Abstract
The rat glucocorticoid induced receptor (rGIR) is an orphan G protein-coupled receptor awaiting pharmacological characterization. Among known receptors, rGIR exhibits highest sequence similarity to the Neuropeptide Y (NPY) - Y2 receptor (38–40%). The pharmacological profile of rGIR was investigated using 125I-PYY3-36, a Y2-preferring radioligand and several NPY analogs. rGIR displayed a similar displacement profile as reported for the Y2 receptor, in that the Y2-selective C terminus fragments of NPY and PYY (NPY3-36 and PYY3-36) showed high affinity binding and activation of rGIR (low nanomolar range). The rank order potency for displacement was NPY3-36 > PYY3-36 = NPY > NPY 13-36 > Ac, Leu NPY24-36 > [D-Trp32]-NPY > Leu31, Pro34- NPY= hPP). NPY and Y2-selective agonists NPY3-36 and PYY3-36 led to significant activation of 35S-GTPγS binding to rGIR transfected cells. BIIE0246, a specific Y2 antagonist, displaced 125I-PYY3-36 binding to rGIR with high affinity (95 nM). Activation of 35S-GTPγS binding by Y2- selective agonist in rGIR transfected cells was also completely abolished by BIIE0246. Our data report, for the first time, an interaction of NPY ligands with rGIR expressed in vitro, and indicate similarities between GIR and the NPY-Y2 receptor.
Keywords: Glucocorticoid induced receptor (GIR), NPY, Y2 receptor, NPY3-36
1. Introduction
The glucocorticoid-induced receptor (GIR), also known as GPR83, GPR72 or JP05, is an orphan G protein-coupled receptor awaiting pharmacological characterization. GIR was originally identified as a stress-responsive element from the murine T-cell line WEHI-7TG and normal thymocytes treated with glucocorticoids and forskolin [3,10]. Mouse GIR mRNA was detected at high levels in the brain and thymus, and its distribution in brain was found to be enriched in the dorsal and ventral striatum, the forebrain limbic structures and the hypothalamic nuclei [22]. The human GIR gene, (87% identical to mouse) also shows high levels of expression in the brain, and has been localized to chromosome 11q21 [5,20]. The rat GIR (rGIR) was cloned from prefrontal cortex in our laboratory, and was shown to be up-regulated following chronic amphetamine [30]. The rat GIR elicits significant sequence identities with mouse GIR (99%) and human GIR (88%). Expression of rGIR is predominant in forebrain limbic and thalamic regions, with a more restricted distribution in hindbrain areas such as the trigeminal nucleus and median raphe nucleus [24]. GIR mRNA distribution in the rat indicates a potential role of this receptor in the control of feeding and ingestive behavior, regulation of stress and emotional behavior, learning and memory, and drug reinforcement and reward.
To date, there are no reports on the pharmacological specificities of GIR. Previous attempts towards ligand binding to human GIR (GPR72) expressed in vitro have been unsuccessful [20]. Sequence comparison of rGIR with other GPCR families indicates similarities with the Neuropeptide Y, Tachykinin and Prolactin Releasing Peptide receptor families (33–38% overall amino acid identity). Within the receptor subtypes of the NPY family, rGIR shows highest homology to the Neuropeptide Y- Y2 receptor (38% – 40% overall), with higher identity in transmembrane (TM) domains 5, 6 and 7 (50–55%). Previous evidence from site-directed mutagenesis and molecular modeling studies with Y2 receptors have shown TM 5 and 6 to be important for binding of agonist and antagonists [4]. The GIR sequence also contains residues that have been implicated in antagonist binding and are specific to NPY- Y2 receptors [25].
The similarity between GIR and NPY- Y2 receptor sequence and the presence of Y2 specific residues in GIR led us to hypothesize that NPY ligands, particularly the Y2 selective compounds may interact with GIR.
In the present study, we tested the binding of several NPY analogs to GIR expressed in vitro. Here, we provide the first evidence that the glucocorticoid-induced receptor can bind NPY and NPY C terminus fragments with nanomolar affinity and shows pharmacological specificity resembling the NPY- Y2 receptor. Data from these studies has been published in abstract form [25].
2. Experimental Methods
2.1 Materials
(125I)-Peptide YY (PYY) (2,200 Ci/mmole) was purchased from Perkin Elmer Life Sciences, Boston, MA. (125I)-Peptide YY3-36 (PYY3-36), 1600–1800 Ci/mole, was either purchased from Phoenix Laboratories (Belmont, CA), or synthesized by iodinating commercial PYY3-36 following the chloramine-T procedure [21]. These preparations of (125I)-PYY3-36 showed very similar competition profiles with Y2-selective Neuropeptide Y (NPY) and Peptide YY (PYY) analogs. All NPY peptide analogs were purchased from American Peptide Company, Sunnyvale, CA. COS-7 cells were procured from ATCC, Manassas, VA. pIRES2-GFP vector was purchased from Clontech, Labs, Mountain View, CA. All other chemicals and tissue culture supplies were obtained from Sigma Chemical Company and GIBCO-BRL respectively.
2.2 Construction of pIRES2-EGFP-rGIR plasmid and transfection of COS-7 cells
The full length rat GIR cDNA (Genbank accession number AY029071 [30] was ligated into expression vector plasmid, pIRES2-EGFP using the primers (5′-CGTGAATTCGCCGCCACCAT GAATGT CCCTCCTG-3′ forward) and (5′-GATGAATTCACTCACGGCCACAGTGGGTTC-3′ reverse). COS-7 cells were transiently transfected with pIRES2-EGFP-rGIR. Plasmid DNA (30 μg/plate) was mixed with Lipofectamine 2000 (60 μl/plate) in OPTI-MEM media and incubated with confluent COS-7 cells grown in 100 mm2 petri plates. Cells were harvested for particulate preparation 24 hr post transfection.
2.3 Detection of rGIR expression by RT-PCR and Immunofluorescence Microscopy
rGIR mRNA expression in COS cells was investigated using reverse transcription-polymerase chain reaction (RT-PCR) experiments. Total RNA was isolated from cells, 24 hour post transfection, by a single step guanidine thiocyanate-phenol extraction using the TRI-REAGENT (Molecular Research Center, Cincinnati, OH) following the manufacturer instructions. The concentrations of RNA samples were determined by spectrophotometric measurements at 260 and 280 nm. First strand cDNA was synthesized from total RNA (5 μg) using a random hexamer primer (Invitrogen, Carlsbad, CA) following manufacturer instructions. rGIR specific primers (5′-TAC TTT GCC TTC CAC TGG TT 3′, 1283–1303 bp and 5′-CTA ACT CAC GGC CAC AGT GGG TT 3′, 1550–1568 bp, GenBank accession number AY029071, were synthesized by Integrated DNA Technologies, Coralville, IA. PCR was run for 1 cycle at 95°C for 4 minutes, 40 cycles at 95 °C for 30 sec., 57 °C for 30 sec., 72 °C for 30 sec, followed by 1 cycle at 72 °C at 10 min. PCR products were electrophoresed on 1.2 % agarose gel and stained with ethidium bromide.
Expression of rGIR protein was assessed by immunofluorescence. Cells plated on poly-D-lysine pre-coated chamber slides were transiently transfected with the rGIR carrying plasmid. After 24 hours, cells were washed twice with phosphate buffered saline (PBS) and fixed for 30 min at 4 °C with 4% paraformaldehyde and 0.1 % Triton-X 100 in PBS. Following a 30 min incubation in blocking solution (3% BSA, 10% Normal Goat serum, 0.1 % Triton X-100 in PBS) and subsequent washes, cells were incubated (overnight, 4 °C) with the anti-GPR72 peptide antibody (1:100, LifeSpan Biosciences, Seattle, WA). After three washes in PBS, cells were incubated with goat anti-rabbit IgG conjugated to Alexa Fluor-595 (1: 200, Molecular Probes, Eugene, Oregon, USA) for 1 hour, RT. Cells were then washed three times in PBS before being mounted on coverslips with gelvatol. Fluorescence microscopy was performed using Spotcam1 CCD camera coupled to a Nikon upright scope (Nikon, Tokyo, Japan).
2.4 Preparation of cell particulates
Cells were harvested 24 hours post-transfection and membranes were prepared as described previously [19], with minor modifications. Briefly, cells were washed twice with PBS and collected in ice cold 50 mM HEPES-KOH, pH 7.4, supplemented with 1X protease inhibitor cocktail (Boehringer Mannheim). The cell suspension was homogenized on ice, then centrifuged (15,000 rpm; 15 min; 4 °C). Following two washes and re-centrifugation (15,000 rpm; 15 min; 4 °C), the final pellet was stored dry at −80 °C until reconstitution before use. Membrane protein content was determined with the Coomassie protein assay kit (Pierce, Rockford, IL).
2.5 125I-pPYY and 125I-pPYY3-36 binding assays
Radioligand binding assays were performed as described before [19] with minor modifications. The assay buffer contained 20 mM HEPES-NaOH (pH 7.4), 4 mM CaCl2, 2 mM MgCl2, 50 μM ATP, 0.025%of bacitracin, 10 μg/ml each of leupeptin, pepstatin, aprotinin, chymostatin and antipain, 1 mM phenylmethylsufonyl fluoride, and 2 mg/ml of proteinase-free bovine serum albumin. Briefly, binding assays were performed in the binding buffer containing 60 μg membrane protein and 100 pM radioligand, in a total volume of 200 μl, incubating for 90 min at room temperature. [125I]-PYY was used in initial experiments for detecting specific binding to rGIR-transfected cells. All the experiments for characterizing competition parameters were performed using [125I]-PYY3-36, as the signal to noise ratio was better for this ligand than [125I]-PYY. Nonspecific binding was determined in the presence of 2 μM of the Y2 antagonist BIIE0246 (Tocris, Ellisville, MO).
Competition binding assays were carried out in the presence of various concentrations of competing peptides (0.01 nM-1 μM). Following 90 min of incubation, unbound radioactivity was separated by centrifugation at 16,000 x g, 15 min, 4° C. After discarding the supernatants, pellets were washed by cold assay buffer prior to counting in a Packard γ counter. All data were analyzed by nonlinear regression analysis (GraphPad Prism software, San Diego, CA).
2.7 35S-GTPγ S Binding Assay
Receptor-linked G protein activation by agonists at rGIR was determined by measuring the stimulation of 35S-GTPγS (1332 Ci/mmole; Perkin Elmer Life Sciences, Norton, OH). Particulates from rGIR-transfected COS-7 were prepared as described above. Pre-activation of the binding sites in the presence of NPY analogs was done for 90 min at 28 °C in the assay buffer containing10 mM HEPES-NaOH pH 7.35, 100 mM NaCl, 0.2% BSA, 1 mM diisopropylfluorophosphate, 4 mM MgCl2, 50 μM EDTA, 0.05% bacitracin and 3 μM GDP. This was followed by addition of GTPγ35S to 0.3 nM in the final volume of 200 μl, and binding for 40 min at 28 °C. The non-specific binding was defined at 10 μM cold GTPγS. The assay was terminated by centrifugation at 4 °C and washing, as for the receptor assay above. The resulting pellets were solubilized in 0.5 ml 2 % SDS, pH 8.8 and counted using BudgetSolve liquid scintillation medium in a Packard liquid scintillation counter.
2.8 Data Analysis
Group data are expressed as mean ± SEM. Statistical analysis was conducted using unpaired Student’s t test or analysis of variance (ANOVA) followed by Tukey’s post hoc analysis (Prism software, GraphPad, San Diego,CA). Saturation and competition binding data was analyzed by using non-linear regression analysis (Prism software, GraphPad, San Diego, CA). A probability of < 0.05 was considered statistically significant. Sequence alignment (Fig 1) was done using the ClustalW alignment algorithm.
Fig 1.
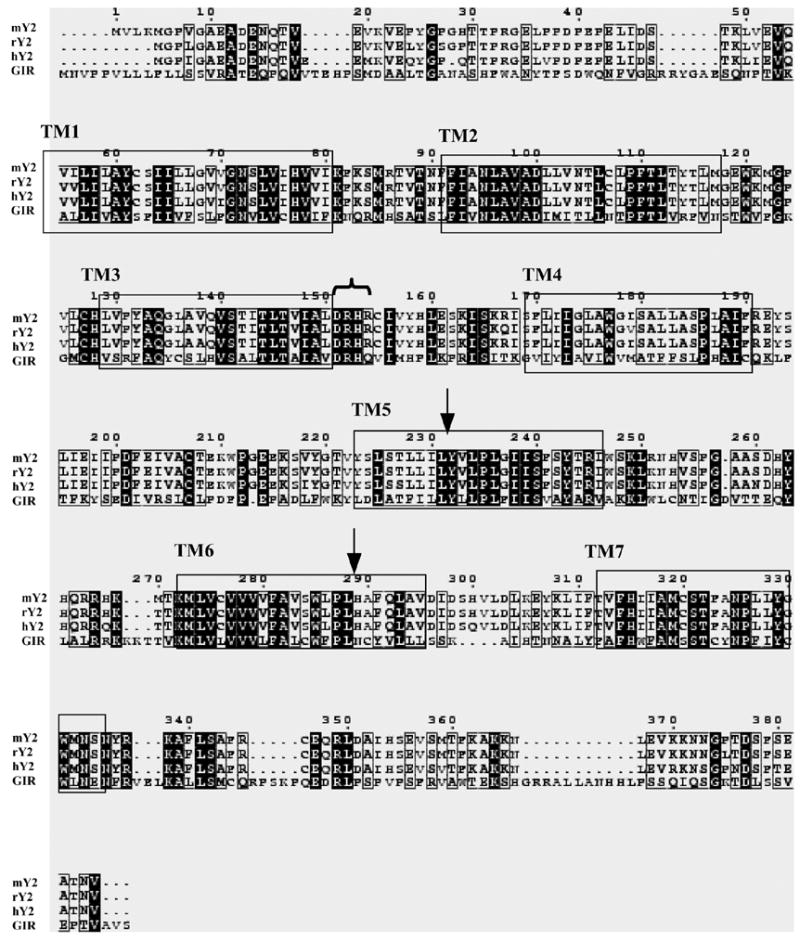
Sequence alignment of rGIR polypeptide (accession number Q8VHD7) with the neuropeptide Y-Y2 receptor sequences from mouse (mY2; accession number P97295), rat (rY2; accession number Q9ERC0) and human (hY2; accession number P49146) subtypes. The alignments were created using multiple sequence alignment software using Clustal W algorithm. Conserved residues between Y2 and rGIR sequences are highlighted in black. Boxes mark the putative TM regions as predicted from comparisons with the crystal structure of bovine rhodopsin. Certain residues conserved in all Y2 receptors and present in rGIR are marked (
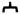 DRH sequence after TM-3 and ↓ for conserved leucine residues in TM5 and TM6 important for Y2 antagonist binding). Dashes show gaps introduced to optimize the alignment.
DRH sequence after TM-3 and ↓ for conserved leucine residues in TM5 and TM6 important for Y2 antagonist binding). Dashes show gaps introduced to optimize the alignment.
3. Results
3.1 Amino acid sequence comparison of rGIR with the NPY-Y2 receptor
rGIR encodes a 422 amino acid protein with sequence similarities with the NPY (31–38%), Neuropeptide FF (36%) and tachykinin (31–34%) receptor families. Among the NPY receptor subtypes, the highest amino acid identity is observed with the NPY- Y2 receptors (38%). Fig 1 shows the amino acid alignment of rGIR with human, rat and mouse NPY- Y2 receptors. Although the overall amino acid identity between Y2 receptors and rGIR is 38%, TM domains 5, 6 and 7 show higher amino acid identity (50–55%). rGIR also contains certain amino acid residues specific to the NPY- Y2 receptors. Among these is the tri-peptide “DRH” motif located after the third transmembrane domain (see Fig 1). In addition, the rGIR sequence also has specific leucine residues (see arrows in Fig 1) corresponding to Leu231 in TM5 and Leu288 in TM6 of the mouse Y2 receptor sequence. These residues are conserved and specific to all mammalian Y2 receptors, but not in avian Y2 receptor [4].
3.2 Detection of rGIR expression in COS-7 cells
Expression of rGIR in transfected cells was detected by RT-PCR and immunocytochemistry. Following RT-PCR, a strong band of expected size, (290 bp) was detected in rGIR-transfected cells (Fig 2a). No signal was observed in vector transfected cells (Fig 2a). A similar sized product was also obtained with rat hypothalamic RNA, that was used as a positive control for rGIR.
Fig 2.
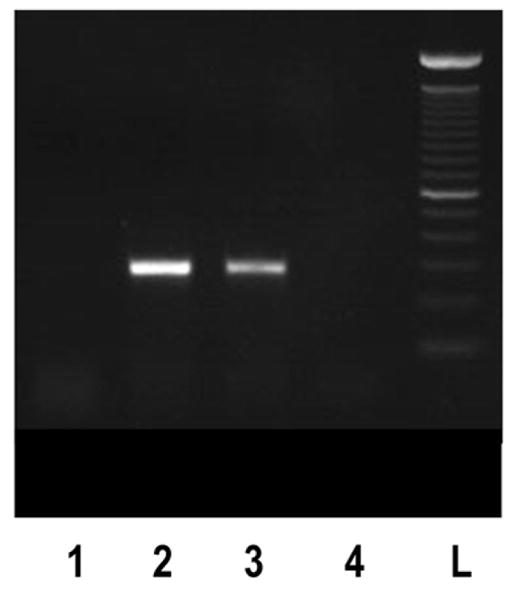
Expression of rGIR mRNA and protein in COS-7 cells. (A) RT-PCR analyses of rGIR expression in COS-7 cells. Total RNA from cells was reverse transcribed and PCR was performed using primers specific to GIR. The expected size of band was 290 bp. Sample lanes are labeled as follows; Lane 1, Mock plasmid transfected COS-7 cells, Lane 2 pIRES2-EGFP-rGIR plasmid transfected COS-7 cells, Lane 3, rat hypothalamus, and Lane 4, blank. L= 100 bp ladder. (B) Detection of GIR protein by immunofluorescence. COS-7 cells grown in chamber slides were transfected with pIRES2-EGFP-rGIR plasmid (Panel A) or mock plasmid (Panel B) and processed for immunochemical analysis 24 hr post-transfection. Experiments were repeated with all conditions run in duplicate wells.
Immunochemical detection of rGIR protein in transfected COS cells was performed using an anti GIR peptide antibody. Strong immunofluorescence was observed only in rGIR-transfected cells whereas low background fluorescence was seen in vector transfected cells (Fig 2b). rGIR expression levels were high following 24 hours of transfection, but decreased after 48 hours (data not shown); therefore all binding and functional studies were performed 24 hours post transfection.
3.3 Pharmacological Characterization of rGIR
3.3.1. Binding Studies
The pIRES2-EGFP-rGIR plasmid was transiently transfected into COS-7 cells, that are null for NPY binding [23] and do not express endogenous NPY-Y2 receptors [27]. Specific, displaceable binding was observed in rGIR transfected cells using 125I-PYY as radioligand and Y2-specific agonist, NPY3-36 as displacer (specific binding between 50–75% of total binding); whereas negligible binding was observed in mock-transfected COS-7 cells (see Fig 3).
Fig 3.
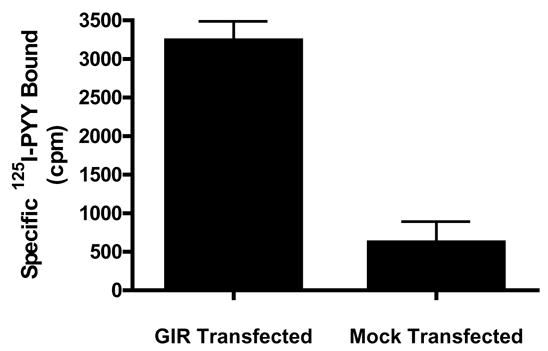
Detection of specific 125I-Peptide YY binding to rGIR-transfected COS-7 cells. 125I-Peptide YY binding was measured in the absence and presence of cold Y2 selective agonist NPY3-36 (1 μM) to particulates from COS-7 cells transiently transfected with pIRES2-GFP-rGIR or mock plasmid. Data are representative of three independent assays.
125I-PYY3-36 binding to rGIR expressing cells was competed with various NPY derivatives and analogs. As shown in Fig. 4, NPY- Y2 selective agonists were the most effective displacers of 125I-PYY3-36 binding with a rank order potency of NPY3-36 > PYY3-36 = NPY > NPY 13-36 > Ac, Leu NPY24-36 > [D-Trp32]-NPY > Leu31, Pro34- NPY= hPP. Interestingly, the Y2-specific antagonist BIIE0246 elicited displacement of 125I-PYY3-36 binding with moderately high affinity (95.139 nM). Table 1 lists the IC50 values for the competition curves depicted in Fig. 4. The displacement profile of rGIR is similar to that observed for the NPY- Y2 receptor expressed in vitro in the, a) observed preference for NPY C-terminal fragments than NPY agonists preferring the Y1 ([Leu31, Pro34]-NPY), Y4 (h-PP) and Y5 ([D-Trp32]-NPY) receptors, and, b) interaction with Y2-specific antagonist BIIE0246.
Fig 4.
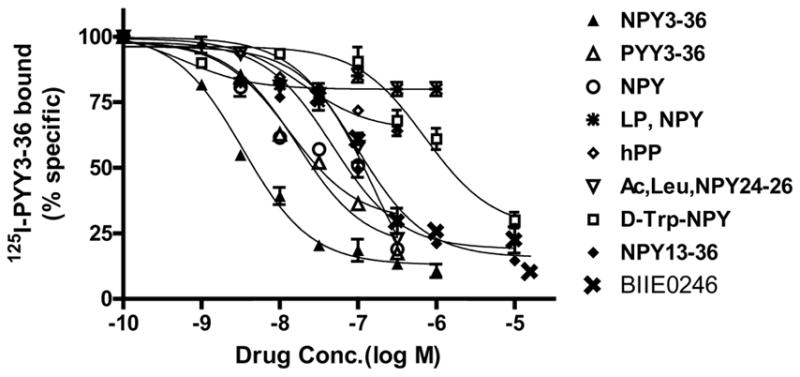
Comparative competition binding profile of NPY derivatives against 125I-pPYY3-36 binding in homogenates from COS-7 cells transiently transfected with rGIR plasmid. Data are from a representative experiment repeated three times. Each point represents mean ± S.E.M. of triplicate determinations. Values are expressed as the percentage of specific binding. IC50 values are shown in Table 1.
Table 1.
Binding properties of NPY and PYY analogs and BIIE0246 to rGIR
| Competing Agent | IC50 values (95% confidence intervals) nM |
|---|---|
| NPY3-36 | 3.36 (2.8–3.96) |
| PYY3-36 | 16.5 (11.4–24.0) |
| NPY | 13.0 (5.17–32.38) |
| NPY13-36 | 46.8 (31.5–65.6) |
| BIIE0246 | 95.14 (66.15–136.83) |
| Ac, Leu NPY24-36 | 164.5 (86.037–313.6) |
| [D Trp32] NPY | 713.49 (433.6–1174.85) |
| Leu,Pro NPY | > 1000 |
| hPP | > 1000 |
IC50 values and 95 % confidence intervals were calculated from competition binding data (3 experiments) using the GraphPad Prism software with a fit to a sigmoidal concentration-response curve.
3.3.2. GTPγ-35S binding upon agonist activation
When membranes from rGIR-transfected COS cells were tested for their capability to bind GTPγ-35S after exposure to NPY agonists, significant specific binding (30–60% over basal levels) was observed (Fig 5 A–C). This indicates that rGIR expressed in COS cells can interact with G proteins upon agonist activation. The ability of various peptides to activate G-protein binding to rGIR was in agreement with their binding interactions with rGIR. Thus, NPY, PYY and their C-terminus fragments showed significant increases in GTPγ-35S binding to rGIR, while the Y1 and Y5 selective peptides that showed poor binding to rGIR were also unable to activate GTPγ-35S binding to rGIR (Fig 5A). Although activation with N-acetyl [Leu(28,31)] NPY 24-36 did not reach statistical significance, the trend for activation of GTPγ-35S binding was evident (see Fig 5A). Exposure to increasing concentrations of PYY3-36 and NPY (3–300 nM) led to a dose dependent increase in GTPγ-35S binding (Fig 5B), IC50 values PYY3-36 = 22.6 ± 0.14 nM; NPY=33.9 ± 0.08 nM). The Y2 specific antagonist, BIIE0246 significantly reversed PYY3-36 induced GTPγ-35S binding to rGIR transfected COS membranes (Fig 5C).
Fig 5.
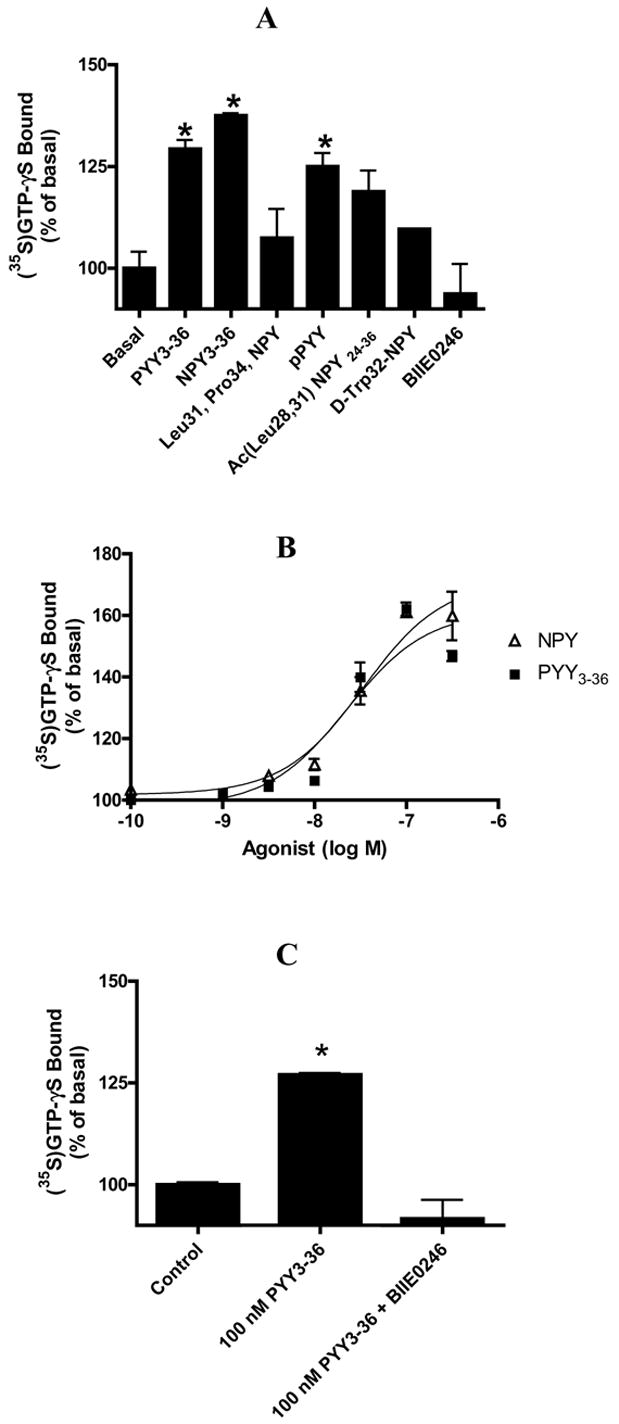
Agonist induced stimulation of rGIR receptor-mediated G protein activation. (A) Effect of NPY and PYY analogs and Y2 antagonist, BIIE0246 on GTPgamma35S binding to particulates from rGIR-transfected COS-7 cells. . (B) Agonist concentration-response curves at particulates from r-GIR-transfected COS-7 cells. (C) Reversal of PYY3-36 induced stimulation of GTPγ35S binding to rGIR-transfected COS-7 cells by Y2 antagonist BIIE0246 (10 μM). Concentration of agonists in A, B and C was 100 nM. GTPγ35S binding is expressed as percentage over basal (no agonist). Points are means of triplicate determinations from representative experiments repeated thrice. * p < 0.05 versus basal.
4. Discussion
This is the first report demonstrating specific, high affinity binding of NPY compounds to rGIR. The rGIR receptor shows structural as well as pharmacological similarity to the Y2 receptor and may be classified as a novel “Y2-like” receptor.
4.1 GIR and NPY- Y2 receptor: structural similarities
High sequence homology between rGIR and Y2 was observed in TM 5 and 6 domains, which are important for binding of agonists and antagonists to NPY receptors, as revealed by site-directed mutagenesis studies [4,26]. The amino acid sequence of rGIR also contains the DRH sequence at the junction of TM-3 and intracellular loop 2, as found in Y2 receptors. Among NPY receptors, the DRH motif is highly specific to all mammalian and non-mammalian Y2 receptors [31]. rGIR sequence also contains Leu227 and Leu288which have been reported to interact directly with specific Y2 antagonist, BIIE0246 [4]. These residues are highly conserved in all mammalian Y2 receptors but not chick Y2 and other NPY receptor subtypes [4].
Although the overall homology between rGIR and Y2 receptor is moderate (38%), the presence of pharmacologically relevant residues in rGIR makes it an attractive candidate for classification as a novel “Y2-like receptor.” This is supported by our pharmacological data as described in section 4.2.
4.2 Pharmacological specificities of rGIR: analogy to the NPY-Y2 receptor
Competition experiments with various NPY ligands revealed a preference of rGIR for NPY C-terminus fragments similar to that reported for NPY- Y2 receptors. The observed IC50 value for Y2 agonist, NPY3-36 in our study (3.36 nM) is similar to affinity values reported for the Y2 receptor expressed in vitro [23,27]. The affinity of NPY for rGIR, however, is 3–30 fold lower than that reported for the Y2 receptors in in vitro expression systems [7–9,23]. Thus, rGIR is distinct from Y2 receptors in its preference for N-truncated fragments of NPY, over the NPY holopeptide. Shorter fragments of NPY such as NPY13-36 and Ac, Leu, NPY24-36 also displaced 125I-PYY3-36 binding to rGIR, albeit with lower affinity than NPY3-36 (46.8 and 165 nM, respectively). The displacement of 125I-PYY3-36 binding by Y2- specific antagonist BIIE0246 lends support to the “Y2-like” profile of rGIR. It may be speculated that the leucine residues that facilitate the interaction of BIIE0246 with the Y2 receptors [4], and are present in rGIR sequence may also be involved in BIIE0246 binding to rGIR. As reported for Y2 receptors, Y1 and Y5 selective agonists, [Leu31, Pro34] NPY and [D-Trp32] NPY had poor affinity for rGIR. Additionally, the poor affinity of Y4 selective compound, pancreatic polypeptide (PP) for rGIR, further supports the “Y2-like” nature of rGIR.
To measure the level of G-protein activation following agonist occupation of rGIR, we conducted the [35S] GTPγS assay. A significant activation of GTP binding by NPY, PYY and their C terminus fragments indicates that these peptides can not only bind to rGIR but also induce functional coupling to G proteins. The peptide selectivity and profile for stimulation of rGIR receptors was similar to that observed for binding. Thus Y1, Y4 and Y5 preferring compounds did not show significant GTP-γS binding to rGIR. Furthermore, complete reversal of agonist induced GTP-γS binding in GIR cells by BIIE0246, a specific Y2 antagonist strongly supports the specificity of peptide stimulation at rGIR and the “Y2-like” pharmacology of rGIR.
4.3 Is rGIR a NPY receptor subtype?
Based on our pharmacological data, rGIR appears distinct from previously characterized NPY receptor subtypes primarily due to its lower affinity for NPY, and poor affinity for hPP. The Y1, Y2, Y3, Y5 and Y7 receptors bind NPY with sub-nanomolar affinities [6,15] in contrast to rGIR (13 nM). The question arises whether NPY can bind rGIR under physiological conditions. Basal extracellular NPY levels in the hypothalamic area range from 10 pg/μl (2.4 nM) measured by microdialysis [28] to as high as 150 pg/μg protein (7 μM) as measured by push-pull cannula (PPC) [13]. Levels are reported to be almost twice as high after depolarization [28]. Although a comparison between ligand affinities measured using in vitro expression systems cannot be directly co-related with in vivo conditions, a physiological interaction between NPY and rGIR appears feasible based on the availability of sufficient phasic NPY levels.
The classification of GIR as a NPY receptor subtype is debatable. Recent phylogenetic analyses have shown no evolutionary links between GIR and the NPY family (Larhammer et. al., unpublished observations), suggesting that rGIR may not represent a novel NPY receptor subtype. Until a specific, higher affinity endogenous ligand is identified for GIR, it can be classified as a distinct “Y2-like” receptor that shows preference for C terminus fragments of NPY.
4.4 Physiological implications of interaction between NPY ligands and rGIR
The high affinity binding of NPY compounds to GIR may have important physiological implications. NPY receptors, particularly the Y2 subtype, have been implicated in regulation of food intake, control of excitatory neurotransmission and epilepsy, circadian rhythms, learning and memory, stress-anxiety and angiogenesis [1,11,12,29]. rGIR is highly expressed in brain regions implicated in the motor as well as sensory aspects of feeding, such as various hypothalamic nuclei and hindbrain regions like the nucleus of solitary tract (NTS), dorsal motor nucleus of vagus nerve (DMX), and motor trigeminal, facial and hypoglossal nuclei [24]. Our pharmacological data supports the morphological evidence on rGIR expression in regions regulating feeding behavior. High affinity binding of PYY3-36, an anorectic peptide, with rGIR may have relevance in satiety signaling. Interestingly, rGIR is expressed in the arcuate and NTS, two areas which are known to mediate the anorectic effects of PYY3-36 [2,16]. High blood-brain barrier permeability been reported for PYY3-36 [18], which may facilitate its interaction with rGIR in a manner similar to Y2 receptors.
Within the hippocampus high expression of rGIR is found in large neurons located in proximity to the pyramidal cells [24], probably represent GABAergic interneurons [32]. NPY expression is also reported to be abundant in GABAergic interneurons and mossy fibers in the hippocampus. A significant role of hippocampal NPY and NPY-Y2 receptors has been recognized in the control of excitatory neurotransmission and epilepsy. An interaction with rGIR may contribute to the anticonvulsant and inhibitory effects of NPY in the hippocampus. However, co-localization studies on NPY, NPY-Y2 and rGIR are needed to support this.
rGIR binds NPY C-terminus fragments with high affinity. Presence of truncated forms of NPY (NPY3-36) and PYY (PYY3-36) have been reported to exist endogenously [9,14,17]. Recent studies have described the importance of N terminal dipeptide cleavage of NPY in regulation of the functional effects of NPY [14]. Thus, NPY3-36 generated from NPY may have preferential binding to Y2, Y5, and rGIR over the Y1 receptor, resulting in the regulation of NPY transmission via Y1 receptor.
The interaction of NPY ligands with rGIR becomes extremely relevant when studying the effects of NPY agonist administration on behavior. Most investigators administer intracerebroventricular (i.c.v.) or regional injections in the range of picomoles to nanomoles (nM – μM concentrations). At these concentrations, NPY agonists are likely to interact with rGIR, in addition to NPY receptor subtypes. Therefore, the contribution of rGIR in the behavioral effects of NPY ligands should be considered.
In conclusion, our studies show an interaction of NPY3-36 and anorectic peptide PYY3-36 with the rat glucocorticoid induced receptor. Our data suggest a potential role for GIR in feeding behavior and in the regulation of NPY neurotransmission. This is the first report to demonstrate high affinity binding and activation of rGIR by NPY-Y2 selective agonists and antagonist, as reported for the NPY-Y2 receptors. The results of our study support the classification of rGIR as a “Y2-like” receptor.
Acknowledgments
This work was supported by grants U01 HD37249, R01 NS39087 and DK 53548. We thank Mr. Jeffery Scott Andrews for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balasubramanium A. Clinical potentials of neuropeptide Y family of hormones. Am J Surg. 2002;183(4):430–434. doi: 10.1016/s0002-9610(02)00803-6. [DOI] [PubMed] [Google Scholar]
- 2.Batterham RL, Cowley MA, Small CJ, Herzog H, Cohen MA, Dakin CL, et al. Gut hormone PYY3-36 physiologically inhibits food intake. Nature. 2002;418:650–654. doi: 10.1038/nature00887. [DOI] [PubMed] [Google Scholar]
- 3.Baughman G, Harrigan MT, Campbell NF, Nurrish SJ, Bourgeois S. Genes newly identified as regulated by glucocorticoids in murine thymocytes. Mol Endo. 1991;5:637–644. doi: 10.1210/mend-5-5-637. [DOI] [PubMed] [Google Scholar]
- 4.Berglund MM, Fredriksson R, Salaneck E, Larhammer D. Reciprocal mutations of neuropeptide Y receptor Y2 in human and chicken identify amino acids important for antagonist binding. FEBS Lett. 2002;518:5–9. doi: 10.1016/s0014-5793(02)02534-6. [DOI] [PubMed] [Google Scholar]
- 5.De Moerlooze L, Williamson J, Liners F, Perret J, Parmentier M. Cloning and chromosomal mapping of the mouse and human genes encoding the orphan glucocorticoid-induced receptor (GPR83) Cyto Cell Gen. 2000;90:146–150. doi: 10.1159/000015650. [DOI] [PubMed] [Google Scholar]
- 6.Fredriksson R, Larson ET, Yan YL, Postlethwait JH, Larhammer D. Novel Neuropeptide Y Y2-like receptor subtype in Zebrafish and Frogs supports early vertebrate chromosome duplications. J Mol Evol. 2004;58:106–114. doi: 10.1007/s00239-003-2529-z. [DOI] [PubMed] [Google Scholar]
- 7.Gehlert DR, Beavers LS, Johnson D, Gackenheimer SL, Schober DA, Gadski RA. Expression Cloning of a Human Brain Neuropeptide Y Y2 Receptor. Mol Pharm. 1996;49:224–228. [PubMed] [Google Scholar]
- 8.Goumain M, Voisin T, Lorinet AM, Ducroc R, Tsocas A, Roze C, et al. The peptide YY-preferring receptor mediating inhibition of small intestinal secretion is a peripheral Y2 receptor: pharmacological evidence and molecular cloning. Mol Pharm. 2001;60:124–134. doi: 10.1124/mol.60.1.124. [DOI] [PubMed] [Google Scholar]
- 9.Grandt D, Schimiczek M, Rascher W, Feth F, Shively J, Lee TD, et al. Neuropeptide Y 3-36 is an endogenous ligand selective for Y2 receptors. Reg Pep. 1996;67:33–37. doi: 10.1016/s0167-0115(96)00104-8. [DOI] [PubMed] [Google Scholar]
- 10.Harrigan MT, Baughman G, Campbell NF, Bourgeois S. Isolation and characterization of glucocorticoids- and cyclic AMP-induced genes in T lymphocytes. Mol Cell Biol. 1989;9(8):3438–3446. doi: 10.1128/mcb.9.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilig M. The NPY system in stress, anxiety and depression. Neuropep. 2004;38:213–224. doi: 10.1016/j.npep.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Kaga T, Fujimiya M, Inui A. Emerging functions of neuropeptide Y Y(2) receptors in the brain. Pept. 2001;22:501–506. doi: 10.1016/s0196-9781(01)00362-x. [DOI] [PubMed] [Google Scholar]
- 13.Kalra SP, Dube MG, Sahu A, Phelps CP, Kalra PS. Neuropeptide Y secretion increases in the paraventricular nucleus in association with increased appetite for food. Proc Natl Acad Sci. 1991;88:10931–10935. doi: 10.1073/pnas.88.23.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karl T, Hoffman T, Pabst R, von Horsten S. Behavioral effects of neuropeptide Y in F44 rat substrains with a reduced dipeptidyl-peptidase IV activity. Pharmacol Biochem Behav. 2003;75:869–879. doi: 10.1016/s0091-3057(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 15.Keire DA, Bowers CW, Solomon TE, Reeve JR., Jr Structure and Receptor binding of PYY analogs. Pept. 2002;23:305–321. doi: 10.1016/s0196-9781(01)00602-7. [DOI] [PubMed] [Google Scholar]
- 16.Koda S, Date Y, Murakami N, Shimbura T, Hanada T, Toshinai K, et al. The role of the vagal nerve in peripheral PYY3-36-induced feeding reduction in rats. Endocrin. 2005;146:2369–2375. doi: 10.1210/en.2004-1266. [DOI] [PubMed] [Google Scholar]
- 17.Mentlein R. Dipeptidyl-peptidase IV (CD26)-role in the inactivation of regulatory peptides. Reg Pept. 1999;85:9–24. doi: 10.1016/s0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 18.Nonaka N, Shioda S, Niehoff ML, Banks WA. Characterization of blood-brain barrier permeability to PYY3-36 in the mouse. J Pharmacol Exp Ther. 2003;306:948–953. doi: 10.1124/jpet.103.051821. [DOI] [PubMed] [Google Scholar]
- 19.Parker EM, Babij CK, Balasubramanium A, Burrier RE, Guzzi M, Hamud F, et al. GR231118 (1229U91) and other analogues of the C-terminus of neuropeptide Y are potent neuropeptide Y-Y1 receptor antagonists and neuropeptide YY4 receptor agonists. Eur J Pharmacol. 1998;349:97–105. doi: 10.1016/s0014-2999(98)00171-x. [DOI] [PubMed] [Google Scholar]
- 20.Parker R, Liu M, Eyre HJ, Copeland NG, Gilbert DJ, Crawford J, et al. Y-receptor-like genes GPR72 and GPR73: molecular cloning, genomic organisation and assignment to human chromosome 11q21. 1 and 2p14 and mouse chromosome 9 and 6. Biochim Biophys Acta. 2000;1491:369–375. doi: 10.1016/s0167-4781(00)00023-3. [DOI] [PubMed] [Google Scholar]
- 21.Parker SL, Parker MS, Crowley WR. Characterization of Y1, Y2 and Y5 subtypes of neuropeptide Y (NPY) receptor in rabbit kidney. Reg Pept. 1998;75/76:127–143. doi: 10.1016/s0167-0115(98)00061-5. [DOI] [PubMed] [Google Scholar]
- 22.Pesini P, Detheux M, Parmentier M, Hokfelt T. Distribution of a glucocorticoid-induced orphan receptor (JP05) mRNA in the central nervous system of the mouse. Mol Brain Res. 1998;57:281–300. doi: 10.1016/s0169-328x(98)00099-0. [DOI] [PubMed] [Google Scholar]
- 23.Rose PM, Fernandes P, Lynch JS, Frazier ST, Fisher SM, Kodukula K, et al. Cloning and functional expression of a cDNA encoding a human type2 Neuropeptide Y receptor. J Biol Chem. 1995;270:22661–22664. doi: 10.1074/jbc.270.39.22661. [DOI] [PubMed] [Google Scholar]
- 24.Sah R, Pritchard LM, Richtand NM, Ahlbrand RL, Eaton K, Sallee FR, et al. Expression of the glucocorticoid-induced receptor mRNA in rat brain. Neurosci. 2005;133:281–292. doi: 10.1016/j.neuroscience.2005.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sah R, Sheriff S, Balasubramanium A, Wang D, Sallee FR. Characterization of the rat glucocorticoid induced receptor (GIR) as a novel neuropeptide Y-Y2 like receptor subtype. Pharmacologist. 2002;44(2):A184. [Google Scholar]
- 26.Sautel M. Neuropeptide Y and the non peptide antagonist BIBP3226 share an overlapping binding site at the human Y1 receptor. Mol Pharmacol. 1996;50:285–292. [PubMed] [Google Scholar]
- 27.Sharma P, Holmberg SKS, Eriksson H, Beck-Sickinger AG, Grundemar L, Larhammer D. Cloning and functional expression of the guinea pig neuropeptide Y Y2 receptor. Reg Pept. 1998;75–76:23–28. doi: 10.1016/s0167-0115(98)00049-4. [DOI] [PubMed] [Google Scholar]
- 28.Thompson AC, Justice JB, McDonald JK. Quantitative microdialysis of neuropeptide Y (NPY) J Neuro Met. 1995;60:189–198. doi: 10.1016/0165-0270(95)00012-j. [DOI] [PubMed] [Google Scholar]
- 29.Vezzani A, Sperk G. Overexpression of NPY and Y2 receptors in epileptic brain tissue: an endogenous neuroprotective mechanism in temporal lobe epilepsy? Neuropept. 2004;38:245–252. doi: 10.1016/j.npep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Herman JP, Pritchard LM, Spitzer RH, Ahlbrand RL, Kramer GL, Petty F, et al. Cloning, expression and regulation of a glucocorticoid-induced receptor in rat brain: effect of repetitive amphetamine. J Neurosci. 2001;21:9027–9035. doi: 10.1523/JNEUROSCI.21-22-09027.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberg DH, Sirinathsinghji DJS, Tan CP, Shiao L, Morin N, Rigby MR, et al. Cloning and expression of a novel Neuropeptide Y receptor. J Biol Chem. 1996;271(28):16435–16438. doi: 10.1074/jbc.271.28.16435. [DOI] [PubMed] [Google Scholar]
- 32.Woodson W, Nitecka L, Ben-Ari Y. Organization of the GABAergic system in the rat hippocampal formation: a quanttative immunocytochemical study. J Comp Neurol. 2002;280:254–271. doi: 10.1002/cne.902800207. [DOI] [PubMed] [Google Scholar]


