Abstract
The T-box transcription factor Tbx5 can interact with Nkx2.5 and Gata4 transcription factors to synergistically regulate heart-specific genes in the nucleus. While a nuclear role for Tbx5 is clearly defined, we have previously shown that Tbx5 shuttles from nuclear to cytoplasmic sites, forming a complex with the PDZ-LIM protein LMP4 on the actin cytoskeleton. In this study, using a developmental series of chicken hearts, we provide the first evidence for differential Tbx5 protein expression and sub-cellular localization during cardiogenesis. At the tissue level, we show temporally and spatially restricted Tbx5 co-expression with LMP4. In cells co-expressing LMP4 and Tbx5 we demonstrate dynamic Tbx5 re-localization from exclusively nuclear to nuclear and cytoplasmic expression in the atrio-ventricular cushion. Furthermore, in coronary vessel development we show exclusive cytoplasmic localization of Tbx5, indicating a function for Tbx5 in the cytoplasm. In addition, we discover unknown regulation of Tbx5 and LMP4 expression in epicardial tissue, suggesting a specific role for Tbx5 in epicardial formation. These studies provide in vivo significance of the LMP4/Tbx5 protein interaction, suggesting both nuclear and cytoplasmic roles for Tbx5. The shuttling between nuclear and cytoplasmic sites reveals a novel mechanism for Tbx transcription factor regulation in chicken heart development allowing new insights for a better understanding of the molecular basis of hand/heart birth defects associated with TBX5 mutations.
Keywords: Tbx5, LMP4, myocardium, epicardium, coronary artery, cardiac myocyte, cytoskeleton, heart development
Introduction
Mutations in the transcription factor TBX5 cause the congenital disease Holt-Oram syndrome, which is characterized by truncations of the upper limbs and heart malformations (Basson et al., 1999; Li et al., 1997). The heart malformations primarily include septal and conduction defects although valvular abnormalities have also been reported (Basson et al., 1999; Mori and Bruneau, 2004). While Tbx5 is critical for forelimb and heart development in all vertebrates, little is known about its regulation (Agarwal et al., 2003; Bruneau et al., 2001). In a protein-protein interaction screen we identified a novel Tbx5 binding protein, LMP4, a member of the PDZ-LIM protein family. When co-expressed with Tbx5 in the cell, the transcription factor relocates from the nucleus to cytoplasmic sites and binds LMP4 along the actin cytoskeleton (Krause et al., 2004). This sequestration of the transcription factor at actin filaments has a negative regulatory effect on Tbx5 target genes (Camarata et al., 2006).
Initial insights into Tbx5 cardiac function during development have come from its mRNA expression profile. In chick, zebrafish and mouse Tbx5 expression is initiated very early in heart development and revealed expression throughout the cardiac crescent and the early heart tube (Begemann and Ingham, 2000; Bruneau et al., 1999; Liberatore et al., 2000). As the heart tube develops, Tbx5 mRNA becomes restricted to the more posterior regions that will later form the sinus venosus, atria, and left ventricle of the four-chambered heart (Bruneau et al., 1999; Liberatore et al., 2000). Whole mount mRNA in situ hybridizations in the chicken revealed ubiquitous and dynamic LMP4 expression in every heart chamber, as well as the outflow tract (Krause et al., 2004). These studies have suggested domains of Tbx5 and LMP4 co-expression as well as domains where LMP4 is expressed in the absence of Tbx5.
Both knock-out and over-expression studies in the mouse and chicken have provided insight into the function of Tbx5 in heart development. Despite expression early in development, knockout Tbx5del/del mice have demonstrated that Tbx5 is not essential for heart tube formation or normal initiation of endocardial, myocardial and epicardial layers (Bruneau et al., 2001). However, Tbx5 is important for proper heart chamber patterning. Heterozygous mutant Tbx5del/+ mice exhibit enlarged hearts with abnormal chamber size, including a dilated right atrium and ventricle as well as atrial septal defects (Bruneau et al., 2001). In contrast, in both chicken and mouse, Tbx5 over-expression in the myocardium results in a reduction of overall heart size and reduced trabeculation of the ventricles (Hatcher et al., 2001; Liberatore et al., 2000). Mis-expression of Tbx5 in the left or right ventricle of the mouse and chicken revealed that the Tbx5 expression gradient between left and right ventricles is critical for patterning of the ventricles and position of the inter-ventricular septum (Takeuchi et al., 2003).
In order to interpret the phenotypes caused by Tbx5 manipulation, an appreciation of the step wise development of the heart and its formation from multiple cell lineages is essential. From the cardiac crescent, a linear tube forms, consisting of myocardium, lined on the inside by endocardium. The posterior portions of the tube will form the sinus venosus, atria and left ventricle while the anterior portions will form the right ventricle and outflow tract. During further development the heart tube undergoes looping, followed by septation to form the mature, four chambered heart. As looping occurs, a third layer, the epicardium, is added, covering the outer surface of the myocardium. The epicardium originates predominantly from the proepicardial organ (PEO), an outgrowth of cells near the septum transversum (Männer, 1992; Viragh and Challice, 1981). Following attachment of the PEO to the myocardium along the inner curvature of the heart, the proepicardial cells migrate to cover the surface of the myocardium. Recent work has demonstrated that cells derived from the aortic sac also contribute to the epicardium; however, these cells cover only the distal portion of the outflow tract (Perez-Pomares et al., 2003).
The heart consists of myocardium, which forms cardiac muscle, sandwiched between epicardial and endocardial layers (Viragh et al., 1989). In addition to forming the endothelial lining of the myocardium, endocardial cells undergo epithelial-to-mesenchymal transition (EMT) to form the atrio-ventricular and outflow tract cushions, which in turn contribute to the formation of the heart valves and septum (De la Cruz et al., 1983; Markwald et al., 1996). Epicardial cells have multiple functions as well. In addition to covering the surface of the myocardium, epicardial cells undergo EMT, giving rise to mesenchymal cells which will migrate into the sub-epicardial space and myocardium. Epicardially derived cells (EPDCs) will form cardiac fibroblasts, the coronary arteries and even contribute to the atrio-ventricular cushion and intraventricular septum (Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Perez-Pomares et al., 2002; Vrancken Peeters et al., 1999).
As an inroad to understand Tbx5 function, many studies have relied largely on Tbx5 mRNA distribution and mis-expression of RNA levels during heart development. Our previous work in biochemical and cell culture experiments demonstrated a regulatory interaction between LMP4 and Tbx5 sub-cellular localization and function (Camarata et al., 2006). To extend the in vitro findings and to provide in vivo validation, this study examines LMP4 and Tbx5 protein co-expression and localization in different cell populations in the developing chicken heart including coronary vessels.
Materials and Methods
Tissue sectioning
Chicken embryos at specified developmental stages were dissected free of membranes and placed into cold PBS. Samples were embedded in Tissue Tek OCT (Sakura Finetek) and frozen in a dry ice/methanol bath. Sections were sliced at 10-12 μm on a Leica CM3050S cryostat (Leica Microsystems) and processed for immunohistochemistry as described.
Immunohistochemistry and imaging
Samples were fixed in 4% paraformaldehyde (PFA) followed by 1% Triton X-100 permeabilization, blocking with 20% normal goat serum (NGS; Invitrogen) and sequential incubation with primary and secondary antibodies in PBS with 0.2% bovine serum albumin (BSA) + 10% NGS. Affinity purified rabbit polyclonal anti-LMP4 (Camarata et al., 2006) and anti-Tbx5 (Khan et al., 2002) were used at a 1:500 dilution. In cases where tissue was double labeled for Tbx5 and LMP4, LMP4 antibodies were directly coupled to rhodamine using the EZ-Label protein labeling kit (Pierce Biotechnology). Anti-sarcomeric myosin (MF20), developed by D.A. Fischman; anti-vimentin (AMF-17B), developed by A.B. Fulton; and quail marker (QCPN), developed by B. Carlson and J. Carlson, were obtained from the Developmental Studies Hybridoma Bank (University of Iowa) and diluted at 1:500. Anti-caldesmon (CALD5; Sigma) was diluted 1:1000. Primary antibodies were detected using Alexa 488- and Alexa 546-conjugated secondary antibodies at 1:1000 dilutions (Molecular Probes). Filamentous actin was detected using Alexa Fluor 488 Phalloidin at 1:300 dilution (Molecular Probes). Nuclei were stained using DAPI (Roche) at 1:1000 dilution. To verify antibody specificity, the Tbx5 and LMP4 antibodies were blocked by pre-incubation with 2mg/ml of the respective peptides against which they were raised, followed by immunohistochemistry as described (Fig. S1). In addition, specificity of the secondary antibodies was confirmed by omitting the Tbx5 or LMP4 antibodies in the reaction (Fig. S1Y-AA). Confocal microscopy was performed using a Zeiss 510 META system (Zeiss, Inc) equipped with a Plan Apochromat 63x/1.4 Oil DIC lens. Overview images were created using the tiling feature, with a Plan Neofluar 25x/0.80 Imm DIC objective. Images were processed in Adobe Photoshop CS2.
Generation of Chimeras
Quail to chick chimeras were generated as follows. PEOs were harvested from HH16 or HH17 japanese quails (Cotournix, cotournix japonicum) and immediately transferred into windowed and inked HH16 or HH17 chick hosts. Grafted quail PEOs were positioned underneath the dorsal side of the heart tube next to the chicken proepicardium and 50μl of a 10x mixture of penicillin, streptomycin and fungizone (Gibco) was pipetted over the yolk sac. Windows were covered with parafilm and embryos allowed to mature until HH29 at 38°C. Following incubation, chimeric embryos were processed for tissue sectioning as described above.
Results
Tbx5 and LMP4 protein expression during chicken heart development
Tbx5 and LMP4 have revealed a dynamic mRNA expression profile throughout heart development (Bruneau et al., 1999; Krause et al., 2004; Liberatore et al., 2000). To appreciate the respective protein distribution at the tissue and cellular levels, we employed immunohistochemistry on a developmental series of chicken embryos. To ensure the specificity of the Tbx5 and LMP4 antibodies in the chicken, we have performed various controls including Western blot, immunohistochemistry on both cultured cells and tissue cryosections (Fig. S1 and Camarata et al., 2006; Khan et al., 2002). Figure 1 provides an overview of four developmental stages, showing adjacent cryosections processed with anti-sarcomeric myosin (MF20), which marks cardiac myocytes, DAPI to counter-stain cell nuclei and antibodies specific for Tbx5 or LMP4. The MF20/DAPI images provide an outline of the developing heart and its respective chambers as well as identify myocardial tissue (Fig. 1A, D, G, J). We found that for Tbx5, the protein expression is in agreement with published mRNA expression data. At Hamburger and Hamilton stage (HH) 20 (Hamburger and Hamilton, 1951), the chicken heart has completed looping, but septation has not occurred, thus the heart consists of only a single atrium and single ventricle. Tbx5 protein was expressed in both the atrium and ventricle, although we observed a gradient of Tbx5 expression with higher expression levels in the outer curvature of the ventricular myocardium. At this stage the proepicardial organ (PEO) included Tbx5 positive cells, although most proepicardial cells did not express Tbx5 (Fig. 1B). This expression will be addressed in greater detail in Figure 3. LMP4 was expressed predominantly in the epicardium covering the surface of the heart but also in the PEO (Fig. 1C). By HH25, Tbx5 expression became restricted to the structures derived from the posterior elements of the heart tube including the left ventricle and both atria (Fig. 1E). However, low Tbx5 expression was observed in the right ventricle. At this developmental stage, a few cells in the atrio-ventricular (AV) cushion began to express Tbx5, while we could not detect any expression in the outflow tract (Fig. 1E and data not shown). Similar to HH20, LMP4 was expressed predominantly in the superficial epicardium with little expression within the myocardium (Fig. 1F). In addition, LMP4 was expressed in both the AV cushion and outflow tract (Fig. 1F and data not shown). At HH29, Tbx5 continued to be expressed in the left ventricle and both atria (Fig. 1H). However, at this later developmental stage Tbx5 was distributed evenly throughout the AV cushion, adopting a new expression domain. Similar to earlier stages, LMP4 was expressed predominantly in the epicardium, although isolated cells within the myocardium expressed LMP4 (Fig. 1I). The latter will be addressed in greater detail in Figure 4. LMP4 remained expressed in the AV cushion at this stage. By HH33, Tbx5 revealed continued expression in the left ventricle and both atria (Fig. 1K). While some Tbx5 expression remained in the trabeculations, overall the expression in the right ventricle was significantly reduced at this developmental stage. Tbx5 was also down-regulated in most of the AV cushion, although some expression remained in the area of the presumptive mitral valve (Fig. 1J-K, double asterisk). Expression in this region will be addressed in more detail in Figure 2. As in earlier stages, LMP4 was expressed in the epicardium and AV cushion. Expression in the myocardium was hardly detectable, except for a small number of isolated LMP4 positive cells (Fig. 1L).
Figure 1.

Tbx5 and LMP4 global expression in the avian heart. Adjacent cryosections from each stage were processed for immunohistrochemistry using antisera specific for either Tbx5 or LMP4. An anti-sarcomeric myosin antibody (MF20, red) was used to outline myocardium, while DAPI (blue) outlines all nuclei. (A-D) Sagittal sections from HH20 chick embryos were stained for MF20 and DAPI (A), Tbx5 (B) and LMP4 (C). (D-F) Transverse sections from HH25 chick embryos were stained for MF20 and DAPI (D), Tbx5 (E) and LMP4 (F). Asterisk in D marks AV cushion. (G-I) Transverse sections from HH29 chick embryos were stained for MF20 and DAPI (G), Tbx5 (H) and LMP4 (I). Asterisk in G marks AV cushion. (J-L) Transverse sections from HH33 embryos were stained for MF20 and DAPI (J), Tbx5 (K) and LMP4 (L). Double asterisk in J marks presumptive mitral valve. Left atria (LA); right atria (RA); left ventricle (LV); right ventricle (RV); proepicardial organ (PEO). Scale bars: C=200μm; F,I=300μm; L=400μm.
Figure 3.
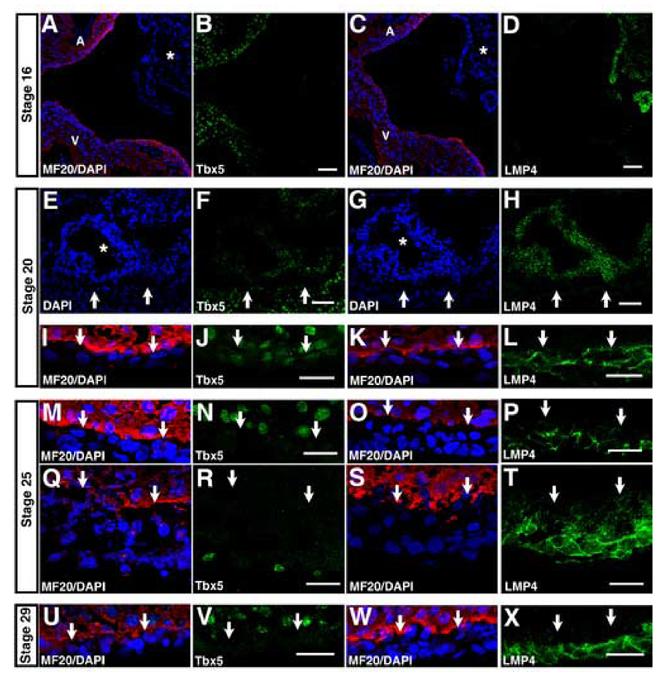
Expression and localization during epicardial formation. Cyrosections were processed for immunohistochemistry using antisera specific for Tbx5 or LMP4, MF20 (red) to outline myocardium and DAPI (blue) to outline all nuclei. (A-D) Adjacent cryosections from HH16 embryos, showing the unattached PEO (asterisk) and neighboring myocardium. (A-B) Section was stained for MF20 and DAPI (A) and Tbx5 (B). (C-D) An adjacent section was stained for MF20 and DAPI (C) and LMP4 (D). (E-L) Cryosections from HH20 embryos are shown. (E-H) Adjacent cryosections show the PEO (asterisk) which has come into contact with the myocardium. Arrows mark border between PEO and myocardium. (E-F) A section was stained for DAPI (E) and Tbx5 (F). An adjacent section was stained for DAPI (G) and LMP4 (H). (I-L) Images show the epicardium covering the ventricle in the HH20 heart. Arrows mark border between epicardium and myocardium. A section was stained MF20 and DAPI (I) and Tbx5 (J). An adjacent section was stained for MF20 and DAPI (K) and LMP4 (L). (M-P) Adjacent cryosections from HH25 show the epicardium over the ventricle. Arrows mark border between epicardium and myocardium. A section was stained MF20 and DAPI (M) and Tbx5 (N). A similar region from an adjacent section shows the epicardium over the ventricle, stained for MF20 and DAPI (O) and LMP4 (P). (Q-T) Adjacent cryosections show the HH25 AV groove. Arrows mark border between epicardium and myocardium. A section was stained for MF20 and DAPI (Q) and Tbx5 (R). An adjacent section was stained for MF20 and DAPI (S) and LMP4 (T). (U-X) Adjacent cryosections from HH29 embryos are show the epicardium over the ventricle. A section was stained for MF20 and DAPI (U) and Tbx5 (V). A similar region is shown, stained for MF20 and DAPI (W) and LMP4 (X). A: atria; V: ventricle. Scale bars: B,D=50μm; F,H,J,L,N,P,R,T,V,X=20μm.
Figure 4.

Tbx5 and LMP4 expression and sub-cellular localization within the myocardium. Cyrosections from HH29 embryos were processed for immunohistochemistry using antisera specific for myocyte marker MF20 and either Tbx5 or LMP4. (A) A region of the left ventricle is shown, stained for Tbx5, demonstrating excusive nuclear localization for the protein. (B-D) A lower magnification of the left ventricle is shown, stained for Tbx5 (B). Merge between Tbx5 and MF20 (C). Overlay with DAPI (D). Arrows mark example MF20 negative regions. (E) A region of the left ventricle is shown, stained for LMP4. (F-I) Higher magnification of a similar region is shown, stained for LMP4 (F) and MF20 (G). Merged image (H). Overlay with DAPI (I). LV: left ventricle; myo: myocardium; epi: epicardium. Scale bars: 20μm.
Figure 2.

Tbx5 and LMP4 sub-cellular localization in the atrio-ventricular cushion. Cyrosections were processed for immunohistochemistry using antisera specific for Tbx5 or LMP4. MF20 marks myocardium, phallodin marks actin and DAPI outlines all nuclei. (A-D) The HH25 atrio-ventricular (AV) cushion is shown, stained for Tbx5 (A), LMP4 (B) and DAPI (C). Merged image (D). (E-H) A region of the HH29 AV cushion is shown, stained for Tbx5 (E), LMP4 (F) and DAPI (G). Merged image (H). (I-L) An adjacent cyrosection, showing a similar region of the AV cushion, is shown, stained for Tbx5 (I), actin marker phalloidin (J), DAPI (K) and merge (L). (M-O) The HH33 presumptive mitral valve is shown. Merge between MF20 (red) and DAPI (blue) outlines myocardium and nuclei respectively (M). Presumptive valve is MF20 negative. Tbx5 was only expressed in limited areas of the valve (N). Asterisk indicates region of enlargement. Enlarged image of Tbx5 demonstrates nuclear only localization (O). (P-R) Images are from an adjacent section showing the presumptive mitral valve. Merge between MF20 and DAPI (P). LMP4 was expressed throughout the presumptive valve (Q). Asterisk indicates region of enlargement. Enlarged image of LMP4 demonstrates filamentous cytoplasmic localization (R). Scale bars: D,H,L,O,R=10μm; N,Q=100μm.
In conclusion, Tbx5 protein reveals dynamic distribution in different cardiac structures during development. It is predominantly expressed in myocardial tissue, initially more broadly and later becomes restricted to the left ventricle and both atria. In addition, Tbx5 proteins are temporally expressed in the AV cushion. LMP4 protein distribution is mostly limited to non-myocardial tissues such as the epicardium and AV cushion. Thus, co-expression of Tbx5 and LMP4 is apparently limited to few domains including the AV cushion.
Tbx5 and LMP4 sub-cellular localization in the AV cushion
Tbx5 and LMP4 were co-expressed in the AV cushion in a limited developmental time window. In the HH25 heart, LMP4 was expressed in the AV cushion but Tbx5 was not, with the exception of a few positive cells (Fig. 1E-F). Higher magnification of the HH25 AV cushion confirmed the overall absence of Tbx5 protein, and revealed that LMP4 was localized along filamentous structures in the cytoplasm (Fig. 2A-D). In the HH29 heart, Tbx5 and LMP4 were both uniformly expressed throughout the AV cushion (Fig. 1H-I). Higher magnification demonstrated that in the presence of LMP4, Tbx5 localized to both the nucleus and cytoplasmic sites (Fig. 2E). Like Tbx5, LMP4 was localized along cytoplasmic filaments (Fig. 2F). The merged image revealed significant co-localization of cytoplasmic Tbx5 with LMP4 (Fig. 2H). In vitro work has shown that the Tbx5/LMP4 complex localizes along actin filaments (Camarata et al., 2006; Krause et al., 2004). Co-stain of an adjacent section for Tbx5 and the actin marker phalloidin revealed strong co-localization between the cytoplasmic Tbx5 and actin, consistent with in vitro findings (Fig. 2I-L).
By HH33, Tbx5 was down-regulated in much of the AV cushion; however, limited Tbx5 expression remained in the area of the presumptive mitral valve (Fig. 1K). High magnification of this region is shown in Figure 2M-O. The myocyte marker MF20 stained the surrounding myocardium, but not the presumptive valve or other AV cushion tissue (Fig. 2M). Tbx5 was expressed in endocardial cells lining the lumen of the cushion (Fig. N, asterisk). Within the cushion tissue, Tbx5 was localized exclusively to the nucleus, with no cytoplasmic localization detectable (Fig. 2O). At this stage, LMP4 remained expressed throughout the AV cushion (Fig. 1L and Fig. 2Q). Comparable to earlier stages, LMP4 was localized to actin filaments (Fig. 2R, data not shown). While we do observe LMP4-mediated re-localization of Tbx5 to the actin cytoskeleton, co-expression is not sufficient for this process to occur.
Tbx5 and LMP4 expression in epicardial formation
The epicardium is formed from the villous proepicardial organ (PEO), an outgrowth of the septum transversum (Manner et al., 2001). In the chick, the PEO is formed during HH14-16 and makes contact with the heart between HH17-18. Once the PEO contacts the heart proper, proepicardial cells migrate onto the myocardium and transition into epicardial cells. These cells form a mesothelial sheet which will eventually envelope the surface of the heart, completing around HH27 (Manner et al., 2001).
To examine Tbx5 and LMP4 expression and localization during epicardial formation, we examined cryosections from HH16-29 embryos. In Figure 3, MF20 marks myocardial tissue, but does not stain the PEO or epicardial tissue. In the HH16 embryo, when the PEO has not come into contact with the myocardium, the PEO expressed LMP4, but not Tbx5 (Fig. 3A-D, asterisk). In contrast, the neighboring atria and ventricle expressed Tbx5 (Fig. 3A-B). However, by HH20, a time point when the PEO has come into contact with the myocardium, some cells of the PEO began to express Tbx5 (Fig. 3E-F, asterisk). Arrows mark the border between PEO and myocardium. The areas of the PEO expressing Tbx5 were those in closest proximity to the neighboring ventricle (Fig. 3F). LMP4 remained expressed throughout the PEO (Fig. 3G-H, asterisk). Both Tbx5 and LMP4 proteins were expressed in the epicardium at this stage (Fig. 3I-L). Interestingly, despite co-expression with LMP4, Tbx5 was localized to the nucleus within epicardial cells, while LMP4 was localized to cytoplasmic sites, suggesting a lack of interaction between the proteins at this stage of heart development. Co-staining of an adjacent section for LMP4 and phalloidin revealed that like in the AV cushion, LMP4 was localized to actin filaments (data not shown and Camarata et al., 2006).
By HH25, epicardial formation is nearly complete. Tbx5 was no longer detected in the majority of epicardial cells (Fig. 3M-N). However, cells expressing Tbx5 were occasionally observed in the superficial epicardium, especially in the vicinity of the AV groove (Fig. 3Q-R). As in HH20 hearts, LMP4 revealed a cytoplasmic localization in epicardial cells (Fig. 3O-P). LMP4 was expressed in both the superficial epicardium and the epicardially derived mesenchymal cells (EPDCs) of the sub-epicardial space (Fig. 3S-T). Notably, we consistently observed a decrease in the intensity of LMP4 expression between cells of the superficial epicardium and sub-epicardial space. This could be due to down-regulation of LMP4 during EMT, but could also be caused by change in morphology due to actin re-organization (Lu et al., 2001). By HH29, we could not observe Tbx5 expression in any epicardial (Fig. 3U-V). However, LMP4 continued to be expressed (Fig. 3W-X).
Thus, during epicardial development Tbx5 expression becomes induced in proepicardial cells after they have contacted the myocardium. The expression is exclusively nuclear and persists until HH25, when Tbx5 protein remains in a small number of cells in the superficial epicardium. LMP4 is expressed in the cytoplasm, localized along actin filaments, throughout the proepicardium, epicardium and EPDCs of the sub-epicardial space at all stages examined.
Tbx5 and LMP4 sub-cellular localization and cell-type specific expression in myocardial tissue
During cardiac development, Tbx5 was initially expressed broadly across the myocardium, and later became restricted to the atria and left ventricle (Fig. 1). At all stages examined, within the myocardium, higher power inspection demonstrated that Tbx5 was localized exclusively in the nucleus of cells (Fig 4A). Within the left ventricular and atrial myocardium, Tbx5 was, however, not expressed in all cells. Figure 4 shows representative regions from the HH29 left ventricle. Co-staining for Tbx5 and MF20, a cardiac myocyte marker, revealed that within the myocardium Tbx5 was expressed only in this cell type (Fig. 4B-D). Moreover, co-staining of Tbx5 and vimentin, a fibroblast marker, indicated that Tbx5 was not expressed in vimentin positive fibroblasts and supported our findings with MF20 (Fig S2). In contrast to Tbx5, the myocardium was predominantly negative for LMP4 except for a few isolated LMP4 expressing cells scattered throughout the interstitium (Fig 4E). Higher magnification demonstrated that these cells did not express MF20, suggesting that they were a distinct population from the Tbx5 expressing cells (Fig. 4F-I). Of note, LMP4 was only expressed in a fraction of the total MF20 negative cells. Only a small number of the LMP4 positive cells co-expressed vimentin and the majority of vimentin positive cells did not express LMP4 (data not shown).
Characterization of LMP4 positive cells within the myocardium
Since LMP4 was expressed in both epicardial cells and EPDCs of the sub-epicardial space, we wondered whether the LMP4 positive cells within the myocardium were epicardially derived. In order to directly test this possibility, we generated quail-chick chimeras, an established technique for tracing epicardial cells (Dettman et al., 1998; Perez-Pomares et al., 1998). Details on the generation of quail-chick chimeras are provided in the supplemental material. Chimeric embryos were incubated until HH29, sectioned, and processed for immunohistochemistry for either LMP4 and the quail-specific antigen QCPN or Tbx5 and QCPN in order to determine distribution and expression of the proteins in epicardially derived cells. To assess LMP4 expression of epicardially derived cells in the myocardium, we counted the number of QCPN single positive and QCPN/LMP4 double positive cells in sections from six separate chimeric embryos. Surprisingly, only a small fraction of the QCPN positive cells expressed LMP4 (29/421, Fig. S4A-D). In contrast, in the same sections, we observed greater than 400 LMP4 positive cells that did not express QCPN. Similar cell counting experiments were performed on sections stained for Tbx5 and QCPN. We did not detect Tbx5 expression in any QCPN positive cells in the myocardium or epicardium (0/213, Fig. S4E-H).
These results demonstrate that only a small number of proepicardially derived cells in the myocardium expressed LMP4 and that the majority of proepicardially derived cells down-regulate LMP4 as they migrate into the myocardium. Furthermore, a small but significant number of non-myocyte cells exist in the myocardium that expressed LMP4 most of which do not appear to be derived from the proepicardium.
Tbx5 expression and localization in outflow tract
Consistent with previous whole mount in situ studies, we could not detect Tbx5 expression in the outflow tract before HH29 (data not shown). However, beginning at HH29, a cross section of the outflow tract revealed a ring of Tbx5 expressing cells lining the lumen (Fig. 5A). Similar expression in cells lining the lumen was observed as late as HH39 (data not shown). LMP4 was expressed throughout the outflow tract at all stages examined (Fig. 5C, and data not shown). Higher magnification of the inner edge of the lumen revealed filamentous cytoplasmic appearance of Tbx5, with no detectable nuclear localization (Fig. 5E-F). The merge between Tbx5 and caldesmon demonstrated that the Tbx5 expressing cells co-expressed this smooth muscle marker (Fig. 5G). An adjacent section stained for LMP4 confirmed co-expression of this protein with its typical cytoplasmic filamentous localization (Fig. 5H-I).
Figure 5.

Tbx5 and LMP4 expression in outflow tract. Cryosections from HH29 chicken embryos were processed for immunohistochemistry using antisera specific for Tbx5, LMP4 or caldesmon. A cross section of the outflow tract (OT) was stained for Tbx5 (A) and DAPI (B). A similar section of the OT was stained for LMP4 (C) and DAPI (D). White boxes indicate regions of enlargement. Enlarged view of the OT stained for Tbx5 (E). Merge with DAPI (F). Merge with caldesmon (G). Enlarged view of the OT stained for LMP4 (H). Merge with DAPI (I).
Tbx5 and LMP4 expression and localization in coronary vessel development
Similar to the outflow tract, Tbx5 exhibited dynamic expression across coronary vessel formation. Presumptive coronary vessels were identified using the smooth muscle marker caldesmon. An example region from the HH29 left ventricle is shown in Figure 6A-H. Tbx5 was not detected in any caldesmon positive cells within the myocardium (Fig. 6A-D). This is consistent with our observation that Tbx5 myocardial expression at this stage was limited to myocytes (Fig. 4). Unlike Tbx5, LMP4 was expressed in all caldesmon positive cells (Fig. 6E-H).
Figure 6.

Tbx5 is localized to the cytoplasm when expressed in coronary smooth muscle. Cryosections from chicken embryos were processed for immunohistochemistry using antisera specific for Tbx5, LMP4 or caldesmon. (A-D) Cryosections from HH29 chick embryos were stained for Tbx5 (A) and smooth muscle marker caldesmon (B). Merged image (C). Overlay with DAPI (D). (E-H) A similar section was stained for LMP4 (E) and caldesmon (F). Merged image (G). Overlay with DAPI (H). (I) A region of the left atrio-ventricular groove from a HH36 heart is shown, containing a portion of the left atria and a coronary vessel (arrow). (J-M) Enlarged view of the coronary vessel stained for Tbx5 (J). Overlay of Tbx5 with DAPI (K). Caldesmon (L). Merge between Tbx5 and caldesmon (M). (N-Q) An adjacent section was stained for LMP4 (N). Overlay of LMP4 with DAPI (O). Caldesmon (P). Merge between LMP4 and caldesmon (Q). (R-Y) A coronary vessel located within the myocardium of a HH39 heart is shown. (R-U) Tbx5 (R), overlay with DAPI (S), caldesmon (T) and merge between Tbx5 and caldesmon (U). (V-Y) An adjacent section is shown, co-stained for Tbx5 (V) and LMP4 (W). Merged image (X). Overlay with DAPI (Y). epi=epicardium; myo=myocardium; LA=left atria; LV=left ventricle; OT=outflow tract. Scale bars=20μm.
Beginning at HH36, Tbx5 expression was identified in sub-epicardial coronary vessels, but not in those of the interstitium. An example vessel located in the left AV groove is shown in Fig. 6I. While Tbx5 had an exclusively nuclear localization in the neighboring left atria, in the coronary vessel Tbx5 displayed different localization. A higher power image showing the merge between Tbx5 and DAPI demonstrated an exclusive filamentous cytoplasmic Tbx5 distribution, with no nuclear localization (Fig. 6J-K). A counter stain of the same section with caldesmon revealed that Tbx5 was expressed in the mesenchyme surrounding the coronary smooth muscle, with little overlap with the caldesmon positive cells (Fig. 6L-M). An adjacent section co-stained for LMP4 and DAPI showed the expected filamentous LMP4 localization in the cytoplasm (Fig. 6N-O). Counter stain of the same section with caldesmon revealed that LMP4 was expressed both in the mesenchyme surrounding the smooth muscle and in the smooth muscle itself (Fig. 6P-Q).
By HH39, Tbx5 was expressed in all coronary vessels, including those located in the interstitium. Unlike the vessels of the HH36 heart, at this later developmental stage the Tbx5 expressing cells also expressed caldesmon (Fig. 6R-U). LMP4 showed continued expression in coronary vessels, and co-staining of sections for Tbx5 and LMP4 demonstrated co-localization of Tbx5 with LMP4 in these mature blood containing vessels (Fig. 6V-Y).
In conclusion, the co-expression of LMP4 and Tbx5 and exclusive cytoplasmic localization of Tbx5 in smooth muscle of both coronary vessels and outflow tract suggest a general, yet unknown role for Tbx5 in smooth muscle differentiation.
Discussion
We have previously demonstrated in both transfected and primary epicardial cells that in the presence of the PDZ-LIM protein LMP4, Tbx5 shuttles dynamically between the nucleus and cytoplasm and, in a complex with LMP4, localizes to actin filaments (Camarata et al., 2006; Krause et al., 2004). The change in Tbx5 sub-cellular localization represses the transcription factor’s ability to activate target gene promoters, introducing a previously unknown mechanism for Tbx5 transcription factor regulation (Camarata et al., 2006).
Here we show experimental evidence for dynamic Tbx5 protein expression as well as differential nuclear and cytoplasmic localization of native Tbx5 during heart development, which provides in vivo significance for the Tbx5/LMP4 regulatory model. While the in vivo studies confirm our previous work with cultured cells, they also reveal that the regulation of Tbx5 is considerably more complex than anticipated.
Tbx5 proteins are localized to the nucleus in myocardial tissue
The heart is composed of multiple cell types and lineages, and in the different tissue layers, we find differential expression of Tbx5 proteins. The myocardium forms cardiac myocytes which differentiate into heart muscle. A range of Tbx5 phenotypes have been observed in this tissue, which provoked extensive studies on Tbx5 expression and function within the myocardium (Bruneau et al., 2001; Liberatore et al., 2000; Takeuchi et al., 2003). We demonstrate the protein distribution in myocardial tissue follows the Tbx5 mRNA expression profile, which indicates overall regulation on a transcriptional rather than translational level. Tbx5 protein is initially expressed broadly across the atrial and ventricular myocardium, and gradually becomes restricted to the atria and left ventricle. Within myocardial tissue, we find Tbx5 expression is limited to myocytes and has a nuclear localization in these cells. LMP4, however, is only expressed in a few isolated non-myocyte cells within the myocardium which do not co-express Tbx5. Thus, in the absence of LMP4 co-expression, Tbx5 has a default nuclear localization, as would be expected for a transcription factor. The possibility existed that the LMP4 positive cells within the myocardium were of epicardial origin; however, lineage tracing studies suggest that the majority of these are not proepicardially derived. Our data further show that while LMP4 is expressed in all epicardial cells and EPDCs of the sub-epicardial space, the majority of EPDCs down-regulate LMP4 as they migrate into the myocardium.
Tbx5 becomes localized to the cytoplasm in non-myocardial tissue
In contrast to the myocardium, endocardial and epicardial tissues do express both Tbx5 and LMP4 proteins. In the atrio-ventricular (AV) cushion, an endocardially derived structure that gives rise to multiple structures, including the valves, Tbx5 and LMP4 are co-expressed. However, while LMP4 is expressed in the AV cushion throughout heart development, Tbx5 expression is dynamic with the highest protein level detected at HH29. At this developmental stage, Tbx5 is distributed to both the nucleus and the cytoplasm, where it co-localizes with LMP4. Interestingly, at HH33 when Tbx5 expression is limited to regions of the presumptive mitral valve, the protein is localized to the nucleus alone, while LMP4 remains cytoplasmic. Thus, co-expression of both proteins is not sufficient for Tbx5 re-localization to the cytoplasm and suggests that either Tbx5 nuclear export or binding to LMP4 is responsive to temporal and/or spatial signals in vivo. This regulated change of nuclear to actin-associated distribution of Tbx5 is supported by our previous in vitro findings (Camarata et al., 2006). We do not see conclusive evidence of LMP4 localization to the nucleus and therefore it seems likely that Tbx5 is shuttled from the nucleus either by a yet unidentified transport protein or by an intrinsic shuttling signal within Tbx5 itself. These possibilities can be experimentally tested, and studies are under way to investigate which cellular mechanism is responsible for altering Tbx5 sub-cellular localization.
Tbx5 is dynamically expressed during epicardium formation
In addition to dynamic sub-cellular localization, we find novel regulation of Tbx5 expression in the formation of the epicardium. The cells of the epicardium originate from the proepicardial organ (PEO). While LMP4 is expressed in the PEO, the PEO is initially Tbx5 negative. When the PEO comes into contact with the myocardium, some cells within the PEO begin to express Tbx5. Of note, the Tbx5 positive cells are those in closest proximity to the myocardium, suggesting that either direct contact or diffusible factors up-regulate Tbx5 expression in proepicardial cells. While epicardial cells at an early developmental stage co-express LMP4 and Tbx5, Tbx5 remains localized in the nucleus, indicating that the interaction of Tbx5 with LMP4 is a regulated process, responsive to local cues. Notably, this co-expression, but lack of interaction is consistent with our observation in cultured epicardial cells that have not undergone EMT (Camarata et al., 2006). LMP4 remains a constant marker for the epicardium; however, Tbx5 expression in the epicardium appears to be transitory and is lost at later developmental stages, corresponding to the time at which epicardial formation is completed (Manner et al., 2001).
This transient expression of Tbx5 suggests that the protein may play a temporally limited role in epicardial formation. Two likely roles for Tbx5 function can be considered: epicardial migration and/or differentiation from proepicardium into epicardium. A role in cell migration has been suggested by Hatcher et al., (2004), who used Tbx5 anti-sense knock-down in cultured proepicardial explants to inhibit cell migration. Interestingly, using retroviral over-expression of Tbx5 in the PEO of chick embryos, the same group reported reduced migration of cells from the PEO onto the heart. While the data seem contradictory, it is possible that while Tbx5 is required for migration of proepicardial cells, premature expression of Tbx5 in the PEO may have similar negative effects, either by direct impairment of migration or triggering premature transition into epicardial cells.
Tbx5 is expressed in the cytoplasm of smooth muscle cells
We found novel Tbx5 expression and localization in smooth muscle of both coronary vessels and the outflow tract. While the outflow tract does not express Tbx5 at earlier stages, at HH29 and beyond, we observed a ring of Tbx5 expressing cells lining the lumen, that co-express the smooth muscle marker caldesmon. LMP4 is co-expressed with Tbx5 in these cells, and of considerable interest, Tbx5 has an exclusively cytoplasmic localization along the actin cytoskeleton. Tbx5 is expressed only in the interior of the outflow tract at these later stages of development, which may account for the fact that this expression has not been detected by whole mount in situ hybridization in other studies.
Tbx5 has a similar cytoplasmic localization in smooth muscle cells of sub-epicardial and interstitial coronary vessels. While Tbx5 is not expressed early, at later developmental stages the protein is co-expressed with LMP4 in the cells of coronary vessels. Similar to the outflow tract, in these vessels Tbx5 is localized exclusively to the cytoplasm, in a complex with LMP4. Initially, Tbx5 is expressed only in sub-epicardial vessels, particularly those in the AV groove and near the aorta. Of interest, at this stage, the Tbx5 expressing cells did not express caldesmon, but instead surrounded the caldesmon expressing smooth muscle. Such cells surrounding coronary vasculature smooth muscle have been hypothesized to be perivascular fibroblasts, cells that have the potential to become vascular smooth muscle or pericytes (Dettman et al., 1998). By HH39, Tbx5 expression is detected in nearly all coronary vessels, including those within the myocardium. At this later developmental stage, the Tbx5 expressing cells are caldesmon positive, suggesting that Tbx5 expression may be involved in the differentiation of cells into smooth muscle. The difference in timing of Tbx5 expression in sub-epicardial versus interstitial vascular structures could reflect the overall progression of maturation of these vessels or deposition of smooth muscle around vessels. Given the unique Tbx5 localization we observe in these smooth muscle cells, it would be interesting to test the impact of factors involved in smooth muscle differentiation such as FGFs and VEGF on Tbx5 expression and sub-cellular localization (Mu et al., 2005).
Surprisingly, in the cells of both the outflow tract and coronary vessels, Tbx5 could not be detected within the nucleus at any stage. While binding of Tbx5 to LMP4 may serve as a method of transcriptional regulation, as we have demonstrated in cultured cells (Camarata et al., 2006), the complete absence of nuclear Tbx5 proteins in differentiating smooth muscle cells of coronary arteries may indicate that Tbx5 has additional functions in the cytoplasm. In this context it is noteworthy that the Tbx5/LMP4 complex is localized to the actin cytoskeleton, which is consistent with other PDZ-LIM protein family members such as Enigma, CLP-36, and Cypher/ZASP that are thought to have roles as regulators of cyto-architecture and cell motility (Guy et al., 1999; Kadrmas and Beckerle, 2004; Vallenius et al., 2000; Zhou et al., 1999). An attractive possibility for the function of LMP4-bound Tbx5 would be a direct role in regulating actin dynamics, particularly during differentiation processes. Future experiments will address the potential transcription independent functions of Tbx5 in the cytoplasm; however, disrupting cytoplasmic Tbx5 without interfering with nuclear Tbx5 will remain a challenge. One approach may involve creation of modified forms of Tbx5, which localize to either the nucleus or cytoplasm.
Nuclear versus cytoplasmic Tbx5 localization and its role in Holt-Oram Syndrome
Our observations with the chicken developing heart and primary epicardial cells (Camarata et al., 2006) reveal a previously unknown developmentally regulated expression of Tbx5, with protein localization to both the nucleus and the cytoplasm. The new findings on dynamic sub-cellular localization of Tbx5 would suggest a function for Tbx5 outside the nucleus.
Cardiac and skeletal malformations known as Holt-Oram syndrome (HOS) in humans are caused by mutations in TBX5. The majority of TBX5 mutations critical for disease manifestation are thought to result in early termination and haploinsufficiency (Basson et al., 1999; Brassington et al., 2003); however, increased TBX5 dosage, such as chromosome 12q2 duplication, has been reported to also lead to HOS (Hatcher et al., 2001; Vaughan and Basson, 2000). These data would suggest that both under- and over-expression could cause fairly similar if not identical phenotypes, making a case that appropriate TBX5 levels are critical. A role for Tbx5 as a transcription factor in the nucleus has been discussed in HOS; however, a potential cytoplasmic function for Tbx5 has not been appreciated in disease manifestation. When impaired, this function, in addition to the nuclear role, may contribute to HOS phenotypes. Arguably, cytoplasmic Tbx5 localization is only observed in a subset of heart tissues and it is not observed in the myocardium, where the most pronounced TBX5 phenotypes have been reported. Of the structures where Tbx5 is localized to the cytoplasm, the AV cushion has an obvious connection to disease manifestation. The AV cushion contributes to the formation of the heart valves as well as the interventricular, atrial and atrioventricular septa, all structures particularly affected when Tbx5 level is altered both experimentally and in HOS (Basson et al., 1999; Bruneau et al., 2001). It is possible that in cells of the AV cushion the amount of Tbx5 in the nuclear/cytoplasmic compartment must be precisely controlled for proper function, and therefore these cells would be particularly sensitive to any alteration of Tbx5 levels. It is not clear what consequences alterations in overall Tbx5 levels would have on the ratio between nuclear and cytoplasmic compartments, but it may not impact them equally. In addition, we have found cytoplasmic Tbx5 distribution in developing coronary arteries and cells of the outflow tract lumen. Although the coronary arteries in HOS have not been well studied, various vascular defects have been recognized in HOS patients including patent ductus arteriosus, persistent left superior vena cava, anomalous pulmonary venous return and aortic coarctation (Hatcher and McDermott, 2006). Given that Tbx5 appears to be exclusively cytoplasmic in the coronary vessels, examination of these structures for defects in HOS patients and Tbx5 compromised animal models may give clues as to the potential cytoplasmic function of Tbx5.
The data presented here indicate a specific role for Tbx5 in epicardial formation, which would have significant implications for the analysis of HOS phenotypes. Cells derived from the epicardium form coronary fibroblasts and contribute to the septum, AV cushion and coronary vasculature and therefore defects in epicardial formation would directly impact these structures. However, potentially of even greater importance, signaling from the epicardium has been demonstrated to be essential as a modulator of myocardial development. Thus, subtle defects in the epicardium can result in significant phenotypes within the myocardium (Kang and Sucov, 2005; Manner et al., 2001; Stuckmann et al., 2003). Chicken embryos in which epicardial formation and/or function is impaired have abnormal chamber size as well as trabeculation and septal defects, myocardial phenotypes which are also observed when Tbx5 cardiac level is experimentally altered (Bruneau et al., 2001; Gittenberger-de Groot et al., 2000; Liberatore et al., 2000; Merki et al., 2005). While the epicardium is reported to be present in mutant Tbx5del/+ mice (Bruneau et al., 2001), it is possible that subtle defects in epicardial formation or function have remained undetected. Considering our new findings, an epicardial specific Tbx5 knock-out mouse would be of particular interest and complement published mutant Tbx5 mouse models.
Supplementary Material
Acknowledgments
The authors sincerely thank Danijela Dokic, Jennifer Yeung, Margaret Miller, Jennifer Krcmery, and Wenjing Huang for their assistance. Drs. J. Topczewski, S. Ahlgren and especially Dr. A. Kulisz and Troy Camarata for critical reading and suggestions on various manuscript drafts, and members of the Developmental Biology Core for stimulating discussions. This work was supported by NIH grant HL085834-01 (to H.-G.S.) and AHA Grant-in-aid 0455612Z (to R.W.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–33. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, Bruzzone R, Quadrelli R, Lerone M, Romeo G, Silengo M, Pereira A, Krieger J, Mesquita SF, Kamisago M, Morton CC, Pierpont ME, Muller CW, Seidman JG, Seidman CE. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci U S A. 1999;96:2919–24. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Ingham PW. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mechanisms of Development. 2000;90:299–304. doi: 10.1016/s0925-4773(99)00246-4. [DOI] [PubMed] [Google Scholar]
- Brassington AE, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, Whitby FG, Jorde LB, Bamshad MJ. Expressivity of Holt-Oram syndrome is not predicted by TBX5 genotype. American Journal of Human Genetics. 2003;73 doi: 10.1086/376436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211:100–8. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–21. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Camarata T, Bimber B, Kulisz A, Yeung J, Chew TL, Simon HG. LMP4 regulates Tbx5 protein sub-cellular localization and activity. Journal of Cell Biology. 2006 doi: 10.1083/jcb.200511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz MV, Gimenez-Ribotta M, Saravalli O, Cayre R. The contribution of the inferior endocardial cushion of the atrioventricular canal to cardiac septation and to the development of the atrioventricular valves: study in the chick embryo. Am J Anat. 1983;166:63–72. doi: 10.1002/aja.1001660105. [DOI] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw W, Jr., Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–81. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res. 2000;87:969–71. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–52. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Guy PM, Kenny DA, Gill GN. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Molecular Biology of the Cell. 1999;10:1973–1984. doi: 10.1091/mbc.10.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:49–92. [PubMed] [Google Scholar]
- Hatcher CJ, Diman NY, Kim MS, Pennisi D, Song Y, Goldstein MM, Mikawa T, Basson CT. A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics. 2004;18:129–40. doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, Kim MS, Mah CS, Goldstein MM, Wong B, Mikawa T, Basson CT. TBX5 transcription factor regulates cell proliferation during cardiogenesis. Dev Biol. 2001;230:177–88. doi: 10.1006/dbio.2000.0134. [DOI] [PubMed] [Google Scholar]
- Hatcher CJ, McDermott DA. Using the TBX5 transcription factor to grow and sculpt the heart. Am J Med Genet A. 2006;140:1414–8. doi: 10.1002/ajmg.a.31256. [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kang JO, Sucov HM. Convergent proliferative response and divergent morphogenic pathways induced by epicardial and endocardial signaling in fetal heart development. Mech Dev. 2005;122:57–65. doi: 10.1016/j.mod.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Khan P, Linkhart B, Simon HG. Different regulation of T-box genes Tbx4 and Tbx5 during limb development and limb regeneration. Dev Biol. 2002;250:383–92. [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–20. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis ARJ, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram Syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nature Genetics. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol. 2000;223:169–80. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- Lu J, Landerholm TE, Wei JS, Dong XR, Wu SP, Liu X, Nagata K, Inagaki M, Majesky MW. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 2001;240:404–18. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- Männer J. The development of pericardial villi in the chick embryo. Anatomy and Embryology. 1992;186:379–85. doi: 10.1007/BF00185988. [DOI] [PubMed] [Google Scholar]
- Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- Markwald R, Eisenberg C, Eisenberg L, Trusk T, Sugi Y. Epithelial-mesenchymal transformations in early avian heart development. Acta Anat (Basel) 1996;156:173–86. doi: 10.1159/000147845. [DOI] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–60. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori AD, Bruneau BG. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol. 2004;19:211–5. doi: 10.1097/00001573-200405000-00004. [DOI] [PubMed] [Google Scholar]
- Mu H, Ohashi R, Lin P, Yao Q, Chen C. Cellular and molecular mechanisms of coronary vessel development. Vasc Med. 2005;10:37–44. doi: 10.1191/1358863x05vm584ra. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–13. [PubMed] [Google Scholar]
- Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. The origin of the subepicardial mesenchyme in the avian embryo: an immunohistochemical and quail-chick chimera study. Dev Biol. 1998;200:57–68. doi: 10.1006/dbio.1998.8949. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Wessels A. Epicardial-like cells on the distal arterial end of the cardiac outflow tract do not derive from the proepicardium but are derivatives of the cephalic pericardium. Dev Dyn. 2003;227:56–68. doi: 10.1002/dvdy.10284. [DOI] [PubMed] [Google Scholar]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–49. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130:5953–64. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- Vallenius T, Luukko K, Makela TP. CLP-36 PDZ-LIM protein associates with nonmuscle alpha-actinin-1 and alpha-actinin-4. Journal of Biological Chemistry. 2000;275:11100–5. doi: 10.1074/jbc.275.15.11100. [DOI] [PubMed] [Google Scholar]
- Vaughan CJ, Basson CT. Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus. Am J Med Genet. 2000;97:304–9. doi: 10.1002/1096-8628(200024)97:4<304::aid-ajmg1281>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–68. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Viragh S, Szabo E, Challice CE. Formation of the primitive myo- and endocardial tubes in the chicken embryo. J Mol Cell Cardiol. 1989;21:123–37. doi: 10.1016/0022-2828(89)90856-0. [DOI] [PubMed] [Google Scholar]
- Vrancken Peeters MP, Gittenberger-de Groot AC, Mentink MM, Poelmann RE. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat Embryol (Berl) 1999;199:367–78. doi: 10.1007/s004290050235. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. Journal of Biological Chemistry. 1999;274:19807–13. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


