressors. BioEssays. 2003;1:9–12. doi: 10.1002/bies.10212. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP Family Proteins: Unique Transcriptional Regulators in the Nucleus with Diverse Cytosolic Functions. In: Chinnadurai G, editor. CtBP Family Proteins. Eurekah.com, Landes Bioscience; Georgetown, Texas: 2005. pp. 1–17. [Google Scholar]
- Corda D, Colanzi A, Luini A. The multiple activities of CtBP/BARS proteins: the Golgi view. Trends Cell Biol. 2006;16:167–173. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, Cadigan KM. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 2006;25:2735–2745. doi: 10.1038/sj.emboj.7601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, Butler PJ, McMahon HT. Endophilin and CtBP/BARS are not acyl transferases in endocytosis or Golgi fission. Nature. 2005;438:675–678. doi: 10.1038/nature04136. [DOI] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;15:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;1:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;4:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;6:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- Lin X, Sun B, Liang M, Liang YY, Gast A, Hildebrand J, Brunicardi FC, Melchior F, Feng XH. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol Cell. 2003;5:1389–1396. doi: 10.1016/s1097-2765(03)00175-8. [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Spanò S, Cericola C, Pesce A, Massaro A, Millo E, Luini A, Corda D, Bolognesi M. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22:3122 – 3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M, Svergun D, Konarev PV, Spano S, Fasano M, Bracco C, Pesce A, Donadini A, Cericola C, Secundo F, Luini A, Corda D, Bolognesi M. The C-terminal domain of the transcriptional corepressor CtBP is intrinsically unstructured. Protein Sci. 2006;15:1042–1050. doi: 10.1110/ps.062115406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 1998a;23:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998b;5360:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;7:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler GM, Firestein BL. Binding of neuronal nitric-oxide synthase (nNOS) to carboxyl-terminal- binding protein (CtBP) changes the localization of CtBP from the nucleus to the cytosol: a novel function for targeting by the PDZ domain of nNOS. J Biol Chem. 2001;51:48262–48268. doi: 10.1074/jbc.M106503200. [DOI] [PubMed] [Google Scholar]
- Schmitz F, Konigstorfer A, Sudhof TC. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron. 2000;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Struffi P, Arnosti DN. Functional interaction between the Drosophila knirps short range transcriptional repressor and RPD3 histone deacetylase. J Biol Chem. 2005;280:40757–40765. doi: 10.1074/jbc.M506819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrias-Grau M, Arnosti DN. CtBP contributes quantitatively to Knirps repression activity in an NAD binding-dependent manner. Mol Cell Biol. 2004;24:5953–5966. doi: 10.1128/MCB.24.13.5953-5966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Crossley M. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays. 2001;8:683–690. doi: 10.1002/bies.1097. [DOI] [PubMed] [Google Scholar]
- Van Hateren N, Shenton T, Borycki AG. Expression of avian C-terminal binding proteins (Ctbp1 and Ctbp2) during embryonic development. Dev Dyn. 2006;235:490–495. doi: 10.1002/dvdy.20612. [DOI] [PubMed] [Google Scholar]
- Weigert R, Silletta MG, Spano S, Turacchio G, Cericola C, Colanzi A, Senatore S, Mancini R, Polishchuk EV, Salmona M, Facchiano F, Burger KN, Mironov A, Luini A, Corda D. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;6760:429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci U S A. 2005;102:2802–2807. doi: 10.1073/pnas.0409373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;5561:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. J Biol Chem. 2006;281:4183–4189. doi: 10.1074/jbc.M509051200. [DOI] [PubMed] [Google Scholar]


