Abstract
DNA copy number changes were investigated in 69 samples of schistosoma-associated (SA) and non-schistosoma-associated (NSA) squamous cell carcinoma (SCC) and transitional cell carcinoma (TCC) of the bladder by comparative genomic hybridization (CGH). DNA copy number changes were detected in 47 tumors. SA tumors had more changes than NSA tumors (mean, 7 vs. 4), whereas the number of changes in SCC and TCC tumors was similar. SA tumors displayed more gains than losses (1.7:1), whereas NSA tumors showed an equal number of gains and losses. Changes that were observed at similar frequencies in SCC and TCC, irrespective of the schistosomal status, included gains and high-level amplifications at 1q, 8q, and 20q and losses in 9p and 13q. These changes may be involved in a common pathway for bladder tumor development and progression independent of schistosomal status or histological subtype. Losses in 3p and gains at 5p were seen only in SCC (P < 0.01) and losses in 5q were more frequent in SA-SCC than in other tumors (P < 0.05). However, changes that were more frequent in TCC than those in SCC included gains at 17q (P < 0.01) and losses in 4q (P < 0.05) and 6q (P < 0.01). Gains and high-level amplifications at 5p were seen only in SA-SCC (P < 0.01), whereas gains and high-level amplifications with minimal common overlapping regions at 11q13 were more frequently seen both in SA-SCC and SA-TCC tumors (P < 0.01). In addition to the above mentioned alterations, several other changes were also seen at lower frequencies. The variations in the DNA copy number changes observed in TCC, SCC, SA, and NSA bladder carcinomas suggest that these tumors have different genetic pathways.
In Western countries, more than 90% of primary bladder carcinomas (BC) are transitional cell carcinoma (TCC), whereas squamous cell carcinoma (SCC) comprises less than 10%. 1 Carcinoma of the urinary bladder is the most common malignancy in many tropical and subtropical countries due to endemic infection by Schistosoma hematobium. Schistosoma-associated bladder carcinoma (SA-BC) defines a characteristic pathology that differs from non-schistosoma-associated bladder carcinoma (NSA-BC). 2
Bladder cancer complicating schistosomiasis constitutes 30.8% of all cancers in Egypt, ranking first among the reported malignancies in Egyptians. 3 Egypt has the highest frequency of bladder cancer in the world. In contrast to Western countries, more than two-thirds of bladder cancer in Egypt are SCC with a peak incidence at around 50 years of age.
The chromosomal alterations in Western TCC have been extensively studied. About 140 tumors have been studied by banding cytogenetics 1,4 and 212 tumors by comparative genomic hybridization (CGH). 5-10 These studies have shown several numerical and structural chromosomal aberrations involving mainly chromosomes 3, 5, 7, 8, 9, 17, and 20. In contrast, only four cases of SCC have been analyzed cytogenetically 11-13 and there are no cytogenetic reports on SA-BC.
The most common genetic alteration identified in TCC is loss of heterozygosity (LOH) on 9p21, where the tumor suppressor gene p16 is located. 14-16 Mutations and nuclear accumulation of p53 are frequently seen in NSA-TCC 17,18 and were demonstrated in SA-SCC. 19,20 However, the proportion of TP53 mutations of basepair substitution at CpG dinucleotides was significantly higher in SA-BC than in NSA-BC. 21
The cytogenetic data available from some studies on Western NSA-TCC have been obtained using fluorescent in situ hybridization on SA-BC with probes for chromosomes 7, 9, and 17. 22,23 Although these studies have shown differences between frequencies of chromosomal changes in invasive SA-SCC and SA-TCC or NSA-TCC, they do not provide an overview of the chromosomal alterations in SA-BC.
CGH enables the screening of entire tumor genomes for gains and losses of DNA copy number and consequent mapping of aberrations to chromosomal subregions. 24,25 So far, only NSA-TCC has been studied by this technique. In this study, we used CGH to compare the DNA copy number changes in SA-TCC, NSA-TCC, and SCC of the bladder.
Materials and Methods
Characterization of Tumors
A total of 69 cases of primary bladder carcinomas were obtained. Thirty-eight cases were SA-BC and 31 NSA-BC. All SA-BC and 16 of the NSA-BC were collected from the files of the Pathology Department, National Cancer Institute (Cairo, Egypt) and 14 cases of the NSA-BC were collected from the files of The Institute of Pathology, The Royal London Hospital (London, UK). All of the material consisted either of cystectomy specimens or surgical biopsies and was obtained either as frozen tissue sections (41 samples) or fixed in 10% buffered formalin and paraffin-embedded (28 samples) as shown in Table 1 ▶ Table 1A ▶ . The diagnosis, classification, and tumor grading were based on light microscopy examination using the criteria of the World Health Organization (WHO) classification of urinary bladder carcinomas. 26 Of the 38 SA-BC, 28 were SCC and 10 were TCC, and of the 31 NSA-BC, 18 were SCC and 13 were TCC. The SCCs had squamous cell differentiation in the entire tumor. SA-BC was histologically verified by the presence of schistosomal cystitis in the bladder mucosa close to the tumor. The stages of paraffin-embedded tumors were comparable to those of frozen tumors. Sixty-three tumors (91%) were staged as pT2-pT4, one tumor as pT1, and five tumors as noninvasive pTa (NSA-TCC; Table 1 ▶ ).
Table 1.
Histopathology and CGH Karyotype in Squamous and Transitional Cell Bladder Carcinomas
| No./age/sex/sample codes | Pathology | CGH karyotype | |||
|---|---|---|---|---|---|
| Tissue | Grade | Stage | Losses | Gains | |
| Schistosoma-associated SCC | |||||
| 1/26/F B11 | PF | I | T3 | 2q31-q33, 3p12-q13, 4q, 8p, 9p21-pter | 2p22-pter, 4p, 6p, 8q, 9q, 11q13, 15q22-qter, 20q12-qter |
| 2/35/M B03 | PF | II | T3 | 3p, 4q11-q28 | 5p15, 8q, 11q13-q22 |
| 3/60/M B05 | PF | II | T3 | 4q24-qter, 13q21-q22 | 5p |
| 4/42/M B14 | PF | II | T3 | 4, 5q, 13q21-q31 | 1q24-qter, 7q |
| 5/38/M B16 | PF | II | T3 | 3p14-q13, 4, 5q15-q23, 9p | 2p22-pter, 2q14-q21, 8q24, 11q11-q21, 15q14-qter |
| 6/52/M B25 | PF | II | T3 | 8p, 13q21-q31 | 1q21-q24, 4p, 8q |
| 7/74/M B32 | PF | II | T3 | 5q11-q22, 6q, 9p21-pter, 13q21.2-q31, 14 | 1q21-q24, 2q12-q21, 3p21-pter, 5p, 7q33-qter, 8q22, 11q11-q13, 12q (12q14-q15), 17q, 20q12-qter |
| 8/45/M B37 | PF | II | T3 | 4q, 13q21.1-q22 | 6p, 7 |
| 9/51/M B53 | PF | II | T3 | 13q13-qter | 11q11-q13 |
| 10/35/M B24 | PF | II | T3 | − | 11q13 |
| 11/55/M BC-22 89-1412A | FZ | II | T3 | 5q | 17q |
| 12/49/F BC-49 89-1294A | FZ | II | T3 | 5q11-q31 | 8q22-qter, 11q11-q13 |
| 13/60/M BC-16 89-1294A | FZ | II | T3 | − | 5p14-pter, 11q13-q14.2 |
| 14/53/M BC-6 89-1394A | FZ | II | T3 | − | 1q21-q24 |
| 15/35/M BC-33 89-1822A | FZ | II | T3 | 3p, 11q22-qter | 11q13 |
| 16/54/M B29 | FZ | III | T3 | 13q21-q31 | 11q11-q13 |
| 17/65/M B56 | PF | III | T3 | 3p, 4, 5q, Xp, 8, 9, 10q22.2-qter, 11p13-pter, 13q13-q32 | 5p, Xq, 10pter-q22.1, 11q11-q14, 12, 17q, 19, 20 (20q), 22 |
| 18/41/M B21 | PF | III | T3 | 9p21-pter | 5p, 6p, 8p12-qter, 11q13, 11q22-q23.2, 20q12-q13.2 |
| 19/51/M BC-50 89-1821A | FZ | I | T4 | 3p, 13, 18q | 3q, 6p22-pter, 8q21-qter, 17q |
| 20/39/M B13 | PF | II | T4 | 18 | 4p15, 6p, 8q (8q24), 11q13, 14q21-qter, 15, 17q, 20q, 22 |
| 21/55/M B34 | PF | II | T4 | 13q13-q31, 9p21-pter | 1q21-q25, 6p, 8p21-pter, 9q34, 12q13-q21 (12q14-q15), 17q, 20q |
| 22/50/M B52 | PF | III | T4 | 5q14-q23, 6q, 13q21-q31 | 5p, 7pter-q21, 9, 11q, 20 |
| Non-Schistosoma-Associated SCC | |||||
| 23/56/M BC-5 SD 71/94 | FZ | I | T2 | 13q | − |
| 24/53/M B20 | PF | I | T3 | − | 8q (8q24) |
| 25/48/M B38 | PF | II | T3 | 6q11-q15, 13q21-q31 | − |
| 26/77/F BC-11 SD-2004/94 | FZ | II | T3 | 13q13-q31, 18q12.2-q22.2 | − |
| 27/45/M B26 | PF | III | T3 | − | 1q, 8q22-qter, 9q22-qter, 20q12-qter |
FZ, frozen tissue; PF, formalin-fixed paraffin-embedded tissue; Ta, papillary, non-invasive; T1, invasion limited to lamina propria; T2, invasion limited to inner half of muscularis propria; T3, invasion into outer half of muscularis propria or the perivesical fat; T4, invasion to contiguous viscera or pelvic organs; SCC, squamous cell carcinoma; TCC, transitional cell carcinoma.
High-level amplifications are in bold type.
Table 1A.
Continued
| No./age/sex/sample codes | Pathology | CGH karyotype | |||
|---|---|---|---|---|---|
| Tissue | Grade | Stage | Losses | Gains | |
| 28/42/F B02 | PF | III | T3 | − | 20q12-qter |
| 29/61/F B01 | PF | II | T4 | − | 8, 17 |
| 30/70/M B19 | PF | II | T4 | 2q31-q35 | 3q, 4p16, 6p22-pter, 8q23-qter (8q24) |
| 31/72/F BC-39 SD 5833/93 | FZ | II | T4 | 4q21-qter, 6q14-q22, 13q14.2-q31.2 | 1q21-q23 |
| 32/85/F BC-13 SD 4773/88 | FZ | III | T4 | 3p, 4, 5q, 9p21-pter, 10p, 18q | 1q, 2p, 3q, 7q, 11q11-q21(11q13), 17q11-q22, 18p, 20q, 22 |
| 33/78/F BC-48 SD 2031/88 | FZ | III | T4 | 3p13-q13, 13q21-q32 | 11q13, 20q |
| Schistosoma-Associated TCC | |||||
| 34/63/M B28 | PF | III | T3 | 4, 6q11-q22, 8q11-q13, 10q11-q21, T1p, 13q11-q32, 18q21-qter | 3p21-pter, 3q, 7, 8q21.3-qter, 9p, 11q11-q14, 11q23-qter, 12q23-qter, 20 |
| 35/60/M BC-43 89-1660A | FZ | III | T3 | − | 7q, 11q13-qter, 17q, 20q |
| 36/46/M BC-41 89-1854A | FZ | III | T3 | 4, 9p | 3p14-pter, 8q22-qter, 17q, 20q |
| 37/66/M BC-21 89-1315A | FZ | III | T4 | 4p | 1q21-q31.2, 10p, 11q11-q13 |
| Non-Schistosoma-Associated TCC | |||||
| 38/90/M BC-31 SD 4194/94 | FZ | II | Ta | 2q23-qter, 4q22-qter, 11p13-pter, 13 | 1q21-q31 (1q22-q23), 4p15-pter, 6p, 10p, 18p |
| 39/70/M BC-15 SD 637/94 | FZ | II | Ta | 8p, 9p21-pter, 11q22.2-qter, 18q | − |
| 40/77/F BC-32 SD 4535/94 | FZ | II | Ta | 2q23-q32,4, 13q21-qter | 1q22-q24, 11q13 |
| 41/76/F BC-17 SD 3401/94 | FZ | II | Ta | 18q | 17q, 20q (20q13.1) |
| 42/78/M BC-30 SD 3158/94 | FZ | II | T1 | 4q13-qter, 9p21-pter, 13q21-q31 | 1q21-q25, 15, 19, 20 (20q13.1-qter), 22 |
| 43/64/F B18 | PF | II | T3 | 2q24-q33 | 2p22-pter, 6p, 8q23-qter, 17q |
| 44/53/M B04 | PF | III | T3 | 6q11-q23 | 11q13 |
| 45/81/F BC-29 SD 5877/94 | FZ | III | T3 | 6q13-q21, 13q21-q31 | 17q, 20 |
| 46/46/M B27 | PF | II | T4 | 6q11-q21 | 7p13-p21, 8q24, 11q13-qter |
| 47/83/F BC-47 SD 3766/94 | FZ | III | T4 | 4q23-qter, 6q16-q23,, 9p, 13q13-q22 | 1q22-q24, 8q23-qter, 17q |
CGH
DNA was extracted from frozen tissue sections following standard methods, whereas DNA from paraffin-embedded tissue sections was extracted as described earlier. 27
CGH was performed according to standard procedures 28 with a modification using a mixture of fluorochromes conjugated to dCTP and dUTP nucleotides for nick translation. 29 Hybridizations, washings, and ISIS digital image analysis (Metasystems GmbH, Altlussheim, Germany) were performed as described elsewhere. 30 Three-color images (red for reference DNA, green for tumor DNA, and blue for counterstaining) were acquired from 8 to 10 metaphases per sample. Only metaphases of good quality with strong uniform hybridization were included in the analysis. Chromosomes not suitable for CGH analysis (ie, chromosomes heavily bent, overlapping, or with overlying artifacts) were excluded. Based on our earlier reports and the control results, we used 1.17 and 0.85 as cut-off levels for gains and losses, respectively.
Controls
In each CGH experiment, a negative control (peripheral blood DNA from a healthy donor) and a positive control were included. The positive control was a gastric tumor with known DNA copy number changes.
Statistical Analysis
All of the CGH results were confirmed using a 99% confidence interval. Briefly, intraexperiment standard deviations for all positions in the CGH ratio profiles were calculated from the variation of the ratio values of all homologous chromosomes within the experiment. Confidence intervals for the ratio profiles were then computed by combining them with an empirical interexperiment SD and by estimating error probabilities based on the t distribution. For the analysis of the frequencies of DNA copy number changes in BC histological subtypes, we used Fisher’s exact two-tailed test. P values <0.05 were considered significant.
Results
Changes in DNA copy numbers were detected in 47 tumors, 26 SA-BC, and 21 NSA-BC. A total of 149 gains and 96 losses was detected. Tumors that had no CGH changes included 6 SA-SCC, 7 NSA-SCC, 6 SA-TCC, and 3 NSA-TCC. SA-BC had more changes than NSA-BC (mean, 7 and 4, respectively), whereas SCC and TCC showed a comparable number of changes. SA-BC displayed more gains than losses (1.7:1), whereas NSA-BC had an equal number of gains and losses. No differences were noticed in the CGH results between DNA extracted from frozen and paraffin-embedded tissue sections.
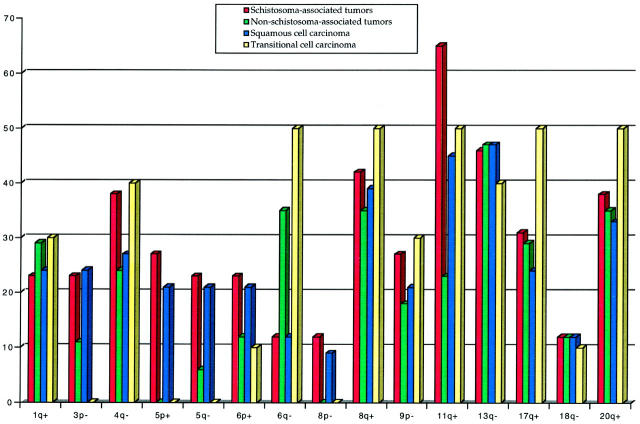
Because CGH sensitivity requires at least 50% of tumor material within a sample, tumors without alterations (Table 1) ▶ were excluded from the interpretation of the results and from the statistical analysis as they may reflect a high contamination by normal cells within the tumor material. In addition, four pTa tumors that had CGH changes (Table 1) ▶ were excluded from the statistical analysis as they may represent an entirely different tumor entity from invasive bladder carcinomas. Among the abnormal cases, the common overlapping regions of the most frequent changes were defined as follows. Gains and high-level amplifications at 11q13 were seen in 65% of SA tumors compared to 23% of NSA tumors (P < 0.01). Gains and high-level amplifications at 5p (21%) and losses in 3p (24%) were only seen in SCC tumors (P < 0.01). The gains at 5p were limited to SA-SCC (32%, 7 tumors, P < 0.01) and losses in 5q were more frequent in SA-SCC (P < 0.05). Changes that were more frequent in TCC than SCC tumors included gains at 17q11-q22 (50% vs. 24%, P < 0.01) and losses in 4q24-qter (40% vs. 27%, P < 0.05) and 6q11-q21 (50% vs. 12%, P < 0.01). Gains and high level-amplifications at 1q, 8q24, and 20q12-q13, and losses in 9p and 13q21-qter were seen equally in both SCC and TCC irrespective of the schistosomal status. Other changes were seen as gains at 1q, 2p, 3q, 7, 9, 12q, 14, 15, and 22, and losses in 2q, 3p, 5q, and 18q.
The details of DNA copy number changes are shown in Table 1 ▶ and Figure 1 ▶ . Figure 2 ▶ shows the relative frequencies of the aberrations among abnormal cases.
Figure 1.
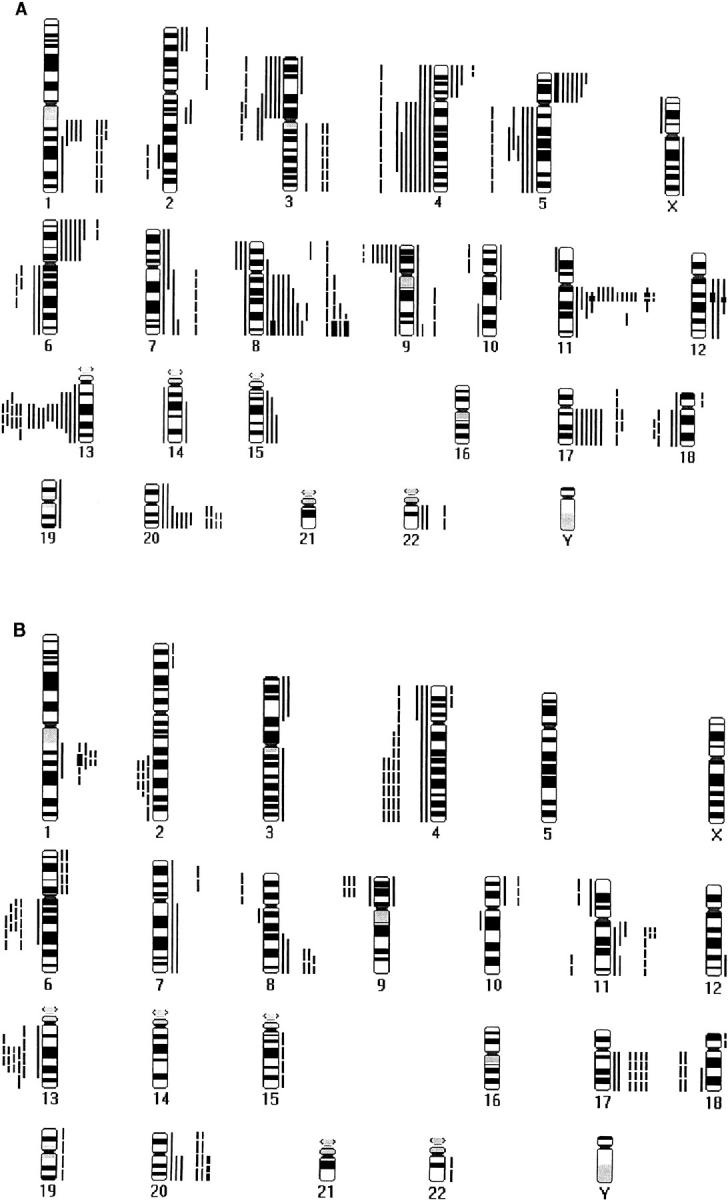
Summary of DNA copy number gains and losses detected by CGH in bladder cancer. A: Squamous cell carcinoma. B: Transitional cell carcinoma. Each bar represents one tumor sample. Gains are on the right hand side, losses on the left. Continuous lines represent schistosoma-associated tumors, broken lines non-schistosoma-associated tumors, and bold lines high-level amplifications.
Figure 2.
Comparative frequencies of common losses (−) and gains (+) detected in bladder tumors. Tumors with normal CGH and pTa tumors are excluded.
Discussion
We undertook to compare, for the first time, the DNA copy number changes in SCC and TCC in both SA and NSA tumors. Although most of the tumors were high-grade/high-stage, our results indicate that some of the recurrent changes were common for all BC subtypes, whereas others were more frequent in a certain histological subtype.
SA-BC versus NSA-BC
The higher number of copy number changes observed in SA-BC than in NSA-BC may be explained by the chromosome instability mediated by either reactive oxygen species or urinary nitrosamines as a result of chronic inflammation and irritation in the urinary bladder by schistosomal infection. 31 Schistosomal infection has been reported to be directly involved in increased chromosomal breakage in the urothelial cells at the micronuclei level. 32
Gains and high-level amplifications at 11q13 were significantly higher in SA-BC than in NSA-BC, indicating that 11q13 gains may be related to the schistosomal status irrespective of the histological subtype. Possible involvement of loci in chromosome 11 in controlling the level of chromosomal breakage caused by oxidative damage due to chronic schistosomal infection has been suggested earlier. 31 Among all reports of CGH studies on TCC, 11q13 gains have been rare (28 tumors, 15%) and the rate is thus comparable to NSA-BC in our material. In one study, 11q13 gains were more frequently seen in pT1 tumors than in pTa tumors. 10 However, all other CGH studies 5-9 have shown this gain to be rare and not associated with tumor stage.
Gains and high-level amplifications at 5p were seen only in SA-SCC. This gain has been less frequently reported (17%, 32 tumors) in CGH studies of TCC 5-10 and has been detected mainly in advanced TCC (pT2 and higher). 9 Cytogenetic data on TCC have shown isochromosome 5p to be the underlying mechanism of 5p gain. 1 One of our cases (no. 17) had a high level-amplification at 5p with a loss of whole 5q, which is likely to be an isochromosome 5p.
Changes involving other chromosomal regions were almost equally distributed among SA-BC and NSA-BC indicating that they may be related to bladder tumors rather than to schistosomal status.
SCC versus TCC
In the present study, gains and high-level amplifications at 5p and losses in 3p were seen only in SCC tumors. Although gains at 5p were exclusively seen in SA-SCC, a similar finding has been reported in advanced TCC. 6 Because secondary SCC can in rare instances develop on top of advanced TCC, gains at 5p may be one of the changes required for SCC differentiation. Alternatively, the high frequency of 5p in SCC may be explained by the higher stage of SCC compared to TCC. Earlier CGH studies have indicated that 3p losses are rare among the abnormal TCC (9 of 186, 5%). 5-9 Losses in 3p had a minimal common overlapping region at 3p12, which coincides with loss of heterozygosity studies that showed that deletions at 3p12 are rare and occur only in invasive TCC tumors. 33 Losses in 5q have rarely been reported in earlier CGH studies and were more frequent in our SA-SCC, indicating that the role of this change is more significant in the development of SA-SCC than in other histological subtypes.
Our results showed that some changes were more frequent in TCC than in SCC, such as gains at 1q21-q24 and 17q11-q22 and losses in 4q24-qter and 6q11-q21. Earlier CGH studies and molecular studies have indicated that these changes are more frequent in advanced TCC. 6,10,34,35 A fluorescence in situ hybridization study has shown that the number of gains at chromosome 17 was significantly higher in TCC than in SCC, 23 which is in agreement with our finding. Other changes have not been investigated in SCC. Almost all our tumors (88%) were histologically above pT2. Therefore, these changes seem to be more related to tumor progression in TCC than in SCC.
Frequent changes that were equally seen in SCC and TCC included gains and high level amplifications at 8q24, 11q13, and 20q12-q13 and losses in 9p and 13q21-qter. Gains at 8q and 20q have been reported in advanced TCC 6,10 and in several malignancies such as breast, colon, and stomach cancer. 25 The five pTa tumors in our material showed no gains at 8q and the frequency of 20q gains was low. Because most of our tumors were histologically advanced, our data together with earlier CGH and cytogenetic reports on TCC suggest that the aforementioned genetic changes are related to advanced invasive bladder tumors and are independent of the schistosomal status or the histological subtype. However, losses in 9p have been reported in both early TCC and advanced TCC on CGH 5-10 and cytogenetic studies. 1 Although chromosomal deletion at 9p was reported to be more frequent in SCC than in TCC, 23 we did not notice a similar trend, which suggests that losses in 9p are equally important for the pathogenesis of both TCC and SCC. However, losses in chromosome arm 9q, which were commonly seen in Western TCC, were rare in our material. This discordance may be attributable to differences in epigenetic and environmental factors in Western and Egyptian cases. A similar observation was made in gastric cancer (GC). Gains and high-level amplifications at chromosome arms 17q and 20q, characteristic of Western GC, 36,37 were rarely seen in Japanese GC. 38 Egypt has the highest frequency of bladder cancer and Japan, similarly, has the highest frequency of GC.
In addition to the gains at 11q13, losses in 13q were the most frequent change observed in our tumors. Losses in 13q occurred equally among SA-BC and NSA-BC. 13q losses have rarely been reported in earlier CGH studies (10%, 18 tumors) 5-10 of pTa to pT2 tumors. CGH studies on SCC of the head and neck 39-41 have shown a pattern of genetic alterations similar to that observed in our bladder SCC, such as frequent gains at 5p and 11q13. Despite the similarities, gains at 3q and 9q, which were among the most frequent changes seen in SCC of the head and neck, 39,41 were rarely seen in our bladder tumors, and losses in 4q and 13q that were common in SCC of the bladder, mainly SA, were rarely observed in SCC of the head and neck. This suggests that the oncogenesis of SCC may require certain genetic alterations, whereas additional tissue-specific alterations are needed for the tumor development.
DNA Copy Number Changes and Genes
Most of the changes observed in bladder tumors have also been reported in other tumors and some of these regions are known to contain oncogenes and tumor suppressor genes. 25 Although 11q13 gains are rare in TCC, DNA amplification of four proto-oncogenes, cyclin D1, FGF3, FGF4, and EMS1, was found in 11% of TCC (5 of 46 tumors). 42 Gains and high-level amplification at 1q21-q24, 8q24, 17q11-q22, and 20q were reported in several tumors such as breast, colon, and stomach cancer, and in osteosarcoma. 25 1q21-q24 contains oncogenes such as SKI 43 and NTRK1, 44 which have not been studied in bladder cancer. CMYC oncogene within 8q24 is known to be amplified in several tumors 45 and has been shown to be overexpressed in bladder cancer. 46 Amplifications and overexpression of oncogene ERBB2 located at 17q21 have been shown to correlate with advanced TCC but not with survival. 34,47 Amplifications at 20q have been observed to correlate with poor prognosis in breast cancer and the region is known to harbor specific amplified genes (AIB1, AIB3, and AIB4). Several candidate genes are located at 20q, eg, the PTP1B/PTPN1 gene (20q12), which is involved in growth regulation, and the MYBL2 gene (20q13), which plays an important role in cell cycle progression. Moreover, the human cellular apoptosis susceptibility gene (CAS) has been mapped to this same region. 25 None of these genes has been studied in bladder cancer.
Losses of DNA copy number in 3p included 3p25, which contains the VHL tumor suppressor gene. Mutations in VHL have been described in the inheritable von Hippel-Lindau disease, sporadic renal cell carcinoma, mesothelioma, and small cell lung carcinoma. 48 However, VHL has not been studied in bladder carcinomas. Losses in 4q have been demonstrated in a large loss of heterozygosity of TCC and correlated with late progression. 35 No tumor suppressor genes have been identified on chromosome 4. The 5q losses detected in SCC span the APC locus (5q21-q22) that plays an essential role in colon cancer as well as in several other tumors. 49 Molecular studies have shown loss of heterozygosity at RB1 locus (13q14) in TCC. 50 In addition, ING1, a candidate tumor suppressor gene, has recently been cloned and mapped to 13q34. 51,52 However, the minimal common overlapping region in our tumors was 13q21; therefore, there is a possibility of an unidentified tumor suppressor gene distal to RB1. Losses in 9p at the locus of tumor suppressor gene p16 (9p21) were more frequent in SA-SCC. 53 Nevertheless, no tumor suppressor genes or oncogenes have been assigned to regions such as 4q and 5p, which are known to be affected in bladder cancer and other malignancies.
Conclusion
Our results show for the first time the genetic changes in SA and NSA tumors of the bladder and highlight the genetic changes underlying the development of SCC. Gains and high-level amplifications at 11q13 and 5p were related to SA-BC irrespective of the histological subtype. Some changes, such as losses in 3p and gains at 5p, were more frequent in SCC, whereas losses in 4q and 6q and gains at 17q were more frequent in TCC. Other changes and high-level amplifications occurred with indistinguishable frequencies between tumor subtypes, indicating that they may be involved in a common pathway for the development and progression of bladder tumors.
Footnotes
Address reprint requests to Sakari Knuutila, Ph.D., Laboratory of Medical Genetics, Helsinki University Central Hospital, P.O.Box 404 (Haartmanink 3, 4th floor), FIN-00029 HUCH, Helsinki, Finland. E-mail: Sakari.Knuutila@Helsinki.fi.
Supported in part by grants from the Finnish Cancer Society and the Helsinki University Central Hospital (grants TYH0049 and -8219) in Finland, the schistosomiasis research project (nos. 03-11-35 and 07-01-63) in Egypt, the National Agency of Scientific and Technological Promotion (Contract grant: BID 802/OC-AR PICT No. 01-00000-00753) in Argentina, and the Gillson Scholarship (to M. S.) from the Worshipful Society of Apothecaries of London, UK.
References
- 1.Gibas Z, Gibas L: Cytogenetics of bladder cancer. Cancer Genet Cytogenet 1997, 95:108-115 [DOI] [PubMed] [Google Scholar]
- 2.Badawi AF: Molecular and genetic events in schistosomiasis-associated human bladder cancer: role of oncogenes and tumor suppressor genes. Cancer Lett 1996, 105:123-138 [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim S: Site distribution of cancer in Egypt: twelve years experience (1970–1981). Cancer Prevention in Developing Countries. 1986, :pp 20-32 Pergamon Press, Oxford [Google Scholar]
- 4.Mitelman F: Catalog of Chromosome Aberrations in Cancer, 5th ed. 1994, Wiley-Liss, New York
- 5.Kallioniemi A, Kallioniemi OP, Citro G, Sauter G, DeVries S, Kerschmann R, Caroll P, Waldman F: Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosomes Cancer 1995, 12:213-219 [DOI] [PubMed] [Google Scholar]
- 6.Richter J, Jiang F, Gorog JP, Sartorius G, Egenter C, Gasser TC, Moch H, Mihatsch MJ, Sauter G: Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res 1997, 57:2860-2864 [PubMed] [Google Scholar]
- 7.Voorter C, Joos S, Bringuier P-P, Vallinga M, Poddighe P, Schalken J, du Manoir S, Ramaekers F, Lichter P, Hopman A: Detection of chromosomal imbalances in transitional cell carcinoma of the bladder by comparative genomic hybridization. Am J Pathol 1995, 146:1341-1354 [PMC free article] [PubMed] [Google Scholar]
- 8.Hovey RM, Chu L, Balazs M, DeVries S, Moore D, Sauter G, Carroll PR, Waldman FM: Genetic alterations in primary bladder cancers and their metastases. Cancer Res 1998, 58:3555-3560 [PubMed] [Google Scholar]
- 9.Bruch J, Wöhr G, Hautmann R, Mattfeldt T, Brüderlein S, Möller P, Sauter S, Hameister H, Vogel W, Paiss T: Chromosomal changes during progression of transitional cell carcinoma of the bladder and delineation of the amplified interval on chromosome arm 8q. Genes Chromosomes Cancer 1998, 23:167-174 [DOI] [PubMed] [Google Scholar]
- 10.Simon R, Burger H, Brinkschmidt C, Bocker W, Hertle L, Terpe HJ: Chromosomal aberrations associated with invasion in papillary superficial bladder cancer. J Pathol 1998, 185:345-351 [DOI] [PubMed] [Google Scholar]
- 11.Lundgren R, Elfving P, Heim S, Kristoffersson U, Mandahl N, Mitelman F: A squamous cell bladder carcinoma with karyotypic abnormalities reminiscent of transitional cell carcinoma. J Urol 1989, 142:374-376 [DOI] [PubMed] [Google Scholar]
- 12.Fadl-Elmula I, Gorunova L, Lundgren R, Mandahl N, Forsby N, Mitelman F, Heim S: Chromosomal abnormalities in two bladder carcinomas with secondary squamous cell differentiation. Cancer Genet Cytogenet 1998, 102:125-130 [DOI] [PubMed] [Google Scholar]
- 13.Vanni R, Scarpa R, Nieddu M, Usai E: Cytogenetic investigation on 30 bladder carcinomas. Cancer Genet Cytogenet 1988, 30:35-42 [DOI] [PubMed] [Google Scholar]
- 14.Reeder JE, Morreale JF, O’Connell MJ, Stadler WM, Olopade OF, Messing EM, Wheeless LL: Loss of the CDKN2A/p16 locus detected in bladder irrigation specimens by fluorescence in situ hybridization. J Urol 1997, 158:1717-1721 [DOI] [PubMed] [Google Scholar]
- 15.Cairns JP, Chiang PW, Ramamoorthy S, Kurnit DM, Sidransky D: A comparison between microsatellite and quantitative PCR analyses to detect frequent p16 copy number changes in primary bladder tumors. Clin Cancer Res 1998, 4:441-444 [PubMed] [Google Scholar]
- 16.Ozen H: Bladder cancer. Curr Opin Oncol 1998, 10:273-278 [PubMed] [Google Scholar]
- 17.Chaturvedi V, Li L, Hodges S, Johnston D, Ro JY, Logothetis C, von Eschenbach AC, Batsakis JG, Czerniak B: Superimposed histologic and genetic mapping of chromosome 17 alterations in human urinary bladder neoplasia. Oncogene 1997, 14:2059-2070 [DOI] [PubMed] [Google Scholar]
- 18.Abdel-Fattah R, Challen C, Griffiths TR, Robinson MC, Neal DE, Lunec J: Alterations of TP53 in microdissected transitional cell carcinoma of the human urinary bladder: high frequency of TP53 accumulation in the absence of detected mutations is associated with poor prognosis. Br J Cancer 1998, 77:2230-2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamel D, Soini Y, Nuorva K, Khalifa A, Mangoud A, Vahakangas K, Paakko P: p53 and c-erbB-2 expression in schistosomal urinary bladder carcinomas and schistosomal cystitis with premalignant lesions. Virchows Arch 1994, 424:349-355 [DOI] [PubMed] [Google Scholar]
- 20.Cooper K, Haffajee Z, Taylor L: Human papillomavirus and schistosomiasis associated bladder cancer. Mol Pathol 1997, 50:145-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren W, Biggs PJ, el-Baz M, Ghoneim MA, Stratton MR, Venitt S: Mutations in the p53 gene in schistosomal bladder cancer: a study of 92 tumours from Egyptian patients and a comparison between mutational spectra from schistosomal and non-schistosomal urothelial tumours. Carcinogenesis 1995, 16:1181–1189 [DOI] [PubMed]
- 22.Ghaleb AH, Pizzolo JG, Melamed MR: Aberrations of chromosomes 9 and 17 in bilharzial bladder cancer as detected by fluorescence in situ hybridization. Am J Clin Pathol 1996, 106:234-241 [DOI] [PubMed] [Google Scholar]
- 23.Pycha A, Mian C, Posch B, Haitel A, El-Baz M, Ghoneim MA, Marberger M: Numerical aberrations of chromosomes 7, 9 and 17 in squamous cell and transitional cell cancer of the bladder: a comparative study performed by fluorescence in situ hybridization. J Urol 1998, 160:737-740 [DOI] [PubMed] [Google Scholar]
- 24.Knuutila S, Aalto Y, Autio K, Björkqvist A-M, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy M, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius V-M, Wolf M, Zhu Y: DNA copy number losses in human neoplasms (review). Am J Pathol 1999, 155:683-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knuutila S, Björkqvist A-M, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius V-M, Vidgren V, Zhu Y: DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998, 152:1107-1123 [PMC free article] [PubMed] [Google Scholar]
- 26.Mostofi FK, Sobin LH, Torloni H: Histological typing of urinary bladder tumours. International Histological Classification of Tumours, No. 1973, World Health Organization, 10. Geneva
- 27.Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988, 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D: Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 29.El-Rifai W, Larramendy ML, Björkqvist A-M, Hemmer S, Knuutila S: Optimization of comparative genomic hybridization using fluorochrome conjugated to dCTP and dUTP nucleotides. Lab Invest 1997, 77:699-700 [PubMed] [Google Scholar]
- 30.El-Rifai W, Sarlomo-Rikala M, Miettinen M, Knuutila S, Andersson LC: DNA copy number losses in chromosome 14: an early change in gastrointestinal stromal tumors. Cancer Res 1996, 56:3230-3233 [PubMed] [Google Scholar]
- 31.Rosin MP, Anwar WA, Ward AJ: Inflammation, chromosomal instability, and cancer: the schistosomiasis model. Cancer Res 1994, 54(suppl.):1929-1933 [PubMed] [Google Scholar]
- 32.Rosin MP, Anwar W: Chromosomal damage in urothelial cells from Egyptians with chronic Schistosoma haematobium infections. Int J Cancer 1992, 50:539-543 [DOI] [PubMed] [Google Scholar]
- 33.Li M, Zhang ZF, Reuter VE, Cordon-Cardo C: Chromosome 3 allelic losses, and microsatellite alterations in transitional cell carcinoma of the urinary bladder. Am J Pathol 1996, 149:229-235 [PMC free article] [PubMed] [Google Scholar]
- 34.Orlando C, Sestini R, Vona G, Pinzani P, Bianchi S, Giacca M, Pazzagli M, Selli C: Detection of c-erbB-2 amplification in transitional cell bladder carcinoma using competitive PCR technique. J Urol 1996, 156:2089-2093 [PubMed] [Google Scholar]
- 35.Ogawa K, Uzvolgyi E, St John MK, de Oliveira ML, Arnold L, Cohen SM: Frequent p53 mutations and occasional loss of chromosome 4 in invasive bladder carcinoma induced by N-butyl-N-(4-hydroxybutyl)nitrosamine in B6D2F1 mice. Mol Carcinog 1998, 21:70-79 [DOI] [PubMed] [Google Scholar]
- 36.El-Rifai W, Harper JC, Cummings OW, Hyytinen ER, Frierson HF, Jr, Knuutila S, Powell SM: Consistent genetic alterations in xenografts of proximal stomach and gastro-esophageal junction adenocarcinomas. Cancer Res 1998, 58:34-37 [PubMed] [Google Scholar]
- 37.Kokkola A, Monni O, Puolakkainen P, Larramendy ML, Victorzon M, Nordling S, Haapiainen R, Kivilaakso E, Knuutila S: 17q12–21 amplicon, a novel recurrent genetic change in intestinal type of gastric carcinoma: a comparative genomic hybridization study. Genes Chromosomes Cancer 1997, 20:38-43 [DOI] [PubMed] [Google Scholar]
- 38.Koizumi Y, Tanaka S, Mou R, Koganei H, Kokawa A, Kitamura R, Yamauchi H, Ookubo K, Saito T, Tominaga S, Matsumura K, Shimada H, Tsuchida N, Sekihara H: Changes in DNA copy number in primary gastric carcinomas by comparative genomic hybridization. Clin Cancer Res 1997, 3:1067-1076 [PubMed] [Google Scholar]
- 39.Liehr T, Ries J, Wolff E, Fiedler W, Dahse R, Ernst G, Steininger H, Koscielny S, Girod S, Gebhart E: Gain of DNA copy number on chromosomes 3q26-qter and 5p14-pter is a frequent finding in head and neck squamous cell carcinomas. Int J Mol Med 1998, 2:173-179 [DOI] [PubMed] [Google Scholar]
- 40.Wolff E, Girod S, Liehr T, Vorderwulbecke U, Ries J, Steininger H, Gebhart E: Oral squamous cell carcinomas are characterized by a rather uniform pattern of genomic imbalances detected by comparative genomic hybridisation. Oral Oncol 1998, 34:186-190 [DOI] [PubMed] [Google Scholar]
- 41.Brzoska PM, Levin NA, Fu KK, Kaplan MJ, Singer MI, Gray JW, Christman MF: Frequent novel DNA copy number increase in squamous cell head and neck tumors. Cancer Res 1995, 55:3055-3059 [PubMed] [Google Scholar]
- 42.Bringuier PP, Tamimi Y, Schuuring E, Schalken J: Expression of cyclin D1 and EMS1 in bladder tumours: relationship with chromosome 11q13 amplification. Oncogene 1996, 12:1747-1753 [PubMed] [Google Scholar]
- 43.Sutrave P, Hughes SH: The ski oncogene. Oncogene 1991, 6:353-356 [PubMed] [Google Scholar]
- 44.Greco A, Villa R, Pierotti MA: Genomic organization of the human NTRK1 gene. Oncogene 1996, 13:2463-2466 [PubMed] [Google Scholar]
- 45.Brison O: Gene amplification and tumor progression. Biochem Biophys Acta 1993, 1165:25-41 [DOI] [PubMed] [Google Scholar]
- 46.Sardi I, Dal Canto M, Bartoletti R, Guazzelli R, Travaglini F, Montali E: Molecular genetic alterations of c-myc oncogene in superficial and locally advanced bladder cancer. Eur Urol 1998, 33:424–430 [DOI] [PubMed]
- 47.Sauter G, Moch H, Moore D, Carroll P, Kerschmann R, Chew K, Mihatsch MJ, Gudat F, Waldman F: Heterogeneity of erbB-2 gene amplification in bladder cancer. Cancer Res 1993, 53:2199-2203 [PubMed] [Google Scholar]
- 48.Beroud C, Joly D, Gallou C, Staroz F, Orfanelli MT, Junien C: Software and database for the analysis of mutations in the VHL gene. Nucleic Acids Res 1998, 26:256-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurent-Puig P, Beroud C, Soussi T: APC gene: database of germline and somatic mutations in human tumors and cell lines. Nucleic Acids Res 1998, 26:269-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto H, Shuin T, Ikeda I, Hosaka M, Kubota Y: Loss of heterozygosity at the p53, RB, DCC and APC tumor suppressor gene loci in human bladder cancer. J Urol 1996, 155:1444-1447 [PubMed] [Google Scholar]
- 51.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K: Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet 1996, 14:415-420 [DOI] [PubMed] [Google Scholar]
- 52.Zeremski M, Horrigan SK, Grigorian IA, Westbrook CA, Gudkov AV: Localization of the candidate tumor suppressor gene ING1 to human chromosome 13q34. Somat Cell Mol Genet 1997, 23:233-236 [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Zulueta M, Shibata A, Ohneseit PF, Spruck CHr, Busch C, Shamaa M, El-Baz M, Nichols PW, Gonzalgo ML: High frequency of chromosome 9p allelic loss and CDKN2 tumor suppressor gene alterations in squamous cell carcinoma of the bladder. J Natl Cancer Inst 1995, 87:1383–1393 [DOI] [PubMed]