Abstract
Background
The prolonged time course of Huntington's disease (HD) neurodegeneration increases both the time and cost of testing potential therapeutic compounds in mammalian models. An alternative is to initially assess the efficacy of compounds in invertebrate models, reducing time of testing from months to days.
Methodology/Principal Findings
We screened candidate therapeutic compounds that were identified previously in cell culture/animal studies in a C. elegans HD model and found that two FDA approved drugs, lithium chloride and mithramycin, independently and in combination suppressed HD neurotoxicity. Aging is a critical contributor to late onset neurodegenerative diseases. Using a genetic strategy and a novel assay, we demonstrate that lithium chloride and mithramycin remain neuroprotective independent of activity of the forkhead transcription factor DAF-16, which mediates the effects of the insulin-like signaling pathway on aging.
Conclusions/Significance
These results suggest that pathways involved in polyglutamine-induced degeneration are distinct from specific aging pathways. The assays presented here will be useful for rapid and inexpensive testing of other potential HD drugs and elucidating pathways of drug action. Additionally, the neuroprotection conferred by lithium chloride and mithramycin suggests that these drugs may be useful for polyglutamine disease therapy.
Introduction
Drug discovery for late onset neurodegenerative diseases is a major challenge. In large part, the complexity of treating these disorders results from our insufficient understanding of the contributions of multiple pathways on disease pathophysiology. Furthermore, since the pathology of these disorders is often only discernable in aged populations, testing the therapeutic value of small molecules in vertebrate disease models requires time consuming and costly experimental designs. The development of rapid and inexpensive in vivo assays to evaluate the numerous candidate compounds identified in high-throughput screens is therefore of paramount importance.
Invertebrate model organisms such as C. elegans provide an attractive alternative for prioritizing lead compounds in the early stages of drug development for age-related diseases [1]–[3]. C. elegans has several characteristics that make it ideal for drug testing- including a short lifecycle, small size and the ease of culturing in liquid. Furthermore, decades of neurobiological and antiparasitic drug studies in C. elegans provide a strong foundation for use of this organism in therapeutic compound identification [1]–[3].
Huntington's Disease (HD) is caused by expansion of a polyglutamine (polyQ) tract in the huntingtin protein leading to neurodegeneration that is age and polyQ tract length dependent [4]. In this study, we use a C. elegans model of polyQ neurotoxicity in which the N-terminal 171 amino acid fragment of human huntingtin protein containing an expanded polyglutamine tract (150Qs) is expressed in neurons. Degeneration and cell death in this model is dependent on both age and polyglutamine tract length, recapitulating these aspects of the human disease [4]–[6].
We tested a collection of compounds that have been previously described to decrease degeneration in cell culture/animal models of polyQ toxicity for their ability to protect C. elegans neurons from the toxic effects of an expanded huntingtin polyglutamine fragment. We developed, optimized and validated new assays for use in rapid assessment of drug efficacy using C. elegans HD models. Of the compounds tested, we found that two FDA approved drugs, mithramycin (MTR) and lithium chloride (LiCl), reduced polyQ toxicity in the C. elegans model.
An important determinant of neurodegenerative diseases is the aging process. However, the mechanistic links between aging and the cellular pathways leading to neurodegeneration are not well understood. The forkhead transcription factor DAF-16, which mediates the effects of the insulin-like signaling pathway on aging, has been shown to play a role in polyQ aggregation. Mutations that reduce insulin signaling, derepress DAF-16 leading to an increase in lifespan and stress resistance [7], [8], whereas RNAi based knockdown of daf-16 accelerates polyQ aggregation and toxicity [9] suggesting that DAF-16 transcriptional targets not only promote longevity but also prevent polyQ aggregation. Despite the pivotal role that growth and aging play in neurodegenerative disease, we found that LiCl and MTR protect C. elegans neurons in the absence of growth and through a daf-16 independent pathway suggesting that these compounds may target pathways that are specific to neurodegeneration. Thus, the integration of pharmacological and genetic examination of drugs, in C. elegans HD models that we describe, should accelerate the identification of interventions for HD along with insight into mechanism of drug action.
Results
Compound concentration range for screening in C. elegans: Food Clearance Assay
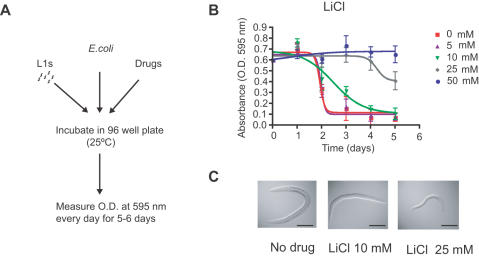
We tested a collection of compounds that have demonstrated therapeutic value in either cell culture and/or animal models of polyQ toxicity for screening in our C. elegans HD models ( Table 1 ). These candidates represent compounds that may protect against polyQ toxicity by affecting a variety of cellular pathways. To efficiently evaluate the effects of these candidates on neurodegeneration and neuronal cell death, we first established a systematic method for selecting optimal drug concentrations to assess in our C. elegans HD models (see Methods). Compounds were tested in a dose dilution series in the food clearance assay ( Figure 1 and Figure S1 ) starting at the highest soluble concentration. Taking advantage of the short life cycle and the ability of C. elegans to grow in liquid culture of E. coli, we evaluated compounds by monitoring the rate at which the E. coli suspension (food source) was consumed. Each adult is capable of producing hundreds of progeny that rapidly consume the limited E. coli supply. As a result, the OD of wells without compound drastically decreases in 3 days. Any drug that decreases C. elegans growth, survival or fecundity would result in a dose dependent reduction of the rate at which food is cleared (consumed) in a well. For example, addition of 5 mM LiCl to the culture showed no effect on food clearance compared to control animals, whereas animals exposed to 10 mM or 25 mM LiCl had delayed food clearance ( Figure 1B ). Visual inspection confirmed that animals treated with 10 mM and 25 mM LiCl were smaller in size compared to untreated animals whereas animals treated with 5 mM LiCl were unaffected ( Figure 1C and data not shown). Furthermore, animals exposed to 50 mM and 100 mM LiCl did not produce progeny over the time course of the experiment (data not shown), which correlated with the lack of clearance of the E coli food source. For all compounds, optimal concentrations for C. elegans were similarly assessed using the food clearance assay ( Table 1 ). This simple assay for determining a compound's concentration range to test in C. elegans uses a small amount of compound and is amenable to high-throughput format, making it applicable to any C. elegans drug study.
Table 1. Compounds screened in C. elegans polyQ model for neuroprotection.
| Drugs tested | Putative Mechanism | TC 50 Food clearance (mM) | Conc. Tested (mM) | EC 50 (mM) | Rescue in C. elegans | Activity reported in HD model(s) |
| LiCl | GSK-3β inhibitor | 25 | 10–100 | 25 | Yes | Cell culture [31] D. melanogaster [32] |
| Mithramycin | DNA binding | 0.003 | 0.01–1 | 0.5 | Yes | M. musculus [33] |
| Trichostatin A (TSA) | HDAC inhibitor | 1 | 0.16–1 | 0.16 | Yes | Cell Culture [34] |
| C. elegans [14] | ||||||
| S. cerevisiae [35] | ||||||
| SAHA* | HDAC inhibitor | 2 | 2 | n.a. | Yes | M. musculus [36] |
| D. melanogaster [37] | ||||||
| Taxol | Cytoskeletal dynamics | none | 0.4 | n.a. | no effect | Cell culture [38] |
| Congo Red | Aggregation inhibitor | 0.5 | 0.12–0.5 | n.a. | no effect | M. musculus [39] |
| Cystamine | Transglutaminase inhibitor | 300 | 150 | n.a. | no effect | M. musculus [40]–[42] |
| D. melanogaster [43] | ||||||
| Cell culture [44] | ||||||
| Budesonide | Glucocorticoid receptor agonist | none | 0.8 | n.a. | no effect | Cell culture [45] |
| Paraquat | Free radical generator | 3 | 3 | n.a. | no effect | Negative control [46] |
All compounds were tested in pqe-1 enhanced background in the presence of food, E. coli.
n.a. = Not applicable as these compounds were not active.
SAHA was only tested and found to be active at a single concentration (2 mM). Therefore, an EC50 was not determined.
Figure 1. Concentration of compounds for testing was assessed using a food clearance assay.
(A) Flow diagram of the food clearance assay. 20 newly hatched L1 animals were incubated at 25°C in E. coli at a final OD (A595) of 0.6 in 96 well microtiter plate wells containing varying drug concentrations. The OD of the microtiter plate was measured daily for 5 days. (B) The OD of E. coli is reported daily for each concentration of LiCl. The mean OD is calculated for each day from triplicate samples and plotted over time. Error bars represent SEM. Food clearance assays were also performed on trichostatin A and mithramycin, (Figure S1). (C) Animals treated with no drug and indicated LiCl concentrations. Animals treated with 50 mM or 100 mM LiCl are alive but concentrations above 100 mM LiCl cause death (data not shown). Scale bar is approximately 75 µm.
Screening neuroprotective compounds in C. elegans polyQ model
In order to test compounds in an efficient manner, we decided to reduce the amount of time required to test drug efficacy. Relatively long assay duration may also be unsuitable for testing compounds that are unstable under assay conditions. We utilized a genetic mutant background (pqe-1 loss of function) that accelerates polyQ mediated neurodegeneration and cell death in C. elegans from 7 days to 2–3 days. We evaluated neuronal death by monitoring loss of expression of a GFP reporter construct in the bilateral ASH sensory neurons in pqe-1;Htn-Q150 animals, a genetically enhanced model of polyQ toxicity [10] ( Figure S2 ). In pqe-1;Htn-Q150 animals, the vast majority (>90%) of ASH neurons undergo cell death in less than three days ( Figure S2 ).
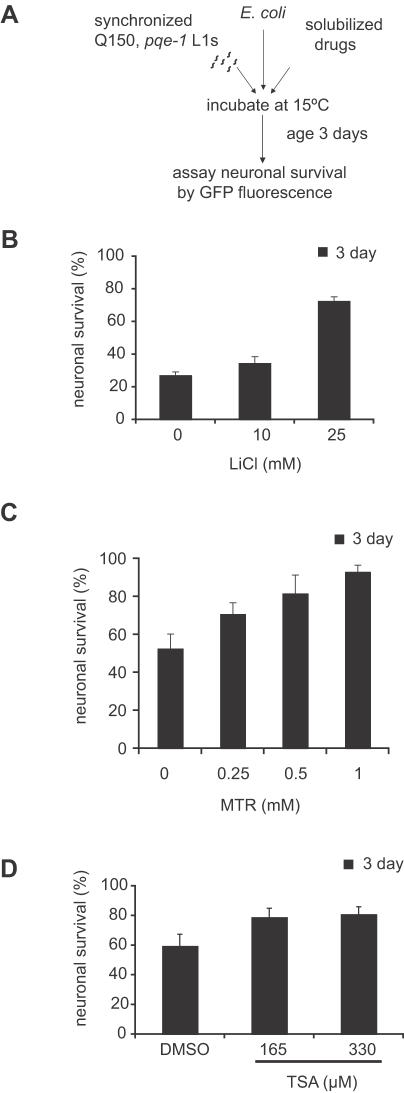
Synchronized pqe-1;Htn-Q150 L1 larvae were treated with and without drug for three days, then GFP expression was monitored to assess ASH neuronal viability ( Figure 2A ). Of the candidates tested, we identified three protective compounds, mithramycin (MTR), trichostatin A (TSA) and lithium chloride (LiCl) ( Table 1 ). All three compounds caused a dose-dependent neuroprotection in the pqe-1 sensitized background ( Figure 2B ). MTR and TSA affect transcriptional activity [11], [12] and LiCl is a drug used clinically to treat individuals with bipolar disorders and is a potential inhibitor of GSKβ [13]. Previous studies in C. elegans demonstrated that TSA reduced polyQ mediated neurodegeneration [14], validating this approach. Htn-Q150 transgene expression was not altered by drug treatment based on RT-PCR ( Figure S3 ). Based on these results, we conclude that a genetically sensitized background (e.g. pqe-1) can be used to dramatically reduce the amount of time required to accurately assess effects of drugs on neuronal cell death.
Figure 2. LiCl, TSA, and MTR decrease polyQ mediated ASH neuronal death.
(A) Flow diagram of the sensitized assay. 20–30 synchronized pqe-1;Htn-Q150 L1s per well were incubated at 15°C in S medium with E. coli at an OD (A595) of 0.6 and varying concentrations of compound to a total volume of 60 µl. Animals were grown in the presence of compound for 3 days. ASH neuron survival was evaluated by the presence or absence of GFP expression. (B–D) LiCl, TSA and MTR increased ASH neuron survival in the pqe-1;Htn-Q150 animals in the presence of E. coli. ASH neuron survival was evaluated by the presence or absence of GFP expression on day 3. 100 neurons were scored for each trial; mean percentage of 3 different trials±SEM is shown. p<0.0001 for 25 mM LiCl. p = .037 for 0.5 mM MTR; p<0.0001 for 1 mM MTR. p = 0.0082 for 165 µM TSA; p = 0.005 for 330 µM TSA.
Neuroprotection in an age-dependent C. elegans polyQ model
To determine if neuroprotection was dependent on pqe-1, we tested candidates in the Htn-Q150 only background that exhibits age-dependent neurodegeneration but no cell death. Late onset degeneration of ASH sensory neurons was monitored by defects in dye uptake, a previously described assay [6] ( Figure S2 ).
One limitation due to the prolonged duration (7 days) of the aging assay is the production of progeny (200–300 per adult) by the aging animals. Since the progeny grow to adulthood in 3 days, they are difficult to distinguish from the aged parent by day 7. A novel genetic strategy was employed to prevent progeny development. We crossed Htn-Q150 into a genetic background (pha-1) in which progeny fail to feed and grow at the restrictive temperature of 25°C [15]. Introducing pha-1(e2123) to eliminate progeny development permits the use of liquid cultures and facilitates any C. elegans aging study (see Methods).
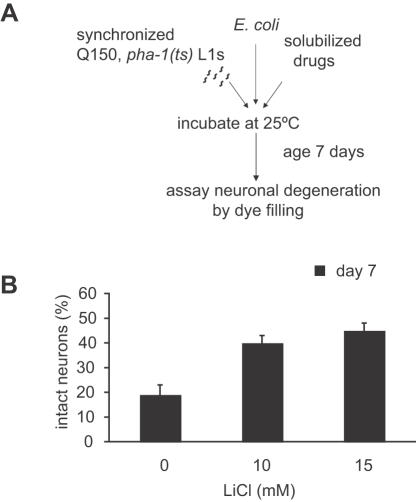
To assay compound efficacy, synchronized pha-1;Htn-Q150 L1 larvae were treated with and without drug for seven days, then scored for degeneration using the dye-filling assay ( Figure 3A ). Introducing the pha-1 mutation had no effect on the dye-filling of ASH neurons or the polyQ dependent neurodegeneration (data not shown and Figure S2 ). LiCl decreased polyQ neurodegeneration in a dose dependent manner ( Figure 3B ) while equivalent concentrations of sodium chloride (NaCl) had no effect (data not shown), showing the specificity of LiCl's effect. Consistent with previous findings [14], the HDAC inhibitor trichostatin A (TSA) also suppressed neurodegeneration in the aging assay format ( Figure S4 ). The confirmation of compound activity in the age dependent Htn-Q150 model validated the utilization of the pqe-1 sensitized strain as a rapid means of identifying HD therapeutic compounds.
Figure 3. Compounds decrease polyQ toxicity in a pqe-1 independent manner in aged animals.
(A) Flow diagram of the aging assay. 30 synchronized pha-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with E. coli at an OD (A595) of 0.5 with varying concentrations of compound to a total volume of 1 ml. On day 7, animals were collected and dye filled using DiD. (B) LiCl increased ASH neuron survival in the pha-1;Htn-Q150 aged animals. ASH neurodegeneration was evaluated by the presence or absence of dye-filled ASH neurons on day 7. 50 neurons were scored for each trial; mean percentage of 3 different trials±SD is shown. p = 0.0033 for 15 mM LiCl.
Dissociating neuroprotection from growth, development and aging
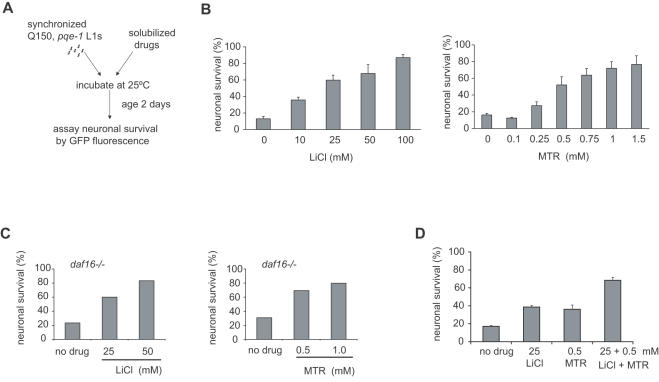
Since the aging process is a critical but poorly understood contributor to late onset neurodegenerative diseases [9], [16], [17], compounds that affect aging may indirectly affect degeneration. To distinguish drug effects on neuronal cell death from those on growth, development and aging rates, we applied two strategies: (i) we modified our assay format to prevent growth of animals for the duration of the assay (ii) we genetically altered the well characterized insulin signaling pathway that affects aging. To modify our assay, we took advantage of the growth arrest induced by starvation of L1 stage C. elegans larvae. When embryos hatch in the absence of food, their development is arrested until food is available [18], [19]. Since starved L1 animals do not grow, the effects of compounds on neuronal cell death can be evaluated independent of their effects on growth and development. A further advantage is that drugs can be tested in the absence of bacteria (E. coli food source) that could degrade or metabolize compounds under analysis. Using a starvation assay format ( Figure 4A ), we found that ASH neurons continue to undergo neuronal cell death in pqe-1;Htn-Q150 animals that are growth arrested by the absence of food (data not shown). This finding suggests that certain pathways that affect polyQ neurodegeneration are distinct from pathways that effect growth and development.
Figure 4. LiCl and MTR decrease polyQ toxicity measuring ASH neuronal death in the absence of growth.
(A) Flow diagram of the starvation assay. 50 synchronized pqe-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with varying concentrations of compound to a total volume of 60 µl. On day 2, the survival of the ASH neuron was determined by visually evaluating the presence or absence of GFP expression. (B) LiCl and MTR increase neuronal survival in starved conditions. 100 neurons were scored for each trial; mean percentage of 3 different trials±SEM is shown. p = 0.0063 for 10 mM LiCl; p<0.0001 for 25 mM, 50 mM and 100 mM LiCl. p<0.0001 for 0.5 mM, 0.75 mM, 1.0 mM, and 1.5 mM MTR. (C) LiCl and MTR increase neuronal survival in a daf-16 independent manner. At least 100 neurons were scored for each trial; percentage of neuronal survival in one representative experiment of 2 independent trials is shown; p<0.0001 for 25 mM and 50 mM LiCl. p<0.0001 for 0.5 mM and 1.0 mM MTR. (D) Combined, MTR (0.5 mM) and LiCl (25 mM) were more effective than either drug alone in protecting ASH neurons from polyQ neurotoxicity. 50 synchronized pqe-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with 0.5 mM MTR and 25 mM LiCl to a total volume of 60 µl. On day 2, the survival of the ASH neuron was evaluated by the presence or absence of GFP. 100 neurons were scored for each trial; mean percentage of 3 different trials±SEM is shown. The excess synergy was 0.14 based on a Bliss Independence analysis. p = 0.046 for 0.5 mM MTR; p = 0.0039 for 25 mM LiCl; p<0.0001 for LiCl+MTR.
We tested LiCl and MTR in this starvation assay format. Both LiCl and MTR protected ASH neurons of starved animals in a dose-dependent manner ( Figure 4B ). Equivalent concentrations of NaCl as that of LiCl had no effect on neuronal cell death demonstrating specificity of the compound's effect (data not shown). Using this assay modification, higher concentrations of compounds that interfere with C. elegans development and growth were tested in the starvation assay ( Figure 1C and Figure 4B ). Therefore, testing drugs in the L1 arrest assay can distinguish contributions of specific drugs on organismal development and aging, providing a clearer demonstration of the neuroprotective properties of compounds.
The insulin signaling pathway, a critical regulator of longevity in C. elegans, is also a modifier of polyQ toxicity. RNAi mediated knockdown of daf-16 decreases lifespan and leads to early onset of polyQ aggregation [9]. To determine if the neuroprotective effects of LiCl and MTR are dependent on components of the insulin signaling pathway, we crossed a daf-16 null mutation into pqe-1;Htn-Q150 animals and tested the effects of LiCl and MTR on neuronal cell death in the starvation assay. We found that both compounds remained neuroprotective in the daf-16 mutant animals in a dose dependent manner ( Figure 4C ), suggesting that LiCl and MTR suppress polyQ mediated neuronal cell death independent of DAF-16 activity. The dissociation of the neuroprotective effects of compounds from growth and specific aging pathways allows a mechanistic classification of therapeutic compounds.
Combinatorial effects of LiCl and MTR
A number of therapies for complex diseases use combinations of drugs that allow targeting of multiple mechanisms and allow therapeutic efficacy at lower compound concentrations, limiting toxicity of single agents. We also investigated the utility of our C. elegans assay in drug combinatorial studies by testing LiCl in combination with MTR on C. elegans polyQ toxicity in the starvation assay. Combining 25 mM LiCl and 0.5 mM mithramycin in the starvation assay format increased ASH neuron survival compared to the protection afforded by either compound alone ( Figure 4D ).
Discussion
In this study, we have devised strategies for efficiently assessing therapeutic efficacy of compounds in C. elegans HD models ( Figure 5 ). Our methods exploit the strengths of C. elegans model for drug discovery, including convenient and precise visualization of live neurons, small-scale liquid cultures that significantly reduce the amount of drug required for testing as well as genetic approaches to implicate specific cellular pathways in a compound's mechanism of action. These assays could be utilized for prioritizing the large numbers of hits already identified by high-throughput screens for HD and other polyQ diseases [20]–[23].
Figure 5. Schematic Diagram of Compound Testing Strategy.
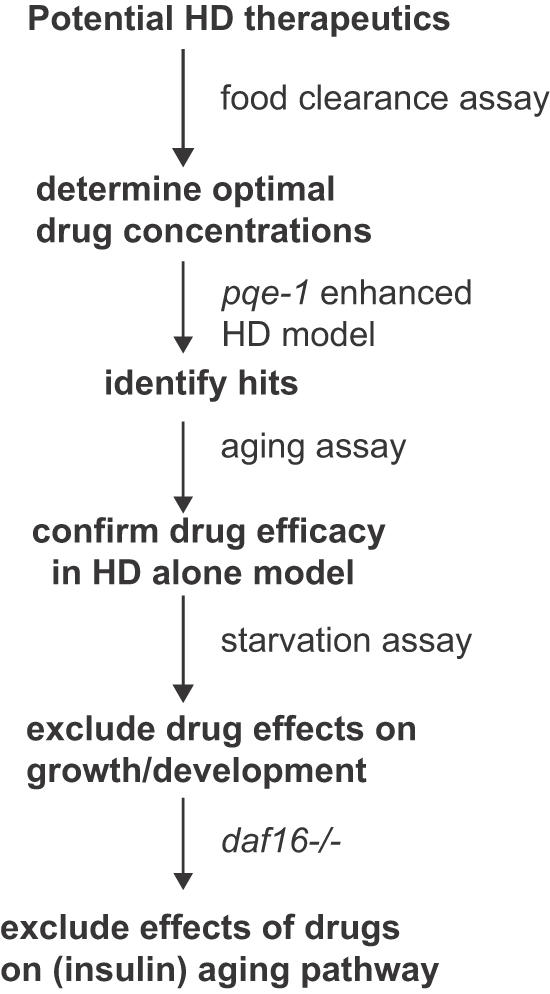
(1) A range of concentrations for each compound for testing in C. elegans was established using the food clearance assay. (2) The protective effects of compounds on polyglutamine neurodegeneration and cell death were assessed in pqe-1;Htn-Q150 animals treated with compounds for 3 days. (3) To determine if neuroprotective effects of compounds were dependent on pqe-1, compounds were retested in animals expressing Htn-Q150 for 7 days. (4) To distinguish drug effects on neuronal cell death versus effects on growth and/or development, synchronized pqe-1;Htn-Q150 L1 animals were incubated in the presence of drugs without food for 2 days. (5) The neuroprotective effects of compounds on the aging process can be tested by introducing mutations (daf-16) in components of specific aging-related (insulin signaling) pathways.
The normal aging process is intricately linked to late onset neurodegenerative diseases. However, the influence of aging on cellular events that impact neurodegeneration is unclear. To dissect the interplay of pathways involved in neurodegeneration from those involved in aging, we combined genetic manipulations with a novel assay that utilizes growth-arrested animals. Our studies demonstrate that LiCl and MTR exert their neuroprotection in a daf-16 independent manner. However, starvation of animals may activate additional stress responsive transcription factors such as heat shock factor (HSF1). HSF1 regulates the heat shock response and also influences longevity [17], [24], [25]. Furthermore, HSF1 RNAi enhances polyQ aggregation [26] raising the possibility that HSF1 or other stress activated pathways may function as molecular targets of these compounds. However, the fact that neurons continue to die under conditions of starvation suggests that the stress responses induced by starvation are insufficient to abrogate the neurodegenerative process. Thus, the combination of strategies outlined in the study allows the separation of drug effects on neurodegeneration from those on growth and stress related aging pathways.
Though we focus on neurodegeneration in HD models, similar strategies may apply to other age dependent phenotypic readouts. For example, deletion of the C. elegans dystrophin gene, the homolog of a gene involved in Duchenne's Muscular Dystrophy, in combination with a second mutation in the myogenic factor MyoD leads to progressive muscle degeneration [27], [28]. Using the strategies described in this study, the protective effect of compounds on muscle degeneration could be conducted rapidly. These C. elegans disease models should be valuable tools for rapid identification of potential therapeutic compounds for aging related human diseases and cellular pathway elucidation of any compounds that are active in these models.
Materials and Methods
Strains
C. elegans N2[wild-type] and GE24[pha-1(e2123)], E. coli food sources OP50 and HB101 (OP50 for compound efficacy and aging assays and HB101 for sensitized and food clearance assays) were obtained from the Caenorhabditis elegans Genetics Center. Htn-Q150(rtIs11) expressing strains with and without pqe-1(rt13) were previously described [6], [10]. daf-16(mgDf47) was crossed into pqe-1(rt13) mutant strain expressing Htn-Q150(rtIs11) marked with dpy-11(e224). All mutations were homozygous in the final strain for analysis. All strains were cultivated on nematode growth media (NGM) plates at 15°C [29].
Food Preparation
E. coli were grown overnight at 37°C in Luria broth (LB) media, pelleted by centrifugation, frozen at −70°C, and then resuspended at a final OD of 0.5 (595 nm) for the aging assay or an OD of 6.6 in nematode S-medium [29] (supplemented with 100× streptomycin/penicillin (Invitrogen, #15140-122) and nystatin (Sigma, #N1638)) for all other food based assays. Note the lower OD (∼0.6) in plates was due to a decreased path length of the 60 µl final suspension, in a well, compared to a 1 cm path length in a spectrophotometer.
Compound Preparation
All compounds were purchased from Sigma (St. Louis, MO). Stock solution of 1 M lithium chloride (LiCl) in water, 10 mM mithramycin (MTR) in PBS and 5 mg/ml trichostatin A (TSA) in DMSO were stored at room temperature (LiCl), 10–14 days at 4°C (MTR) and at −20°C (TSA). Compounds were diluted into E. coli suspension or S-medium (starvation assay) to the desired concentrations. 1 ml of the final mixture was added per well of a 24-well polystyrene plate for the aging assay. For the remaining assays, 50 µl of the final mixture was added per well in a 96-well polystyrene plate.
C. elegans Preparation
We used newly hatched C. elegans, collected as L1 larvae at 15°C. Specifically, adults were lysed, fertilized eggs were collected by hypochlorite treatment of gravid adults and suspended in S-medium for 24–30 hours at 15°C and L1 larvae were allowed to hatch overnight in the absence of food [29]. pha-1(e2123) is a temperature sensitive loss of function allele that affects normal pharyngeal development [15]. A normal pharynx forms at the permissive temperature of 15°C in newly hatched L1 animals while the pharynx fails to develop in pha-1(e2123) animals hatched at 25°C. C. elegans L1 animals do not initiate growth in the absence of food, resulting in a synchronous population of arrested L1 animals. Introduction of a food source (E. coli) results in growth; animals become adult hermaphrodites within 48 hours at 25°C. This L1 growth arrest and recovery provides an efficient means of collecting large numbers of synchronized animals for assays.
Food Clearance Assay
The effect of compounds on C. elegans physiology is monitored by the rate at which the E. coli food suspension was consumed, as a read out for C. elegans growth, survival or fecundity. Approximately 20–30 L1 synchronized animals per 10 µl of S-media were added to an E. coli suspension. Microtiter plates containing animals and drugs were incubated at 25°C. The absorbance (OD 595nm) was measured daily using a Vmax Kinetic microplate reader (Molecular Devices). We used N2 animals when determining compound concentration. Introduction of pha-1(e2123), pqe-1(rt13), daf-16(mgDf47) or Htn-Q150(rtIs11) had no effect on overall growth or health of the animal compared to N2 in the presence of drug at assay temperatures.
Scoring Degeneration and Cell Death
At time points indicated, animals were collected by centrifugation, washed in S-media and immobilized with 5 mM sodium azide on a 2% agarose pad. GFP fluorescence was scored using an Axioplan2 fluorescence microscope (n = 100 ASH neurons). For the aging assay, degeneration was assessed by dye-filling [6]. Animals were incubated for 2 hours in 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD) (Molecular Probes) and immobilized as above for scoring ASH dye filling (n = 50 ASH neurons). To determine statistical significance, we compared the results from parallel experiments using 2×2 contingency tables comparing the number of GFP+ and GFP- neurons in control vs. specific concentrations of a compound. We determined the statistical significance using a Fisher's exact test for each of the three independent experiments and reported the largest p value of the three trials to provide a conservative estimate. In Figure 4d, Bliss Independence was assessed [30]. Neuronal survival was converted to a fractional scale with no drug treatment (17%) arbitrarily set to 0 and 100% survival set to 1. Survival for each drug and the combination was converted to a fraction and the Bliss Independence score was calculated as C = A+B−AB, where A and B are the fractional survival for LiCl and MTR and C is the predicted effect for the combination. The difference between C and the actual value (fractional) was calculated to assess synergy. LiCl had a fractional survival of 0.22, MTR 0.19 and the combination 0.51. The expected value for the combination was 0.37. The excess 0.14 was a measure of synergy.
Quantitative Real Time PCR Analysis
qRT-PCR analyses were carried out using the SYBR Green method (BioRad) and the SmartCycler 1600 System by Cepheid Innovation. Total RNA was extracted using Trizol (Invitrogen), DNase treated (Ambion), followed by cDNA synthesis (BioRad) according to manufacturer's instructions. The primers used for amplification of the actin gene, act-1, are 5′-atcaccgctcttgccccatc-3′ (for) and 5′-ggccggactcgtcgtattcttg-3′ (rev). Primers for Htn transgene were previously described [3].
Supporting Information
Concentration of compounds for testing in C. elegans was assessed using a food clearance assay. The OD of E. coli is reported daily for each concentration of TSA (A) and MTR (B). The mean OD is calculated for each day from triplicate samples and plotted over time.
(1.17 MB EPS)
C. elegans model of polyQ neurodegeneration and cell death [6], [10]. (A) An N-terminal fragment of human huntingtin with 150 glutamines (Htn-Q150) was expressed in C. elegans ASH neurons as previously described [6]. (B) A GFP reporter expressed in the ASH neurons was used to assess cell death; loss of GFP expression in ASH neurons indicates cell death as shown here in young pqe-1;Htn-Q150 animals (bottom panel). (C) The ASH neuron and 5 other sensory neurons in Htn-Q150 young (top panel) or aged (bottom panel) animals were visualized by dye filling. Sensory endings of ASH neurons expressing Htn-Q150 degenerate in aged animals and fail to take up dye (bottom panel). (D) ASH neuronal survival decreases dramatically over time based on GFP fluorescence. By day 3, 75% of ASH neurons are dead in pqe-1;Htn-Q150 animals. Error bars represent SEM. Scale bar is approximately 20 nm. (E) Only 21% of ASH neurons remain intact in aged (7 day) pha-1 animals expressing Htn-Q150 raised at 25°C compared to young (3 day) animals based on dye filling.
(2.94 MB EPS)
HtnQ150 transgene levels are not decreased in the presence of LiCl or MTR. Relative HtnQ150 transgene levels were determined using the ΔΔCt method and normalized to an act-1 control. Results are from two independent RNA collections from untreated animals (C) and animals treated with 25 mM LiCl or 0.5 mM MTR in the starved assay format for 24 hours at 15°C. The error bars indicate the standard deviation from two replicates of qRT-PCR experiments comparing untreated and drug treated samples.
(0.63 MB EPS)
HDAC inhibitor trichostatin A (TSA) decreases polyQ toxicity measuring ASH neurodegeneration or neuronal death. At least three trials for each efficacy assay were performed. Representative experiments are depicted. (A) TSA decreases polyQ toxicity in a pqe-1 independent manner in pha-1;Htn-Q150 aged animals. 30 synchronized pha-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with E. coli at an OD at A595 of 0.5 with varying concentrations of TSA to a total volume of 1 ml. On day 7, animals were collected, dye filled and 100 neurons were scored for degeneration. (B) TSA increases neuronal survival in the absence of growth using the starvation assay. 50 synchronized pqe-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with varying concentrations of TSA to a total volume of 60 µl. On day 2, the survival of 100 ASH neurons was evaluated by the presence or absence of GFP. Animals were treated with final concentration of TSA or vehicle (2% DMSO).
(0.66 MB EPS)
Acknowledgments
We would like to thank Sally McFall, Patrick McMullen, Daniel Stouffer, Mike Stringer and Wyndham Lathem for helpful discussions, and Gary Ruvkun and the C. elegans Genetics Center for strains.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: We gratefully acknowledge the support of the Hereditary Disease Foundation (C.V. and B.R.S.), Cure Huntington Disease Initiative (B.R.S.), the High Q Foundation (H.V. and B.R.S.), the Ellison Medical Foundation (A.C.H.) and the NIH (NS-040048 for A.C.H. and T32 AG20506 for C.V.). Brent R. Stockwell is supported in part by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
References
- 1.Geary TG, Thompson DP. Caenorhabditis elegans: How good a model for veterinary parasites? Vet Parasitol. 2001;101:371–386. doi: 10.1016/s0304-4017(01)00562-3. [DOI] [PubMed] [Google Scholar]
- 2.Rand JB, Johnson CD. Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. Methods Cell Biol. 1995;48:187–204. doi: 10.1016/s0091-679x(08)61388-6. [DOI] [PubMed] [Google Scholar]
- 3.Bargmann CI. Neurobiology of the Caenorhabditis elegans genome. Science. 1998;282:2028–2033. doi: 10.1126/science.282.5396.2028. [DOI] [PubMed] [Google Scholar]
- 4.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.Gusella JF, MacDonald ME. Huntington's disease: CAG genetics expands neurobiology. Curr Opin Neurobiol. 1995;5:656–662. doi: 10.1016/0959-4388(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 6.Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci U S A. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 8.Lin K, Dorman JB, Rodan A, Kenyon C. DAF-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 9.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc Natl Acad Sci U S A. 2002;99:17131–17136. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 12.Jones DE, Jr, Cui DM, Miller DM. Expression of beta-galactosidase under the control of the human c-myc promoter in transgenic mice is inhibited by mithramycin. Oncogene. 1995;10:2323–2330. [PubMed] [Google Scholar]
- 13.Phiel CJ, Klein PS. Molecular targets of lithium action. Annu Rev Pharmacol Toxicol. 2001;41:789–813. doi: 10.1146/annurev.pharmtox.41.1.789. [DOI] [PubMed] [Google Scholar]
- 14.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granato M, Schnabel H, Schnabel R. Genesis of an organ: molecular analysis of the pha-1 gene. Development. 1994;120:3005–3017. doi: 10.1242/dev.120.10.3005. [DOI] [PubMed] [Google Scholar]
- 16.Tang BL. SIRT1, neuronal cell survival and the insulin/IGF-1 aging paradox. Neurobiol Aging. 2006;27:501–505. doi: 10.1016/j.neurobiolaging.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 18.Fukuyama M, Rougvie AE, Rothman JH. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr Biol. 2006;16:773–779. doi: 10.1016/j.cub.2006.02.073. [DOI] [PubMed] [Google Scholar]
- 19.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Gines S, MacDonald ME, Gusella JF. Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 2005;6:1. doi: 10.1186/1471-2202-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Duan W, Igarashi S, Morita H, Nakamura M, et al. Compounds blocking mutant huntingtin toxicity identified using a Huntington's disease neuronal cell model. Neurobiol Dis. 2005;20:500–508. doi: 10.1016/j.nbd.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 22.Varma H, Voisine C, Demarco CT, Cattaneo E, Lo DC, et al. Selective inhibitors of death in mutant huntingtin cells. Nat Chem Biol. 2007;3:99–100. doi: 10.1038/nchembio852. [DOI] [PubMed] [Google Scholar]
- 23.Piccioni F, Roman BR, Fischbeck KH, Taylor JP. A screen for drugs that protect against the cytotoxicity of polyglutamine-expanded androgen receptor. Hum Mol Genet. 2004;13:437–446. doi: 10.1093/hmg/ddh045. [DOI] [PubMed] [Google Scholar]
- 24.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamberlain JS, Benian GM. Muscular dystrophy: the worm turns to genetic disease. Curr Biol. 2000;10:R795–797. doi: 10.1016/s0960-9822(00)00768-5. [DOI] [PubMed] [Google Scholar]
- 28.Gieseler K, Grisoni K, Segalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr Biol. 2000;10:1092–1097. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- 29.Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 30.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 31.Carmichael J, Sugars KL, Bao YP, Rubinsztein DC. Glycogen synthase kinase-3beta inhibitors prevent cellular polyglutamine toxicity caused by the Huntington's disease mutation. J Biol Chem. 2002;277:33791–33798. doi: 10.1074/jbc.M204861200. [DOI] [PubMed] [Google Scholar]
- 32.Berger Z, Ttofi EK, Michel CH, Pasco MY, Tenant S, et al. Lithium rescues toxicity of aggregate-prone proteins in Drosophila by perturbing Wnt pathway. Hum Mol Genet. 2005;14:3003–3011. doi: 10.1093/hmg/ddi331. [DOI] [PubMed] [Google Scholar]
- 33.Ferrante RJ, Ryu H, Kubilus JK, D'Mello S, Sugars KL, et al. Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, et al. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci U S A. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes RE, Lo RS, Davis C, Strand AD, Neal CL, et al. Altered transcription in yeast expressing expanded polyglutamine. Proc Natl Acad Sci U S A. 2001;98:13201–13206. doi: 10.1073/pnas.191498198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 38.Hazeki N, Nakamura K, Goto J, Kanazawa I. Rapid aggregate formation of the huntingtin N-terminal fragment carrying an expanded polyglutamine tract. Biochem Biophys Res Commun. 1999;256:361–366. doi: 10.1006/bbrc.1999.0337. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 40.Van Raamsdonk JM, Pearson J, Bailey CD, Rogers DA, Johnson GV, et al. Cystamine treatment is neuroprotective in the YAC128 mouse model of Huntington disease. J Neurochem. 2005;95:210–220. doi: 10.1111/j.1471-4159.2005.03357.x. [DOI] [PubMed] [Google Scholar]
- 41.Karpuj MV, Becher MW, Springer JE, Chabas D, Youssef S, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med. 2002;8:143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 42.Dedeoglu A, Kubilus JK, Jeitner TM, Matson SA, Bogdanov M, et al. Therapeutic effects of cystamine in a murine model of Huntington's disease. J Neurosci. 2002;22:8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apostol BL, Kazantsev A, Raffioni S, Illes K, Pallos J, et al. A cell-based assay for aggregation inhibitors as therapeutics of polyglutamine-repeat disease and validation in Drosophila. Proc Natl Acad Sci U S A. 2003;100:5950–5955. doi: 10.1073/pnas.2628045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, et al. Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 45.Diamond MI, Robinson MR, Yamamoto KR. Regulation of expanded polyglutamine protein aggregation and nuclear localization by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2000;97:657–661. doi: 10.1073/pnas.97.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochem J. 1993;292(Pt 2):605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentration of compounds for testing in C. elegans was assessed using a food clearance assay. The OD of E. coli is reported daily for each concentration of TSA (A) and MTR (B). The mean OD is calculated for each day from triplicate samples and plotted over time.
(1.17 MB EPS)
C. elegans model of polyQ neurodegeneration and cell death [6], [10]. (A) An N-terminal fragment of human huntingtin with 150 glutamines (Htn-Q150) was expressed in C. elegans ASH neurons as previously described [6]. (B) A GFP reporter expressed in the ASH neurons was used to assess cell death; loss of GFP expression in ASH neurons indicates cell death as shown here in young pqe-1;Htn-Q150 animals (bottom panel). (C) The ASH neuron and 5 other sensory neurons in Htn-Q150 young (top panel) or aged (bottom panel) animals were visualized by dye filling. Sensory endings of ASH neurons expressing Htn-Q150 degenerate in aged animals and fail to take up dye (bottom panel). (D) ASH neuronal survival decreases dramatically over time based on GFP fluorescence. By day 3, 75% of ASH neurons are dead in pqe-1;Htn-Q150 animals. Error bars represent SEM. Scale bar is approximately 20 nm. (E) Only 21% of ASH neurons remain intact in aged (7 day) pha-1 animals expressing Htn-Q150 raised at 25°C compared to young (3 day) animals based on dye filling.
(2.94 MB EPS)
HtnQ150 transgene levels are not decreased in the presence of LiCl or MTR. Relative HtnQ150 transgene levels were determined using the ΔΔCt method and normalized to an act-1 control. Results are from two independent RNA collections from untreated animals (C) and animals treated with 25 mM LiCl or 0.5 mM MTR in the starved assay format for 24 hours at 15°C. The error bars indicate the standard deviation from two replicates of qRT-PCR experiments comparing untreated and drug treated samples.
(0.63 MB EPS)
HDAC inhibitor trichostatin A (TSA) decreases polyQ toxicity measuring ASH neurodegeneration or neuronal death. At least three trials for each efficacy assay were performed. Representative experiments are depicted. (A) TSA decreases polyQ toxicity in a pqe-1 independent manner in pha-1;Htn-Q150 aged animals. 30 synchronized pha-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with E. coli at an OD at A595 of 0.5 with varying concentrations of TSA to a total volume of 1 ml. On day 7, animals were collected, dye filled and 100 neurons were scored for degeneration. (B) TSA increases neuronal survival in the absence of growth using the starvation assay. 50 synchronized pqe-1;Htn-Q150 L1s per well were incubated at 25°C in S medium with varying concentrations of TSA to a total volume of 60 µl. On day 2, the survival of 100 ASH neurons was evaluated by the presence or absence of GFP. Animals were treated with final concentration of TSA or vehicle (2% DMSO).
(0.66 MB EPS)