Summary
The gene encoding the ABCC6 protein, an ABC transporter of the multidrug resistance-associated protein (MRP), is mainly expressed in liver and kidney. Mutations in ABCC6 are responsible for the development of the pseudoxanthoma elasticum (PXE) phenotype. PXE is a recessive disease characterized by the calcification of elastic fibers resulting in dermal, vascular and ocular clinical manifestations. The physiological function of ABCC6 and the rodent orthologs Abcc6 is unknown and their precise relationship to elastic fibers is only a matter of speculation. Despite several studies focused on the transcriptional regulation of ABCC6/Abcc6, the molecular signals conferring the tissue-specificity to the ABCC6/Abcc6 expression are not well defined. In this report, we determined the level of the mouse Abcc6 promoter methylation in tissues with low level of expression (tail extremity and skin), intermediate (kidney) and high level of expression (liver). We observed that high and moderate levels of methylation correlated with low levels of Abcc6 expression. Moreover, we determined that CpG methylation of the Abcc6 proximal promoter region was interfering with the binding of the Sp1 transcription factor thereby inhibiting Sp1-dependent transactivation. Thus, our data provides the first direct evidence that an epigenetic mechanism regulates the binding of the transcription factor Sp1 to the proximal promoter and participates in the tissue-specific expression control of the mouse Abcc6 gene.
Introduction
ABCC6 encodes an ATP-binding cassette (ABC) transporter of the sub-family C previously referred to as multidrug-associated resistance proteins 6 or “MRP6” [1]. ABCC6 expression is primarily found in liver and kidneys but can also be detected in other cell types albeit at lower levels [2–4]. The ABCC6 protein is a transmembrane protein located in the basolateral side of polarized cells and exports metabolite(s) of unknown nature [2, 3]. However, vesicular studies have demonstrated that leukotriene-C4 and N-ethylmaleimide-S-glutathione can be actively transported by the human ABCC6 while the BQ123 cyclic peptide is only efficiently transported by the rat Abcc6 [5]. The overall function of ABCC6 is unclear but relates in some ways to elastic fibers maintenance or integrity. Indeed, mutations in the ABCC6 gene were associated with the development of pseudoxanthoma elasticum (PXE, OMIM #264800, 177850) [6–9]. PXE is a heritable disorder characterized by mineralization of elastic fibers and other connective tissue alterations [10, 11]. The genetics of PXE are now well defined, however, the pathomechanism that links the deficiency of ABCC6 activity to calcification of elastic fibers remains to be elucidated. After the identification of the causative gene (ABCC6), it became evident that PXE was not a connective tissue disease stricto sensu but rather a metabolic disorder [12, 13] and several recent studies provided support to this notion [14–16]. Because the tissue distribution of ABCC6 was a major argument in suggesting PXE as a metabolic disease, there has been much interest in determining the transcriptional regulation signals governing the tissue-specificity of ABCC6 expression. The regulation of gene expression of both human and mouse ABCC6/Abcc6 was found to depend in part on the binding of the Sp1 transcription factor to the proximal promoter in vivo and in vitro [17, 18]. In addition, the ABCC6 proximal promoter was also described as transcriptionally dependent on DNA-methylation in cell lines [19].
In this report, we describe evidences of an association between the DNA methylation status of the mouse Abcc6 proximal promoter and Sp1 binding in tissues with high, intermediate and low level of expression. We notably found that the methylation of the mouse Abcc6 proximal promoter inversely correlates with Sp1 binding and transcriptional activation and that DNA methylation repressed the mouse Abcc6 transcriptional activity in liver and kidney cell lines. This work provides the first direct evidence that concerted effects of genetic and epigenetic factors plays an essential role in the tissue-specific expression of the mouse Abcc6 gene.
Materials and methods
Cell culture and transient transfection
TIB-73 cells from the American Type Culture Collection (ATCC, Manassas, VA) were maintained in Dulbecco’s modified Eagle medium (DMEM), complemented with 10% (v/v) fetal bovine serum. Transient transfections were performed using the GeneJammer transfection reagent (Stratagene, La Jolla, CA) as described by the manufacturer. Cells were co-transfected with the specific Abcc6 promoter::firefly luciferase reporter construct and the renilla luciferase plasmid (pRL-SV40, Promega, Madison, WI) as a control of transfection efficiency.
In vivo methylation promoter studies
Tissues collected from 10 month-old mice were rapidly frozen and stored at −80°C before experiments. Genomic DNA was extracted from tissues using the Qiagen DNeasy kit (Qiagen, Valencia, CA). Methylated DNA was isolated using the Promoter Methylation PCR kit according to the manufacturer’s instructions (Panomics, Redwood city, CA). The methylated genomic DNA was analyzed by PCR using primers specific to the mouse Abcc6 promoter: 5′- CAC CAG CTC CAC CTC TGT AT –3 and 5′- GTT TGT CTT ACA GCT TCC CG –3′. A positive PCR amplification indicates that the promoter is methylated and as the level of amplification is directly proportional to the concentration of (methylated) DNA isolated, the quantification of the PCR products reflected the level of DNA methylation.
Quantification of methylation
The PCR products were amplified, electrophoresed on a 2% agarose gel and stained with SYBRGreen I (Molecular Probe, Carlsbad, CA). After staining, the intensity of each band was measured using the Kodak Gel Logic 200 and associated software Molecular Imager v4.0. As standard controls, the PCR fragment was amplified using the input from total genomic DNA (prior using the Promoter Methylation PCR kit). The ratio of methylation was calculated by comparing the intensity of the methylated fragment with that present in the total genomic DNA extract (input).
Chromatin Immunoprecipitation Assays (ChIP) and quantitative PCR
ChIPs were performed from skin, kidney and liver isolated from 10 month-old mice, as previously described [17]. Immunoprecipitated DNA was purified and analyzed for specific enrichment by quantitative PCR analysis using SYBRGreen™ qPCR SuperMix (Invitrogen, Carslbad, CA) and primers specific to the mouse Abcc6 proximal promoter (see above). As the elution volume of the immunoprecipitated chromatin samples may vary, we used the neomycine-resistance gene present on the pCMV plasmid, which was added in each sample prior to performing the ChIP assays, to account for deviation in the measurement of DNA levels. The sequence of the oligonucleotides specific to the neomycine-resistance gene is as follow: mNeo (5′-GAACAAGATGGATTGCACGCAGG-3′) and aNeo (5′-CGCTGACAGCCGGAACACG-3′). The quantification of co-immunoprecipitated promoter fragments was based on Chakrabarti et al. [20]. Samples were quantified in triplicate from three independent immunoprecipitations.
Electromobility shift assay (EMSA)
20ng of purified Sp1 protein was incubated for 20 minutes at room temperature with a DNA fragment corresponding to the −152/+162bp region of the mouse Abcc6 promoter that was either methylated or not. The DNA methylation was performed as described below. DNA/protein complexes were resolved on a 4% polyacrylamide gel in 0.5X TBE at 120V and detected by SYBR Green/SYPRO Ruby staining (Molecular Probes, Carlsbad, CA).
In vitro methylation of the proximal promoter sequence
A region encompassing the −152/+162bp domain of the Abcc6 gene has been excised from its plasmid vector (pGL3-Basic, Promega, Madison, WI) using KpnI and EcoRI restriction enzymes. The excised DNA fragment was gel-purified and subsequently methylated with the SsI DNA methyltransferase (New England Biolabs, Beverly, MA) according to the manufacturer's instructions. Full methylation was confirmed by resistance to HpaII digestion (New England Biolabs, Beverly, MA). The methylated promoter fragment was re-inserted into the pGL3-Basic vector. The resulting construct was transfected into TIB-73 and RAG cell lines and the luciferase activity was measured as described below. The same unmethylated DNA fragment was used as positive control.
Luciferase activity assays
Cells were washed in phosphate-buffered saline (PBS), scraped from the flasks and resuspended in a Passive Lysis Buffer (PLB from the Promega Dual-Luciferase Reporter Assay System) for luciferase reporter assays. Both firefly and renilla luciferase activities were measured using Turner Designs Luminometer Model TD-20/20 Genetic Reporter System. The transfection efficiency was normalized to renilla luciferase activity.
Statistical analysis
Values were used in paired two-tailed Student’s t-tests to determine statistical significance of difference between 2 groups. Differences were considered significant when p<0.05.
Results
Methylation of the mouse Abcc6 proximal promoter
To investigate the level of in vivo methylation of the proximal promoter, genomic DNA samples were isolated from two groups of tissues. Tail extremity and skin were chosen for their very low level of Abcc6 expression whereas liver and kidney were selected because these tissues present the highest levels of expression. One should note that in spite of being second to the liver in Abcc6 expression levels, the kidney expression is relatively low and somewhat comparable to the expression levels found in tail extremity. Indeed, when compared to liver, kidney, tail extremity and skin expression levels amounted to 5.2%, 0.9% and 0.1% respectively. However, we described in this report the kidney expression of Abcc6 as intermediate between the liver expression and that of the tail extremity and skin.
Methylated genomic DNA was isolated using a methylation promoter PCR kit (Panomics). We found that 88% and 57% of the DNA isolates corresponding to the −152bp/+162bp promoter region was methylated in skin and tail extremity respectively (Figure 1A). In contrast, the proximal region was remarkably hypomethylated in DNA isolates from liver and kidney with values of 4% and 18% respectively (Figure 1A). These results showed an inverse correlation between methylation of the proximal promoter and the level of expression. It is noteworthy that a relatively low and moderate levels of methylation such as those found in kidney (18%) and even in tail extremity tissues (57%) equated to a sharp drop in expression level with 95 to 99% decrease, which clearly underscored the importance of the −152bp to +162bp region for the expression of Abcc6 (Figure 1B). Interestingly, the analysis of DNA regions upstream of the proximal promoter did not reveal any significant methylation in all DNA sample eluates (data not shown) indicating that the distal region of the promoter is not subject to significant methylation.
Figure 1. In vivo methylation of the proximal Abcc6 promoter.
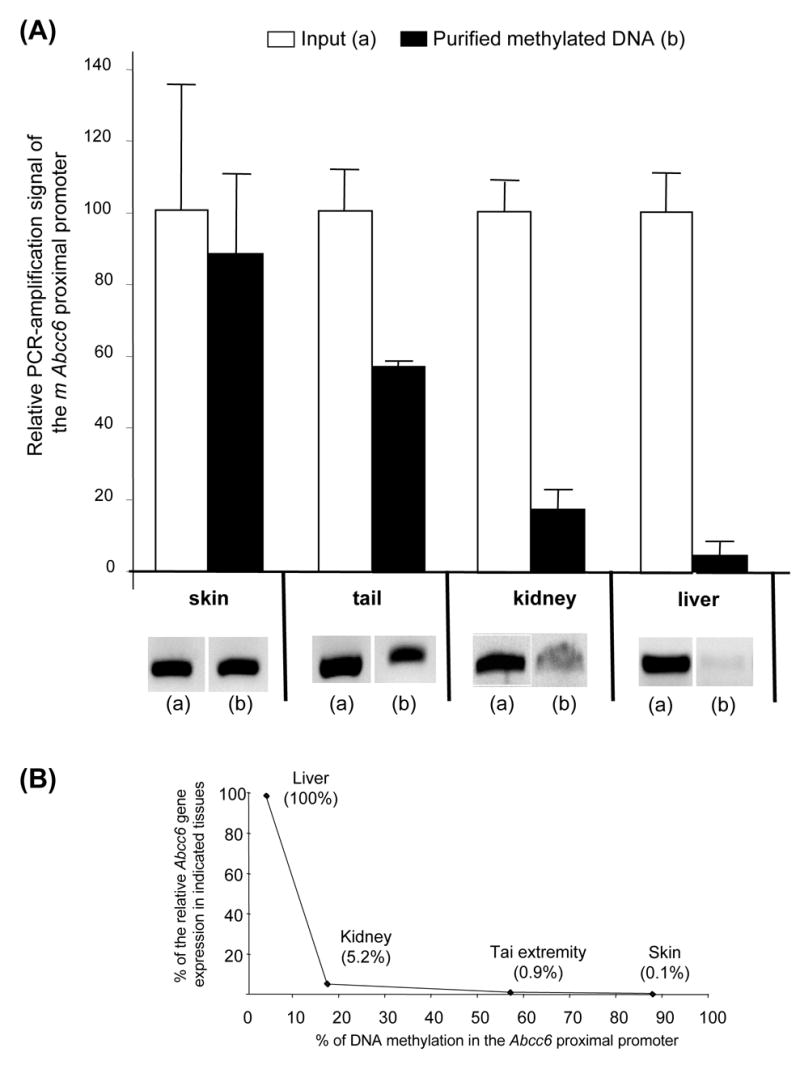
(A) Genomic DNA from tail extremity, skin, kidney and liver was analyzed with the Promoter Methylation PCR kit (Panomics). Methylated DNA fragments isolated from genomic DNA were submitted to PCR-amplification with primers specific to the proximal promoter region (−152/+162bp). The presence of a PCR product reflects the methylation status in the genomic DNA sample. After staining, the intensity of each band was measured using the Kodak Gel Logic 200 imager and associated Molecular Imager software 4.0. The ratio of methylation was calculated by comparing the intensity of the methylated fragment to that present in the total genomic DNA extract (input). The experiment was performed in triplicate. (B) Correlation between the level of Abcc6 gene expression (% of liver expression) and DNA methylation.
Effect of methylation on Abcc6 transcription
To analyze the effect of DNA methylation on the expression of Abcc6 gene in vitro, a reporter plasmid (pGL3) bearing either a CpG methylated proximal promoter sequence or an unmethylated copy was used in transient transfection with hepatocytes (TIB-73) and kidney (RAG) cell lines. DNA fragments corresponding to the −152/+162bp region were excised from the vector, methylated in vitro with the SssI enzyme and inserted into the reporter plasmid. One microgram of the resulting constructs was directly transfected into TIB-73 cells without further bacterial amplification to preserve the methylation profile. We compared the relative luciferase activity of the constructs with the methylated and unmethylated fragments (Figure 2). As previously shown [17], the reporter construct carrying the −152/+162bp fragment (without methylation) induced a high transcriptional activity both in TIB-73 and RAG cells as compared to an empty vector with a 8.8-fold and 7.8-fold induction level, respectively. In contrast, the presence of a methylated fragment in the reporter construct resulted in a sharp decrease of luciferase activity in TIB-73 (99.6%) and in RAG (85.2%) (Figure 2). Similar results were obtained when higher amounts of plasmid constructs (4 μg) were used for transfection (data not shown). These results clearly indicated that the DNA methylation of the Abcc6 proximal promoter region affected the Abcc6 gene transcription efficiency.
Figure 2. Effect of DNA methylation on the Abcc6 proximal promoter activity.
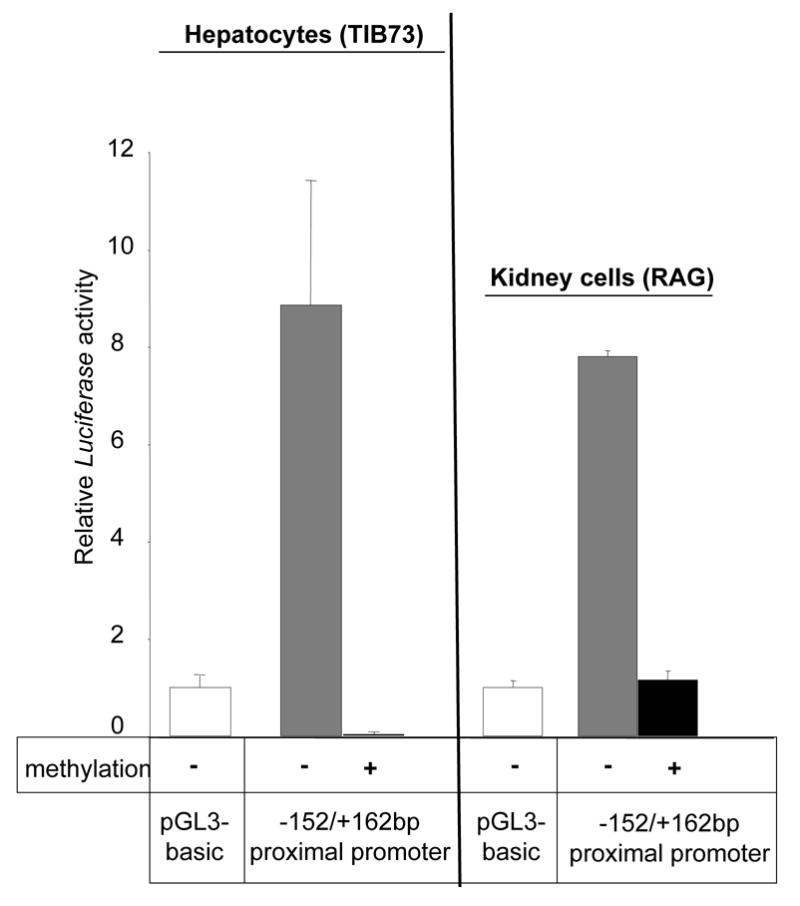
A DNA fragment corresponding to the–152/+162bp fragment was methylated in vitro and inserted into a reporter vector (pGL3). This plasmid construct was transfected into hepatocytes (TIB-73) and renal adenocarcinoma (RAG). These cell lines were transfected with 1μg of plasmid (as indicated) and 0.1 μg of pRL-SV40 plasmid for control purposes, and cultured for 24h. As a positive control, an identical unmethylated DNA fragment was used. Luciferase activity was normalized to the renilla luciferase activity. Each value represents the mean ± standard deviation of at least three independent transfection experiments, each performed in triplicate. The luciferase activities are represented as a ratio of the promoter-less plasmid (pGL3-basic) activity, which was given a value of 1.
Sp1 binding
The Sp1 transcription factor was previously shown to be essential for both human and mouse ABCC6/Abcc6 gene expression [17, 18]. To assess whether the methylation level of the proximal promoter influenced Sp1 binding, the relative association of Sp1 to the Abcc6 proximal promoter was examined by chromatin immunoprecipitation assays (ChIP) in samples derived from liver, kidney and skin. Quantitative PCR (qPCR) was performed as previously described [17] to determine the relative level of association. Our results indicated that the relative Sp1 association with the proximal promoter in liver is more than twice that found in kidney genomic DNA while the level of association in skin was minimal with values similar to the negative control (Figure 3).
Figure 3. Relative association of the Abcc6 promoter with the Sp1 transcription factor in vivo.
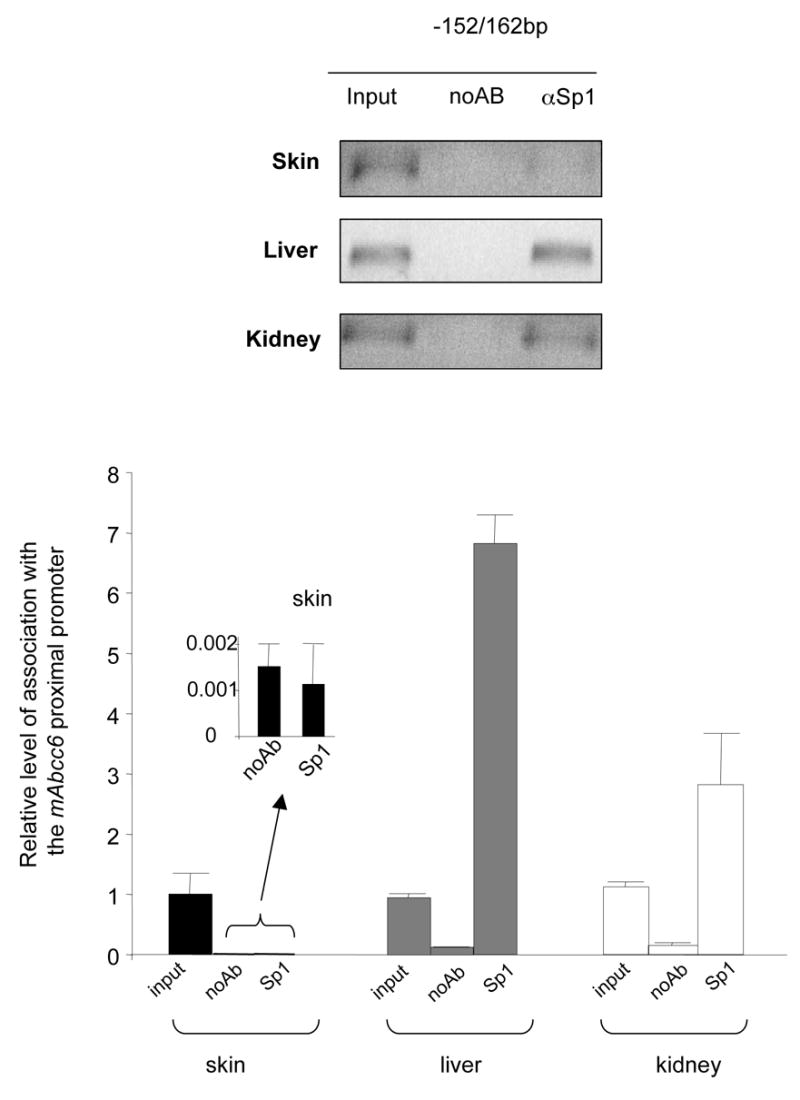
Fragments of chromatin from skin, liver and kidney were immunoprecipitated with anti-Sp1 antibodies and quantified by qPCR using primers specific to the proximal region of the Abcc6 promoter (−152/+162bp). The data was normalized to the total input of DNA used prior to immunoprecipitation and to the control assay (noAB). The differences in association are shown as relative to the total DNA input (input). Data represents the mean of 3 independent immunoprecipitations quantified in duplicate assays. Standard errors are indicated.
Effect of methylation on Sp1 binding
Since the Abcc6 proximal promoter region that binds Sp1 exhibits a low level of methylation in high expressing tissues, we investigated whether Sp1 binding might directly depend on DNA methylation. To test this possibility, electromobility mobility shift assay was carried out using DNA fragments corresponding to the Abcc6 proximal promoter (−152/+162bp fragment). The DNA fragments were amplified by PCR, subjected to in vitro methylation (See Methods) and incubated in the presence of purified Sp1 protein. Non-methylated fragments were used as controls. The control fragments were able to bind Sp1 while the methylated DNA could not as documented on Figure 4. This suggested that CpG dinucleotide methylation decreased the ability of Sp1 to bind to this promoter region.
Figure 4. Sp1 binding to methylated Abcc6proximal promoter.
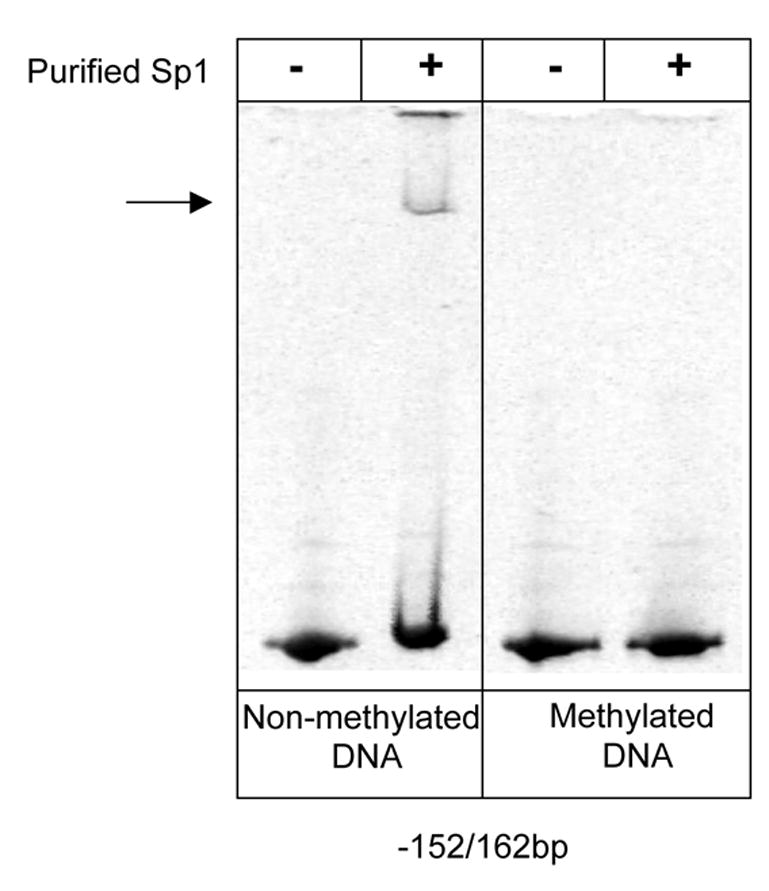
Electromobility shift assays (EMSA) performed with methylated and unmethylated DNA fragments corresponding to the Abcc6 proximal promoter (−152/+162bp) and purified Sp1 proteins. The arrow indicate the Sp1-induced shift.
Discussion
After a comprehensive series of reports on the genetics of pseudoxanthoma elasticum focusing notably on identifying mutations in the ABCC6 gene [21–25] and the generation of mouse models [14, 26], there is now a growing interest in the transcriptional regulation of ABCC6 expression. Indeed, ABCC6 and its rodent orthologs Abcc6 are primarily expressed in liver and kidneys whereas the PXE phenotype affects dermal, vascular and ocular tissues. The apparent discrepancy between the sites of ABCC6 expression and affected tissues as well as recent data has prompted some investigators to suggest that the etiology of PXE might originate in liver and/or kidneys [13, 15, 27]. To date, a few studies focusing on the ABCC6/Abcc6 promoters have been initiated [17–19]. In this report, we present evidences that the tissue-specificity of the mouse Abcc6 gene expression strongly depends on synergy between CpG methyaltion and Sp1 binding on the proximal promoter region (−152/+162bp).
We previously reported that the proximal region of the mouse Abcc6 gene corresponds to a TATA-less promoter containing Sp1 binding sites. These sites may participate in the tissue-specific gene expression along with a liver-specific enhancer region located in the distal promoter region [17]. The proximal promoter region is GC-rich and binds Sp1 which can trans-activate both human and mouse ABCC6/Abcc6 gene in vitro [17, 18]. CpG methylation levels of the human proximal promoter of ABCC6 was recently found inversely correlated to the transcriptional activity in cell lines [19]. Although, no CpG Island per se could be predicted in the mouse proximal promoter sequence, this GC-rich region requires the binding of Sp1 for its basal activity [17]. As DNA methylation can occur on CpG islands as well as on non-CpG sequences at the binding sites of Sp-type transcription factor [28–32], we hypothesized that the transcription of Abcc6 could depend on the methylation status of the proximal promoter in a tissue-specific manner. Indeed, we found such correlation in mouse tissues between high, moderate and low levels of expression (liver, kidney, tail extremity and skin) and the degree of methylation (Figure 1). In addition, we observed that the Abcc6 proximal promoter transcriptional activity in hepatocytes and kidney cells is significantly repressed following in vitro methylation (Figure 2). These results clearly suggested that this epigenetic mechanism is involved in the control of tissue-specific expression of the Abcc6 gene.
Furthermore, as several studies have previously reported that CpG and non-CpG methylation do inhibit the binding of Sp1 proteins [33–35], methylation of the proximal promoter of ABCC6/Abcc6 that contains Sp1 binding sites [17, 18] could directly prevent Sp1 from binding and promoting gene transcription. Our results indeed showed that the Sp1 transcription factor binds to the proximal Abcc6 promoter only in tissues where the promoter is hypomethylated, that is liver and to a lesser extent in kidney (Figure 3). Moreover, we clearly demonstrated that de novo methylation of the proximal promoter blocked the binding of Sp1 to the proximal promoter (Figure 4) strongly suggesting that both Sp1 and methylation synergistically regulate the basal transcriptional activity of the Abcc6 gene thereby controlling tissue-specificity.
In summary, we report the first evidences that the proximal promoter region of the Abcc6 gene is a critical domain that determines the tissue-specific transcription by way of DNA methylation and binding of Sp1 transcription factor. Together with previously published data, one could now describe the transcriptional regulation of ABCC6/Abcc6 as dependent of several distinct genetic and epigenetic mechanisms involving an ubiquitous regulatory protein (Sp1) and DNA methylation determining tissue-specificity in addition to liver and erythroid-specific enhancers (HNF-4a and NF-E2) [17] and at a different level, pro-inflammatory cytokines [18].
Acknowledgments
This study was supported by the National Institutes of Health grant RR16453 and the McKee and Straub trust funds of the Hawaii Community Foundation (20041635, 20060395).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kool M, van der Linden M, de Haas M, Baas F, Borst P. Expression of human MRP6, a homologue of the multidrug resistance protein gene MRP1, in tissues and cancer cells. Cancer Res. 1999;59:175–182. [PubMed] [Google Scholar]
- 2.Madon J, Hagenbuch B, Landmann L, Meier PJ, Stieger B. Transport function and hepatocellular localization of mrp6 in rat liver. Mol Pharmacol. 2000;57:634–641. doi: 10.1124/mol.57.3.634. [DOI] [PubMed] [Google Scholar]
- 3.Beck K, Hayashi K, Nishiguchi B, Le Saux O, Hayashi M, Boyd CD. The distribution of Abcc6 in normal mouse tissues suggests multiple functions for this ABC transporter. J Histochem Cytochem. 2003;51:887–902. doi: 10.1177/002215540305100704. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki Y, Nakano A, Jiang QJ, Pulkkinen L, Uitto J. Tissue-specific expression of the ABCC6 gene. J Invest Dermatol. 2005;125:900–905. doi: 10.1111/j.0022-202X.2005.23897.x. [DOI] [PubMed] [Google Scholar]
- 5.Ilias A, Urban Z, Seidl TL, Le Saux O, Sinko E, Boyd CD, Sarkadi B, Varadi A. Loss of ATP-dependent transport activity in pseudoxanthoma elasticum-associated mutants of human ABCC6 (MRP6) J Biol Chem. 2002;277:16860–16867. doi: 10.1074/jbc.M110918200. [DOI] [PubMed] [Google Scholar]
- 6.Bergen AA, Plomp AS, Schuurman EJ, Terry S, Breuning M, Dauwerse H, Swart J, Kool M, van Soest S, Baas F, ten Brink JB, de Jong PT. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet. 2000;25:228–231. doi: 10.1038/76109. [DOI] [PubMed] [Google Scholar]
- 7.Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, Pasquali-Ronchetti I, Pope FM, Richards A, Terry S, Bercovitch L, de Paepe A, Boyd CD. Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nat Genet. 2000;25:223–227. doi: 10.1038/76102. [DOI] [PubMed] [Google Scholar]
- 8.Ringpfeil F, Lebwohl MG, Christiano AM, Uitto J. Pseudoxanthoma elasticum: mutations in the MRP6 gene encoding a transmembrane ATP-binding cassette (ABC) transporter. Proc Natl Acad Sci U S A. 2000;97:6001–6006. doi: 10.1073/pnas.100041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struk B, Cai L, Zach S, Ji W, Chung J, Lumsden A, Stumm M, Huber M, Schaen L, Kim CA, Goldsmith LA, Viljoen D, Figuera LE, Fuchs W, Munier F, Ramesar R, Hohl D, Richards R, Neldner KH, Lindpaintner K. Mutations of the gene encoding the transmembrane transporter protein ABC-C6 cause pseudoxanthoma elasticum. J Mol Med. 2000;78:282–286. doi: 10.1007/s001090000114. [DOI] [PubMed] [Google Scholar]
- 10.Gheduzzi D, Sammarco R, Quaglino D, Bercovitch L, Terry S, Taylor W, Ronchetti IP. Extracutaneous ultrastructural alterations in pseudoxanthoma elasticum. Ultrastruct Pathol. 2003;27:375–384. [PubMed] [Google Scholar]
- 11.Neldner KH. Pseudoxanthoma elasticum. Int J Dermatol. 1988;27:98–100. doi: 10.1111/j.1365-4362.1988.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 12.Uitto J. The gene family of ABC transporters--novel mutations, new phenotypes. Trends Mol Med. 2005;11:341–343. doi: 10.1016/j.molmed.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Uitto J, Pulkkinen L, Ringpfeil F. Molecular genetics of pseudoxanthoma elasticum: a metabolic disorder at the environment-genome interface? Mol Med Today. 2001;7:13–17. doi: 10.1016/s1471-4914(00)01869-4. [DOI] [PubMed] [Google Scholar]
- 14.Gorgels TG, Hu X, Scheffer GL, van der Wal AC, Toonstra J, de Jong PT, van Kuppevelt TH, Levelt CN, de Wolf A, Loves WJ, Scheper RJ, Peek R, Bergen AA. Disruption of Abcc6 in the mouse: novel insight in the pathogenesis of pseudoxanthoma elasticum. Hum Mol Genet. 2005;14:1763–1773. doi: 10.1093/hmg/ddi183. [DOI] [PubMed] [Google Scholar]
- 15.Le Saux O, Bunda S, Vanwart CM, Douet V, Got L, Martin L, Hinek A. Serum factors from pseudoxanthoma elasticum patients alter elastic fiber formation in vitro. J Invest Dermatol. 2006;126:1497–1505. doi: 10.1038/sj.jid.5700201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Near S, Young K, Connelly PW, Hegele RA. ABCC6 gene polymorphism associated with variation in plasma lipoproteins. J Hum Genet. 2001;46:699–705. doi: 10.1007/s100380170003. [DOI] [PubMed] [Google Scholar]
- 17.Douet V, VanWart CM, Heller MB, Reinhard S, Le Saux O. HNF4alpha and NF-E2 are key transcriptional regulators of the murine Abcc6 gene expression. Biochim Biophys Acta. 2006;1759:426–436. doi: 10.1016/j.bbaexp.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Q, Matsuzaki Y, Li K, Uitto J. Transcriptional regulation and characterization of the promoter region of the human ABCC6 gene. J Invest Dermatol. 2006;126:325–335. doi: 10.1038/sj.jid.5700065. [DOI] [PubMed] [Google Scholar]
- 19.Aranyi T, Ratajewski M, Bardoczy V, Pulaski L, Bors A, Tordai A, Varadi A. Identification of a DNA methylation-dependent activator sequence in the pseudoxanthoma elasticum gene, ABCC6. J Biol Chem. 2005;280:18643–18650. doi: 10.1074/jbc.M501139200. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 21.Chassaing N, Martin L, Calvas P, Le Bert M, Hovnanian A. Pseudoxanthoma elasticum: a clinical, pathophysiological and genetic update including 11 novel ABCC6 mutations. J Med Genet. 2005;42:881–892. doi: 10.1136/jmg.2004.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu X, Plomp A, Wijnholds J, Ten Brink J, Van Soest S, Van Den Born LI, Leys A, Peek R, De Jong PT, Bergen AA. ABCC6/MRP6 mutations: further insight into the molecular pathology of pseudoxanthoma elasticum. Eur J Hum Genet. 2003;11:215–224. doi: 10.1038/sj.ejhg.5200953. [DOI] [PubMed] [Google Scholar]
- 23.Le Saux O, Beck K, Sachsinger C, Silvestri C, Treiber C, Goring HH, Johnson EW, De Paepe A, Pope FM, Pasquali-Ronchetti I, Bercovitch L, Terry S, Boyd CD. A spectrum of abcc6 mutations is responsible for pseudoxanthoma elasticum. Am J Hum Genet. 2001;69:749–764. doi: 10.1086/323704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miksch S, Lumsden A, Guenther UP, Foernzler D, Christen-Zach S, Daugherty C, Ramesar RK, Lebwohl M, Hohl D, Neldner KH, Lindpaintner K, Richards RI, Struk B. Molecular genetics of pseudoxanthoma elasticum: type and frequency of mutations in ABCC6. Hum Mutat. 2005;26:235–248. doi: 10.1002/humu.20206. [DOI] [PubMed] [Google Scholar]
- 25.Ringpfeil F, McGuigan K, Fuchsel L, Kozic H, Larralde M, Lebwohl M, Uitto J. Pseudoxanthoma elasticum is a recessive disease characterized by compound heterozygosity. J Invest Dermatol. 2006;126:782–786. doi: 10.1038/sj.jid.5700115. [DOI] [PubMed] [Google Scholar]
- 26.Klement JF, Matsuzaki Y, Jiang QJ, Terlizzi J, Choi HY, Fujimoto N, Li K, Pulkkinen L, Birk DE, Sundberg JP, Uitto J. Targeted ablation of the abcc6 gene results in ectopic mineralization of connective tissues. Mol Cell Biol. 2005;25:8299–8310. doi: 10.1128/MCB.25.18.8299-8310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Q, Uitto J. Pseudoxanthoma elasticum: a metabolic disease? J Invest Dermatol. 2006;126:1440–1441. doi: 10.1038/sj.jid.5700267. [DOI] [PubMed] [Google Scholar]
- 28.Lee LT, Tan-Un KC, Pang RT, Lam DT, Chow BK. Regulation of the human secretin gene is controlled by the combined effects of CpG methylation, Sp1/Sp3 ratio, and the E-box element. Mol Endocrinol. 2004;18:1740–1755. doi: 10.1210/me.2003-0461. [DOI] [PubMed] [Google Scholar]
- 29.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 30.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi BZ, Cedar H. DNA methylation represses transcription in vivo. Nat Genet. 1999;22:203–206. doi: 10.1038/9727. [DOI] [PubMed] [Google Scholar]
- 32.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 33.Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene. 1997;195:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 34.Inoue S, Oishi M. Effects of methylation of non-CpG sequence in the promoter region on the expression of human synaptotagmin XI (syt11) Gene. 2005;348:123–134. doi: 10.1016/j.gene.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 35.Kitazawa S, Kitazawa R, Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J Biol Chem. 1999;274:28787–28793. doi: 10.1074/jbc.274.40.28787. [DOI] [PubMed] [Google Scholar]


