Abstract
Semaphorins/collapsins are a family of secreted and membrane-associated proteins involved in nerve growth cone migration. However, some are expressed widely in adult tissues suggesting additional functions. SEMA3F/H.SemaIV was previously isolated from a 3p21.3 homozygous deletion region in human lung cancer. We studied SEMA3F cellular localization using our previously characterized anti-SEMA3F antibody. In normal lung, SEMA3F was found in all epithelial cells at the cytoplasmic membrane and, to a lesser extent, in the cytoplasm. In lung tumors, the localization was predominantly cytoplasmic, and the levels were comparatively reduced. In non-small-cell lung carcinomas, low levels correlated with higher stage. In all tumors, an exclusive cytoplasmic localization of SEMA3F correlated with high levels of vascular endothelial growth factor and was related to the grade and aggressiveness. This suggests that vascular endothelial growth factor might compete with SEMA3F for binding to their common receptors, neuropilin-1 and -2 and might contribute to SEMA3F delocalization and deregulation in lung tumor. In parallel studies, SEMA3F distribution was examined in cell cultures by confocal microscopy. Marked staining was observed in pseudopods and in the leading edge or ruffling membranes of lamellipods or cellular protrusions in motile cells. SEMA3F was also observed at the interface of adjacent interacting cells suggesting a role in cell motility and cell adhesion.
SEMA3F, previously named H.SemaIV, 1 was isolated from a 3p homozygous deletion involving three small-cell carcinoma (SCLC) cell lines. 2,3,4 It is a member of the semaphorin/collapsin family, a large group of transmembrane, membrane-associated and secreted proteins containing a characteristic ∼500 amino acid sema domain. 5-10 Originally identified as repulsive molecules for nerve growth cones, 5,11 more recent studies indicate that semaphorins are bifunctional 12-16 and can have repulsive and attractant properties as a function of cGMP levels. 14 The widespread expression of semaphorins in adult tissues suggested they might have other functions as well. This was confirmed by a Sema3A (M-semaIII/D) knockout that resulted in abnormal development of somite-derived and visceral tissues in addition to neural abnormalities. 17,18
Receptors for class 3 semaphorins, including SEMA3F, have been cloned. 19,20 These include two related proteins: neuropilin-1 21,22 and neuropilin-2. 23 Interestingly, neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor VEGF165, 24 the most potent endothelial cell mitogen of five VEGF isoforms. In addition to neuropilin-1, neuropilin-2 was isolated in the expression cloning experiment in the search for VEGF165 receptor. Neuropilin-1 also binds additional proteins including a VEGF-like protein from orf virus, 25 placental growth factor-2 (a VEGF family member), 26 and putative cell adhesion ligands. 19,27 Neuropilin-1 is widely expressed in adult human tissues including lung, 24 and neuropilin-2 is expressed in mouse embryonic lung. 23 During development of the rat embryo, neuropilin-2 and SEMA3F are expressed in complementary patterns, 28 and SEMA3F binds with higher affinity to neuropilin-2. 23 In the hippocampus, neuropilin-2 is the functional receptor for SEMA3F. 28
Loss of chromosome 3p is a consistent event in human lung cancer. Frequent loss of heterozygosity (LOH) and the finding of a common homozygous deletion in three small-cell lung cancer (SCLC) cell lines suggested that 3p21.3 contains a tumor suppressor gene. 29 In support of this, it was shown that an 80-kb genomic DNA fragment from this region suppressed tumorigenic growth of mouse A9 cells in vivo. 30 This fragment was localized to a region of gene duplication containing two GTP-binding proteins (GNAI-2 and GNA-T) and two genes coding for SEMA3F and SEMA3B (previously known as H-SemaV). 2,3,4 Based on sequence-tagged sites reported by Daly et al, 31 along with available genomic sequence data covering a majority of the deletion region (http://genome.wustl.edu/gsc/human/chrom3.shtml), one could predict that SEMA3F was contained in the suppressing fragment. This was recently confirmed in abstract form (Naylor et al, abstract 433, American Society for Human Genetics, Denver CO, 1998) showing that SEMA3F was responsible for the inhibition of mouse A9 tumorigenesis.
However, despite the apparent involvement of SEMA3F in the mouse A9 system and homozygous deletions in three cell lines, other alterations of SEMA3F have not been reported. By Northern blots, SEMA3F was found to be expressed in most lung cancer cell lines 3,4 but with marked differences in expression levels including 4/23 SCLCs and 3/16 non-small-cell lung carcinomas (NSCLCs) without SEMA3F transcripts. 4 In no studies have SEMA3F protein levels been examined. To further elucidate the role of SEMA3F in lung cancer, we screened tumors for protein expression using a specific polyclonal antibody raised against a unique peptide. 32 In addition, neuropilin-1 and -2 expression patterns were examined by reverse transcriptase-polymerase chain reaction (RT-PCR) along with immunohistochemical analysis of VEGF in the same tumor samples. Although Sema3A (SemaIII) can effectively compete with VEGF for binding to neuropilin-1, 28,33 whether similar competition exists between SEMA3F and VEGF165 and neuropilin-1 or/and neuropilin-2 is unknown. Our findings imply that this does occur. Last, indirect immunofluorescence with laser-scanning confocal microscopy was used to analyze the cellular distribution of endogenous SEMA3F in tumor cell lines.
Materials and Methods
Patients and Samples
Ninety-five lung tumors were obtained at the time of tumor resection in NSCLC and carcinoids, in three large-cell neuroendocrine carcinomas (LCNEC), and in one SCLC. Tumor samples from seven LCNEC and nine SCLC were obtained by diagnostic lymph node resection. The tumors were classified according to the 1999 World Health Organization (WHO) histological classification of lung tumors 34 as follows: 64 non-small-cell lung carcinomas (non-neuroendocrine tumors) comprising 29 squamous carcinomas and 35 adenocarcinomas; and 31 neuroendocrine tumors comprising 10 SCLCs, 10 LCNEC (another high grade neuroendocrine tumor) and 11 carcinoids (7 typical and 4 atypical) according to newly defined criteria (WHO 1999). According to TNM-derived stage classification, 37 of 64 patients with NSCLC were at stage I, 10 at stage II, 17 at stage III, and 1 at stage IV at the time of tumor resection. Nine of 10 carcinoid tumors were at stage I and one (atypical) at stage II. Among patients with LCNEC, 4 were at stage I, 4 at stage III, and 4 at stage IV. Six patients with SCLC were at stage III and 4 at stage IV. Tumor samples were quickly frozen for immunohistochemical and RT-PCR analysis in nitrogen liquid-cooled isopenthane. Other samples were used for histological classification.
Cell Lines
A series of 18 lung cancer cell lines including NCI-H661 (large-cell NSCLCs), NCI-H1450 and NCI-H740 (SCLC) were obtained from the Lung Cancer SPORE Tumor Bank Core of the University of Colorado Cancer Center. The SCLC cell line GLC20 has been described previously 35 and was kindly provided by Dr. Charles Buys (University of Gronigen, The Netherlands). Human acute myeloid leukemia Kasumi-1 cell line, 36 K562 cell line (from American Type Culture Collection (ATCC), Manassas, VA) in addition to the human cervical tumor cell line HeLa, were grown in RPMI-1640 containing 10% fetal calf serum. The human neuroblastoma cell line SK-N-SH (HTB-11), obtained from Dr. M. O. Jauberteau (University de Limoges, France) was grown in Dulbecco’s modified Eagle’s medium (DMEM) with 10% calf serum. The human lung adenocarcinoma cell line Calu-3 (ATCC: HTB-55) was grown in DMEM/F-12 with 10% calf serum.
RNA Expression
Total RNA was prepared with the SV Total RNA isolation kit (Promega, Madison, WI), and RT-PCR was performed with SuperscriptII reverse transcriptase (Life Technologies, Inc.) using the procedure supplied by the manufacturer. Alternatively spliced 915- and 1008-bp fragments from the SEMA3F cDNA were amplified by PCR with the 59H8 and 39G5 primers as previously described. 2 A 154-bp fragment of neuropilin-2 cDNA was amplified with the following primers: 5′-AAGAGCGAAGAGACAACCAC-3′ and 5′-CATACATCCAACCACAGGG-3′ with an annealing temperature of 55°C. A 373-bp neuropilin-1 cDNA fragment was amplified with the primers 5′-GAAAGATAGCCCCTCCTCC-3′ and 5′-CCACAGTAACGCCCAATG-3′ with touch-down conditions starting from 64°C with a decrease of 1°C for every two cycles to the final temperature of 55°C (which was followed by 15 more cycles). The specificity of the PCR reaction was checked by sequencing the amplified fragment.
Quantitative PCR
We assessed levels of SEMA3F transcription relative to G3PDH in lung tumors by quantitative PCR carried out with the GeneAmp 5700 (PE Biosystems, Norwalk, CT) quantitative PCR system with syber-green chemistry. The cycle at which a particular sample reaches an arbitrary threshold fluorescence level (Ct) is indicative of input quantity of that template. Two different sets of primers were used to quantify SEMA3F cDNA: one set was SEMA3F-r 5′-GAG TCA GGG AAG GGC AAG-3′ and SEMA3F-f 5′-CAA GTG TGC GGA AGA TGG-3′ giving a 130-bp product and the other one was SEMA3F-7/21f 5′-AGC AGA CCC AGG ACG TGA G-3′ and SEMA3F-7/21r 5′-AAG ACC ATG CGA ATA TCA GCC-3′ giving a 114-bp product. G3PDH cDNA was amplified with primers G3PDH For 5′-TGC ACC ACC AAC TGC TTA GC-3′ and G3PDH Rev 5′-GGC ATG GAC TGT GGT CAT GAG-3′ giving a 87-bp product. The PCR was carried out in 50-μl reaction volumes consisting of 1× PCR SYBR Green 10X buffer, 0.25 μM primers, 200 μM each dNTP and 0.03 units/μl AmpliTaq Gold (PE). cDNA was amplified as follows: 50°C for 2 minutes, 95°C for 10 minutes followed by 40 cycles at 95°C × 15 sec, 60°C × 1 minute.
Immunohistochemical Analysis on Normal Lung and Tumors
SEMA3F immunostaining was performed with a rabbit polyclonal, affinity-purified, anti-SEMA3F antibody (3443–3AP) that was raised against a 16-amino acid peptide located in the C-terminal domain of the protein. 32 Five μm thick frozen sections were fixed in cold acetone (−20°) for 10 minutes and allowed to dry at room temperature for 3 h. The anti-SEMA3F antibody was applied overnight at a 1:50 dilution. VEGF immunostaining was performed on paraffin and frozen sections from the same tumors. Endogenous peroxidase was quenched using 0.3% hydrogen peroxide in distilled water for 5 minutes, at room temperature, before incubation with anti-VEGF antibodies. Polyclonal anti-VEGF A20 (Santa Cruz Biotechnology, Santa Cruz, CA) (1:400 dilution on paraffin sections; 1:800 on frozen sections) and monoclonal antibody mAb 293 (R&D Systems, Oxon, UK) (1:25 dilution) were incubated overnight at 4°C. Slides were washed in phosphate-buffered saline (PBS) and then exposed to the secondary biotinylated donkey F(ab′)2 anti-rabbit antibody (Ab) (1:1000 dilution; Jackson Laboratory, West Grove, PA) or anti-mouse Ab (1:500 dilution; Jackson Laboratory), for 1 h at room temperature. All slides were then washed in PBS and incubated with the streptavidin, biotin, and peroxidase complex (1:400 dilution; DAKO, Copenhagen, Netherlands) for 1 h at room temperature. The chromogenic substrate of peroxidase was a solution of 0.05% 3.3′-diaminobenzidine tetrahydrochloride, 0.03% H2O2, and 10 mM imidazole in 0.05 mol/L Tris buffer (pH 7.6). The slides were counterstained with Harris’ hematoxylin. Normal rabbit or mouse IgG at the same concentration as the primary antibodies served as a negative control. SEMA3F and VEGF immunostaining were scored, taking into account the percentage of positive cells (from 1 to 100%) and intensity of staining (from 1 to 3), as compared with internal controls. Internal controls for SEMA3F were normal bronchi and alveolar epithelial cells as described in Results and considered as intensity 3. Normal controls for VEGF were normal bronchi and smooth muscle cells, both were recorded as intensity 2. The total score was established by multiplying the percentage of stained cells by the intensity of staining, giving scores from 0 to 300. Final scores of 0, 1, 2, and 3, were given for total scores of 0, 10 to 30, 30 to 150, and 150 to 300, respectively. Scores were assessed by two independent observers (EB and JR), and a consensus was obtained.
To assess the specificity of the polyclonal antibodies, we performed a blocking experiment with both anti-VEGF A20 and anti-SEMA3F antibodies by incubating the primary antibody with a 10 times excess weight of immunizing peptide: VEGF A20 blocking peptide (Santa Cruz) and the 16-amino acid SEMA3F peptide, 32 respectively, at room temperature for 2 h before incubation on sections. The remainder of the incubation and detection were performed as above. No immunostaining with these preabsorbed antibodies was obtained.
Immunostaining and Confocal Microscopy on Cell Lines
Cells in culture were fixed in 1% paraformaldehyde in Tris-buffered saline solution (TBS: 20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2 mM EGTA, 2 mM MgCl2) for 10 minutes and permeabilized during 6 minutes in cold methanol for immunostaining of endogenous SEMA3F. For double-staining of SEMA3F and F-actin, cells were fixed in 1% paraformaldehyde in TBS for 10 minutes and permeabilized for 5 minutes in 0.1% Triton X-100/TBS. F-actin intracellular organization was visualized by staining with 1 μg/ml tetramethylrhodamine B isothiocyanate (TRITC)-conjugated phalloidin. Immunostaining for SEMA3F was performed with the anti-SEMA3F antibody 3443–3AP 32 diluted at 1:50, combined with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin secondary antibody, diluted at 1:200. All antibodies were diluted in TBS containing 1% bovine serum albumin (BSA) and 2% goat serum. Negative controls were performed by preabsorbing the antibody with the immunizing peptide. 32 Stained samples were mounted and viewed with ×63 oil immersion objectives using a Zeiss epifluorescence microscope (Axiovert 135 mol/L) equipped with HBO 50 W mercury short-arc lamp. The fine distribution of SEMA3F and actin was analyzed by means of laser-scanning confocal microscopy (Biorad 1024) with an inverted microscope (Olympus IX70) equipped with ×63 and ×100 Neofluar oil immersion objectives. Samples were excited with the visible lines 488 or 568 nm of an argon/krypton laser beam for green and red fluorescence, respectively. Fluorescence emissions were collected via two photomultiplier tubes through a band pass filter (522DF35) for green FITC fluorescence, or a band pass filter (605DF32) for red TRITC fluorescence.
Results
SEMA3F Level and Cellular Localization in Normal Lung and Lung Tumors
SEMA3F was detected with a polyclonal antibody raised against a 16-amino acid peptide as previously described. 32 This affinity-purified antibody is useful for immunochemistry but does not work for immunoprecipitation and Western blot. The specificity was demonstrated by the following observations. First, in three separate lung cancer cell lines with homozygous deletions involving SEMA3F, no signals were observed. In contrast, in all other cell lines where SEMA3F mRNA was detected, the immunohistochemical stain was positive. Second, blocking with the immunizing peptide eliminated the signal. Third, the antibody recognized the wild-type protein in transfected insect cells, whereas no signal was observed in untransfected cells. Last, in support of these observations the immunizing peptide is a unique epitope among semaphorins as determined from an analysis of sequences present in GenBank.
Normal Lung Structures
In normal lung, SEMA3F was detected in all epithelial cells (Figure 1A and B) ▶ . The use of frozen section (instead of paraffin section) does not allow expansion of lung parenchyma; thus, it gives a false impression of interstitium thickening (Figure 1B) ▶ . In large bronchi (cartilaginous bronchi), basal bronchial cells were stained more intensely than ciliated or goblet cells. There was a stronger membrane staining in addition to mild diffuse cytoplasmic staining. Mucous and serous gland cells displayed the same staining pattern. In bronchioles, SEMA3F was essentially restricted to basal epithelial cells with mild cytoplasmic and strong membrane staining. All types of alveolar lining cells displayed only a membranous staining pattern that was restricted to the apical intra-alveolar surface except in areas of type II cell hyperplasia where a mild cytoplasmic staining was observed in addition to membranous staining. This pattern of staining resembled that observed in basal bronchial and bronchiolar cells. Endothelial cells of the alveolar capillary bed did not express SEMA3F, whereas about 20% of vessels more than 100-μm in diameter (arterioles and venules) displayed endothelial staining with the typical apical membrane pattern. There was no apparent staining in lymphatic vessels, however the lymphatic bed is quite obscured in normal lung.
Figure 1.
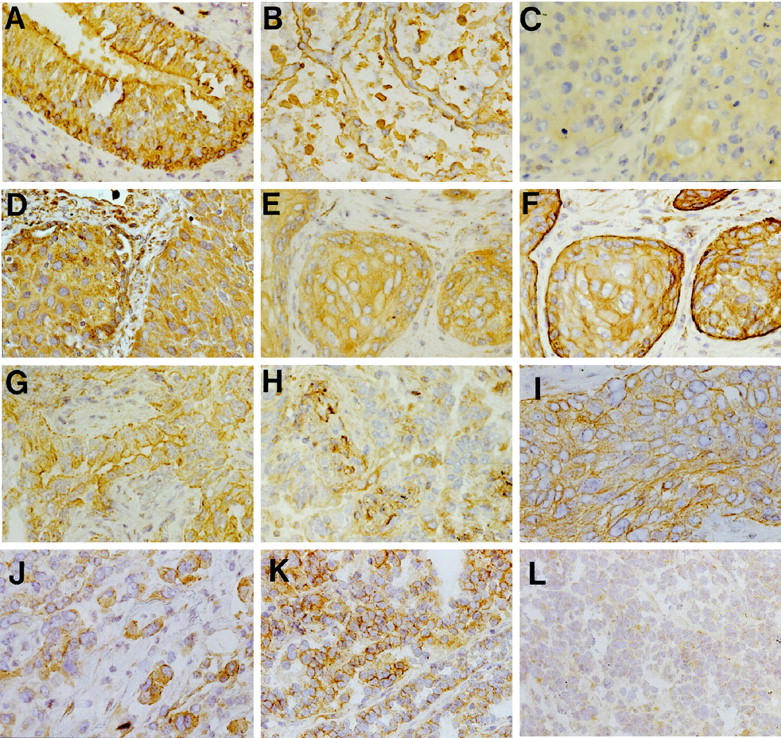
SEMA3F and VEGF levels and localization in normal lung and lung tumors. SEMA3F (A–C, E, G–L) and VEGF (D, F, H) were detected with a secondary biotinylated anti-rabbit antibody. In normal lung (A, B), SEMA3F was detected in all epithelial cells (score 3) with strong membranous staining in bronchial epithelial cells (A) and type II pneumocytes (B). Three examples are given for lung tumors (C–H) for: a squamous cell carcinoma with SEMA3F score 0 to 1 (C) and VEGF score 3 (D); for another squamous cell carcinoma with SEMA3F score 2 (E) and VEGF score 3 (F); and for an adenocarcinoma with SEMA3F score 3 (G) and VEGF score 0 to 1 (H). Note in C and E the diffuse cytoplasmic localization of SEMA3F, and in G the membranous localization of SEMA3F. SEMA3F was located at the basally oriented membrane toward tumor stroma in carcinoma suggesting a function in cellular interactions (I). Cells disseminated in the stroma expressed higher levels of SEMA3F (J) than in lobules. SEMA3F staining was intense and membranous in low grade neuroendocrine carcinoid (K) but diffuse and cytoplasmic in high grade neuroendocrine SCLC tumors (L).
Lung Tumors
Before performing SEMA3F immunodetection, we first verified by RT-PCR that SEMA3F mRNA was expressed in 18 lung tumors. We did not have enough material to perform Northern blots. The two alternative spliced forms of SEMA3F cDNA described before 2 were present in all samples. In addition, neuropilin-1 and neuropilin-2 mRNAs were expressed in the same samples. Although contamination by normal cells could have explained these findings, we found that a variety of 16 lung tumor cell lines expressed SEMA3F and neuropilin mRNAs by themselves. The GLC20 SCLC cell line used as a negative control for SEMA3F expression did not express SEMA3F as expected but was positive for neuropilin-2.
A total of 95 lung carcinomas, histologically classified according to 1981 and 1999 WHO criteria, were studied for SEMA3F immunostaining on frozen sections. These included 29 squamous cell carcinomas, 35 adenocarcinomas, 20 high grade neuroendocrine (NE) lung tumors consisting of 10 SCLC and 10 LCNEC, 37 and 11 carcinoids (8 typical and 3 atypical). Because surgical samples were used in this study, there was a higher number of low stage tumors (stage I to II: 47 cases) than high stage tumors (stage III to IV: 17 cases). Low grade NE tumors (carcinoids) as well as high grade NE tumors (SCLC and LCNEC) were included in attempt to correlate differences in SEMA3F levels with tumor aggressiveness. The results are summarized in Table 1 ▶ .
Table 1.
Semaphorin SEMA3F Expression in Lung Cancer: Level and Localization According to Different Histological Types
| Histological type | n | SEMA3F expression scores | SEMA3F localization pattern | ||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | Membranous | Cytoplasmic | ||
| Squamous cell carcinoma | 29 | 2 | 9 | 8 | 10 | 6 | 21 |
| Adenocarcinoma | 35 | 2 | 3 | 14 | 16 | 12 | 21 |
| Small cell carcinoma | 10 | 1 | 4 | 5 | 0 | 0 | 9 |
| LCNEC | 10 | 3 | 2 | 5 | 0 | 0 | 7 |
| Carcinoid | 11 | 0 | 0 | 4 | 7 | 8 | 2 |
| Total | 95 | 8 (8%) | 18 (19%) | 36 (38%) | 33 (35%) | 26 (27%) | 60 (63%) |
Scores of immunostaining were calculated by multiplying the number of labeled cells (0 to 100%) by the level of staining intensity (1 to 3). Final scoring grades were: 0 (no cell stained), 1 (score <30), 2 (score 30 to 150), and 3 (score >150). n = number of cases in each histological type. Pattern of semaphorin staining: membranous pattern = membranous reinforcement of staining in addition to cytoplasmic staining always present; cytoplasmic pattern = diffuse cytoplasmic staining without membranous outline.
In squamous cell carcinoma, SEMA3F immunostaining was negative or low (scores 0 to 1) in 11/29 (38%) of cases and intermediate or intense (scores 2 to 3) in 62% (Figure 1, C and E ▶ , respectively). Fewer adenocarcinomas were negative or expressed low levels of SEMA3F (5/35: 14%), whereas the majority (86%) expressed intermediate or high levels (scores 2 to 3) (Figure 1G) ▶ . Thus, in NSCLC tumors overall, 16/64 (25%) had SEMA3F scores of 0 to 1 compared with staining of normal lung epithelial cells (score = 3). In NE lung tumors, 50% of high grade tumors (SCLC and LCNEC) showed an absence or low level of SEMA3F expression in contrast to carcinoid tumors, which all exhibited intermediate or high expression levels (P = 0.005). Overall, when all histological types were considered, 27% of tumors displayed no or low SEMA3F, and 38 and 35% showed intermediate and high levels, respectively.
We then examined SEMA3F localization according to histological NSCLC subtypes. Membrane localization of SEMA3F was infrequent in squamous cell carcinoms (6/27: 22%). However, when membrane staining was observed, it gave an intercellular mosaic pattern reminiscent of desmosomal junction, and it was often prominent at the basally oriented membrane toward the tumor stroma consistent with the possible involvement of SEMA3F in cell-cell adhesion or migration (Figure 1I) ▶ . In adenocarcinomas, most acinar and solid types (11/15) displayed diffuse cytoplasmic staining, whereas 4 of 15 showed the basally oriented pattern observed in squamous carcinomas. In contrast, 8/17 well differentiated adenocarcinomas of bronchioloalveolar and papillary types displayed the characteristic membrane pattern of apical or luminal membrane staining observed in their candidate normal progenitor cells, ie, type II alveolar and Clara cells. In some NSCLC tumors, small clusters of cells disseminated in the stroma expressed higher levels of SEMA3F than cells in large lobules (Figure 1J) ▶ . In high grade NE tumors, SCLC and LCNEC never displayed membranous staining, which was in marked contrast to that observed in carcinoid neoplasms.
Neovasculature in tumor stroma were essentially unstained except in 12 of 95 tumors in which 10 to 50% of the vessels were SEMA3F positive (2 squamous cell carcinomas, 3 adenocarcinoms, 2 LCNEC, 1 SCLC, 4 carcinoids). Lymphatic vessels, which appeared more developed and dilated than in the normal lung, were likewise unstained. However since fibroblasts were stained in approximately one-half of the tumors, we cannot exclude that some SEMA3F-positive fusiform cells in the stroma were actually endothelial cells lining obscured lymphatic channels.
SEMA3F Scores Correlate Inversely with Tumor Stage and Clinical Aggressiveness
In NSCLC, the TNM stage (comprised of tumor size, node status, presence of metastasis) is presently the most useful means of predicting prognosis. SEMA3F levels were evaluated in individual tumors and compared with their stage (Table 2) ▶ . An intermediate or high level of SEMA3F (scores 2 to 3) was observed in 87% of limited stages NSCLCs (stage I to II) compared with 41% in tumors of higher stages (III to IV) (P = 0.0005). Thus, in NSCLC, high SEMA3F scores correlated with low stage. In addition, membrane staining correlated with low stages when all tumor types were considered (P = 0.05). In NE tumors, clinical aggressiveness and survival are highly dependent on histological type as recently described in a large survival analysis of 200 cases. 37 We observed that 50% (10/20) of high grade NE tumors expressed no or low levels of H.SemaIV, whereas all carcinoids (typical and atypical) expressed high levels (Table 1 ▶ and Figure 1, K and L ▶ ). Thus, high SEMA3F scores were significantly more frequent in low or intermediate grade NE tumors than in high grade tumors.
Table 2.
SEMA3F Scores Correlate Inversely with Tumor Stage in NSCLC Lung Tumors
| Stages | SEMA3F level (scores) | ||
|---|---|---|---|
| 0–1 | 2–3 | n | |
| Stages I to II | 6 | 41 (87%) | 47 |
| Stages III to IV | 10 | 7 (41%) | 17 |
| P = 0.0005 |
SEMA3F Membranous Localization Correlates Inversely with VEGF Levels
The results of quantitative VEGF immunostaining are presented in Table 3 ▶ and examples are given in Figure 1 ▶ (D, F, H). Both anti-VEGF antibodies (polyclonal A20 and monoclonal mAb 293) gave concordant results in 90% of tumors, and the highest score was retained in the 10% cases with discordant immunostaining. Overall, only 18/95 tumors (19%) showed no or low VEGF immunostaining, whereas 29 and 41% exhibited intermediate or high levels, respectively. When the mean VEGF score was chosen as a threshold value, 63% of NSCLCs displayed high VEGF levels. VEGF immunostaining was cytoplasmic and diffuse on paraffin sections, but distinct membrane staining was observed on frozen sections in all cases of NSCLC expressing VEGF and normal lung. Surprisingly, however, most high grade NE tumors and one-half of the carcinoids did not display membranous localization despite high VEGF levels. No significant correlation could be established between the intensities of SEMA3F and VEGF immunostaining using the 0 to 3 scoring values (exact Fisher test P = 0.6 and Spearman test P = 0.7). However, when the localization of SEMA3F (membranous or only cytoplasmic) was analyzed in relation with VEGF (Table 4) ▶ , a significant correlation was found between scores for VEGF immunostaining less than the mean value and the membrane localization of SEMA3F. We noted that all bronchioloalveolar and papillary adenocarcinomas (well differentiated types) lacking a membranous localization for SEMA3F exhibited high levels of VEGF (more than the mean value). In small clusters of cells disseminated in NSCLC tumor stroma and expressing higher levels of SEMA3F than cells in large lobules, VEGF staining was also enhanced (data not shown).
Table 3.
VEGF Expression in Lung Cancer: Scores of Immunostaining According to Histological Types
| Histological type | n | VEGF expression score | ||
|---|---|---|---|---|
| 0–1 | 2 | 3 | ||
| Squamous cell carcinoma | 29 | 2 | 16 | 11 |
| Adenocarcinoma | 35 | 11 | 10 | 14 |
| Small cell carcinoma | 10 | 3 | 3 | 4 |
| Large cell carcinoma | 10 | 0 | 3 | 7 |
| Carcinoid | 11 | 2 | 6 | 3 |
| Total | 95 | 18 (19%) | 28 (29%) | 39 (41%) |
Scores of VEGF immunostaining were calculated in multiplying the percentage of labeled cells (0 to 100) by the staining intensity (1 to 3). Final scoring grades were 0 (no cell stain): 1 (score <30); 2 (score 30–150); and 3 (score >150).
Table 4.
Relationship between VEGF Expression and SEMA3F Localization: SEMA3F Membranous Localization Correlates Inversely with VEGF Levels
| SEMA3F localization | VEGF expression (scores) | ||
|---|---|---|---|
| <2 | ≥2 | n | |
| Cytoplasmic and membranous | 19 (56%) | 15 | 34 |
| Cytoplasmic | 14 (26%) | 40 | 54 |
| P = 0.007 |
Relative Expression of SEMA3F mRNA in Lung Tumors
As SEMA3F protein levels were found variable in lung tumors, one hypothesis was that SEMA3F transcription efficiency would be responsible for these differences. To investigate this, we assessed levels of SEMA3F mRNA in selected tumors using quantitative real-time RT-PCR (Figure 2) ▶ . Compared to normal lung, the levels of SEMA3F mRNA in tumors ranged from 68% to as low as 8% (Table 5) ▶ . The tumor containing the lowest level of SEMA3F mRNA (8%) was negative for SEMA3F protein expression by immunochemistry. However, only a loose correlation existed between SEMA3F mRNA levels and SEMA3F protein scores. In addition to competition by VEGF165 for binding to a common receptor (discussed below), another variable could be the amount of contaminating stromal cells in the tumor that ranged from 10 to 30% (Table 5) ▶ .
Figure 2.
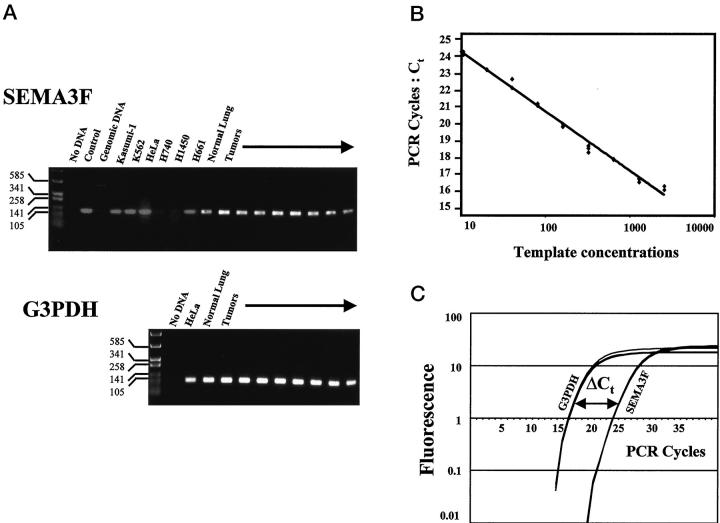
Real-time quantitative RT-PCR analysis for SEMA3F mRNA. A: Electrophoresis using a 2% agarose gel showing PCR products after 40 cycles, with SEMA3F-f and -r primers from cDNA of positive cells (Kasumi-1, K562, HeLa, H661), negative cells (H740 and H1450, which are homozygously deleted for SEMA3F), along with normal lung and lung tumors. The positive control was SEMA3F cDNA cloned into pBS 2 and amplified with the primers. The DNA size is indicated in bp on the left. A specific product was obtained for all positive samples. The corresponding G3PDH products are shown in bottom panel. B: Linearity of the SEMA3F amplification was tested using various dilutions of the cloned SEMA3F cDNA. The PCR cycle (Ct) at which the product level (determined by fluorescence) crossed an arbitrary threshold during the exponential phase of amplification was measured as function of template concentration (in arbitrary units). The Ct for SEMA3F-f and -r primers over nine serial template dilutions demonstrated a linear fit on a semilog plot, indicating the quantitative nature of the process. For each concentration, the amplifications were performed in triplicate. C: Fluorescence versus cycle number for three replicate reactions each using G3PDH and SEMA3F-f and -r primers in one tumor. The difference between the Ct for G3PDH and the Ct for SEMA3F corresponds to the relative expression level of SEMA3F.
Table 5.
SEMA3F and VEGF Results in Selected Tumors
| Sample | Histological type | % Stroma cells | SEMA3F protein score | VEGF protein score | SEMA3F protein localization | ΔCt | Relative amount of SEMA3F mRNA |
|---|---|---|---|---|---|---|---|
| Normal lung | 3 | 2 | Memb | 4.72 | 100 | ||
| Tumor 1 | Sq cell carc | 30 | 3 | 2 | Cyto | 5.28 | 68 |
| 2 | Adenocarc | 20 | 3 | 0 | Memb | 7.03 | 20 |
| 3 | Adenocarc | 20 | 1 | 1 | Cyto | 5.71 | 50 |
| 4 | Sq cell carc | 20 | 1 | 2 | Cyto | 6.79 | 23.8 |
| 5 | Sq cell carc | 10 | 1 | 2 | Cyto | 7.17 | 18 |
| 6 | Sq cell carc | 30 | 0 | 3 | 8.42 | 7.6 |
Normal lung, 4 squamous cell carcinoma (sq cell carc) and 2 adenocarcinoma (adenocarc) were analyzed for SEMA3F and VEGF expression. The histological type and the percentage of stroma cells are given for each tumor. SEMA3F localization is membranous (Memb) or cytoplasmic (Cyto). The relative amount of SEMA3F mRNA is compared with normal lung (100%). ΔCt is the difference between the SEMA3F Ct and the G3PDH Ct. An increase of one cycle in ΔCt corresponds to 50% less SEMA3F mRNA.
Cellular Localization of SEMA3F in Tumor Cells: A Confocal Microscopy Analysis
Indirect immunofluorescence in conjunction with laser-scanning confocal microscopy was used to analyze the cellular distribution of endogenous SEMA3F in four malignant cell lines: NCI-H661 (large-cell NSCLC), Calu-3 (lung epithelial adenocarcinoma), HeLa (cervical carcinoma), and SK-N-SH (neuroblastoma). Paraformaldehyde- and methanol-fixed cells displayed a granular cytoplasmic fluorescence signal as shown by confocal images obtained in horizontal (Figure 3) ▶ and vertical sections (not shown). No signal was detected in nuclei of the four cell lines and in peripheral vesicles in NCI-H661 cells (Figure 3, A and B) ▶ . The SCLC cell line, GLC20, used as a negative control, displayed a negligible nonspecific fluorescence signal (not shown). Similarly, no signal was detected in NCI-H661, Calu-3, HeLa, and SK-N-SH cells when the anti-SEMA3F antibody was preabsorbed with the immunizing peptide.
Figure 3.
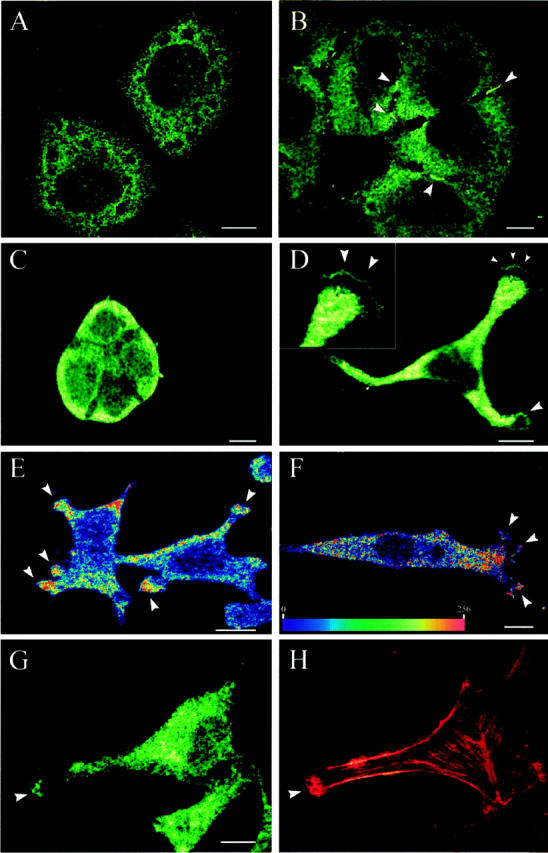
SEMA3F immunostaining in tumor cells detected by confocal microscopy. SEMA3F was detected with a FITC-conjugated goat-anti-rabbit IgG secondary antibody. Detection of SEMA3F in NSCLC NCI-H661 cells showed a localization in the cytoplasm (A, B) and at the interface of adjacent cells (B: arrows). Polarized lung adenocarcinoma Calu-3 cells, growing in islets, confirmed the major distribution of SEMA3F in the apical domain of lung epithelial cells (C). In cervical carcinoma HeLa cells (D, E) and neuroblastoma SK-N-SH cells (F), SEMA3F was detected in the cytoplasm but also in motile regions (D, E, F: arrows). In D, the inset showed the edge of lamellipodia. In E and F, the intensity of SEMA3F staining was quantified by an arbitrary color scale from blue (no SEMA3F) to red (highest level of SEMA3F). In G and H, SEMA3F and F-actin were double stained in HeLa cells. SEMA3F was detected with a FITC-conjugated goat-anti-rabbit IgG secondary antibody (G), and F-actin was labeled with TRITC-conjugated phalloidin (H). SEMA3F colocalized with actin microfilaments in ruffling regions. Scale bar, 10 μm.
NCI-H661 cells show an epithelial phenotype without locomotion activity, and they grow in monolayers of multicellular islets of closely apposed cells. In a significant proportion of cells, SEMA3F was localized as a bright fluorescent signal at the interface of adjacent interacting cells in addition to mild diffuse cytoplasmic staining (Figure 3B ▶ : arrows). These dense intercellular structures suggest the possibility that SEMA3F could be associated with cell adhesion domains reminiscent of adherens junctions. However, SEMA3F was not observed at the basal membrane of Calu-3 cells that are also growing in islets. On the contrary, SEMA3F was concentrated at the apical domain of polarized cells (Figure 3C) ▶ . This localization is reminiscent of that observed in lung epithelia sections.
Cells with locomotion activity, like HeLa (Figure 3, D, E, G, and H) ▶ and SK-N-SH (Figure 3F) ▶ cells, migrated by forming a large lamellipodium in the direction of the movement. In cells displaying this morphological aspect, a spatial concentration of fluorescence was observed in motile regions like leading edges or ruffling membranes at the margin of lamellipodia or cellular protrusions (Figure 3, D ▶ -F). Arbitrary representations of the fluorescence intensity by color scale, showed that SEMA3F staining is sometimes more intense at the edge of all protrusions that in other parts of the cytoplasm (Figure 3, E and F) ▶ . When F-actin was labeled in the same cells with FITC-conjugated phalloidin, SEMA3F colocalized with actin microfilaments in ruffling regions but was neither associated with ventral stress fibers or cortical ring of microfilaments (Figure 3, G and H) ▶ . Migrating cells showed a prominent actin-rich region at the edge of lamellipodium, followed by the lamella, a transition zone with low actin content (Figure 3H) ▶ . SEMA3F showed the same kind of distribution in this motile region as in actin-rich membrane ruffles at the edge of cell protrusions. This suggests a role of SEMA3F in cell motility.
Discussion
With the exception of SEMA3F homozygous deletions in a few SCLC cell lines, 2,3,4 there have been no reports in lung cancers of point mutations or other alterations excluding polymorphisms of SEMA3F. 3,4 However, by Northern blot analysis, marked differences in expression levels were observed in lung cancer cells. 3,4 Therefore, it was important to look at the protein level for differences in expression levels or subcellular localization in tumors. To address this issue, we recently described the development of a specific anti-SEMA3F polyclonal antibody and the analysis of SEMA3F protein in normal and Alzheimer’s brains. 32 In this study, SEMA3F protein was found in all epithelial cells from normal lung and was localized predominantly at the apical membrane and, to a lesser extent, in the cytoplasm. These results are in agreement with previous Northern blots, which found SEMA3F mRNA, and its murine homologue, in adult lung. 2,3,4,38 In lung tumors by contrast, SEMA3F levels were not infrequently undetectable or considerably reduced (27% overall) compared with normal. Analysis of SEMA3F mRNA expression by real-time quantitative RT-PCR showed a marked decrease in the tumors compared with normal lung. However, there was only partial correspondence between SEMA3F mRNA levels and SEMA3F protein scores. There are several possibilities for this including contamination by stroma cells (10 to 30%), SEMA3F secretion, and the likely competition by VEGF165 for binding to cell surface SEMA3F receptors (see below). Subjectively, it is difficult by immunochemistry to quantitate the amount of cytoplasmic staining as opposed to membrane staining. Also, the processing and stability of the mRNA could have been differentially affected in tumors. Alternatively, different post-translational regulation cannot be excluded. With regard to histological correlations, the characteristic membrane staining of SEMA3F in normal cells was retained in mixed cases of bronchioloalveolar and papillary adenocarcinomas (well differentiated types), whereas it was usually lost in poorly differentiated adenocarcinomas. Similarly, there were no observable SEMA3F alterations in carcinoids (low grade NE tumors) in contrast to its loss in 50% of high grade NE tumors and in extended stage NSCLCs. Similar results have been reported recently for BRCA1 in high grade breast cancers despite the absence of demonstrable mutations. 39
The mechanism and specificity of altered SEMA3F localization is presently unknown. We observed, by immunohistochemistry on frozen sections, that other membrane proteins (EGF receptor, IGF-1 receptors, NCAM molecules) 40 were appropriately positioned in these tumors excluding the possibility that fixation conditions or other artifacts were responsible. Defects in the secretion of extracellular proteins are frequently observed in cancer cells 41 (and references therein). Whether SEMA3F post-translational modifications such as cleavage by furin, 42,43 N-glycosylation or homodimer formation 43,44 can affect its localization is unknown. Another explanation could be that SEMA3F is unable to bind its receptor either because of a component deficiency or because of a competitor. We found by RT-PCR that neuropilin-1 and -2, the SEMA3F receptors, are expressed in all lung tumors and tumor cell lines. SEMA3F binds neuropilin-2 in vitro with higher affinity than neuropilin-1, kd ∼ 0.1 and 1 nM, respectively, 23 although the respective concentrations of these proteins in normal lung and tumors are unknown. Interestingly, lung cancer cells and especially poorly differentiated adenocarcinomas that had lost membrane staining of SEMA3F showed high levels of VEGF. Because neuropilin-1 and possibly neuropilin-2 are common receptors for both VEGF165 and SemaIII, 24 VEGF might be a competitor for SEMA3F binding. This hypothesis is in agreement with other data showing that Sema3A (SemaIII) and SEMA3A (Collapsin-1) compete with VEGF for binding to neuropilin-1. 28,33 On frozen sections of NSCLCs, we found VEGF localized at the plasma membrane, which is consistent with this possibility. It is thus tempting to attribute a function of SEMA3F in angiogenesis, because recent studies demonstrated that exogenous SEMA3A inhibited the capillary sprouting of endothelial cells from rat aortic ring segments. 33 However, few vessels of median caliber are positive for SEMA3F, and lymphatic vessels in tumors are negative. This is in contrast with VEGF staining of stroma capillaries in the vast majority of tumors. Moreover, our observation that neither SEMA3F nor VEGF exhibited membrane localization in most high grade NE tumors renders this simple competition model more elusive and favors the possibility that one of the receptor components might be limiting. This is conceivable, because there is evidence that neuropilins form a complex with a promiscuous accessory molecule necessary for signal transduction 10,28,45-47 recently identified as plexin. 48,49 Alternatively, high turnover of the receptor with rapid internalization in these tumors has not been ruled out.
Immunolocalization of SEMA3F by confocal microscopy in Calu-3 cells (lung epithelial adenocarcinoma), confirmed that the protein is predominantly distributed in the apical domain of polarized cells, as for normal lung epithelia sections. However, in a large proportion of NCI-H661 cells (large cell NSCLC), showing no or little locomotion activity, SEMA3F was at the interface of adjacent cells reminiscent of adherens junctions. Also, in 22% of squamous cell carcinomas, SEMA3F was localized at cell junction interfaces and at the basally oriented membrane toward the tumor stroma supporting the hypothesis of SEMA3F involvement in cell adhesion or migration. Cell-cell adhesion through homotypic cadherin binding, as well as mobilization of actin-containing undercoats at adherens junctions, are inversely correlated with invasiveness and tumorigenicity. 50 Disruption of this system is frequent in cancer. For example, E-cadherin-catenin complexes were delocalized to the cytoplasm in invasive clusters of 32 out of 44 primary lung tumors. 51
In contrast, in more motile cell lines (HeLa and SK-N-SH), SEMA3F was localized in both the cytoplasm and in motile regions such as leading edges or ruffling membranes of lamellipodes or cellular protrusions where actin microfilaments are colocalized. Likewise in NSCLC tumors, small clusters of scattered cells presumably emanating from large tumor lobules showed enhanced cytoplasmic SEMA3F expression. These two observations suggest that SEMA3F could be involved in cell migration. Actin-rich membrane ruffles and lamellipodia are known to be involved in the dynamic actin reorganization necessary for extension of cell protrusions and cell motility, 52 and semaphorins are believed to induce a net loss of F-actin in the leading edge of growth cones 53,54 rather than inducing microtubule polymerization or depolymerization. These observations are relevant, because motility factors are not only involved in physiological events like embryogenesis or wound healing, but also in pathological processes such as tumor invasion and metastasis. A recent study 55 demonstrated the involvement of SEMA3A in neuronal crest migration in the chick and in morphological changes induced to crest cells in vitro after exposure to SEMA3A suggesting that this semaphorin might operate to control the movement of cells. Similarly, other results that relate to neuropilin function indirectly involve semaphorins in migration. Neuropilin-1 is also a placental growth factor-2 receptor, 26 which induces migration of endothelial cells. Neuropilin-1 expression, like VEGFR-2, is up-regulated by tumor necrosis factor-α, 56 which enhances migration and wound healing triggered by VEGF165.
The localization of SEMA3F at motile regions in HeLa and SK-N-SH cells seems, at first, contrary to the findings in NCI-H661 cells where SEMA3F was detected at the interface of adjacent cells. However, SEMA3F could be involved with membrane-associated proteins that regulate both adhesion and motility. It clearly appears that SEMA3F localization is highly dependent on the cell type, the tumor class, and/or the cell phenotype. These observations suggest that the cell transformation process could affect one or several partners involved in SEMA3F compartmentalization.
Our results support the involvement of SEMA3F in the pathogenesis of human lung cancer and its possible role in VEGF pathway. We also suggest that SEMA3F may be involved in cell-cell adhesion or migration and that SEMA3F distribution may be affected by changes in cell activity. We hypothesize that the reduction of SEMA3F expression in tumors (manifested by the loss of SEMA3F membrane staining) facilitates VEGF receptor activation and an enhanced migratory response to VEGF in part through the permeabilization of stroma extracellular matrix during this migratory process. 57 Either of these potential pathways might be sufficient to explain how reduced expression of SEMA3F or, conversely, its overexpression in mouse A9 cells could lead to altered tumorigenicity. Whether other human semaphorins will function in a similar manner remains to be determined. For example, SEMA4D (CD100) a leukocyte transmembrane class 4 semaphorin, 58 which induces B cells to aggregate and would improve their viability in vitro, 59 is lost in follicular B cell lymphomas. 60 Another example includes a semaphorin related to SEMA4D located at a chromosomal region, 5q21–22, which is frequently deleted in advanced lung cancers. 61 The role of semaphorins in cancer is not limited to loss of expression, however. For example, expression of another member of the class 3 family, Sema3E (M.semaH), correlates with the metastatic ability of mouse tumor cells. 62 Likewise, Yamada et al 63 identified SEMA3C (H-sema E) as a non-multidrug-resistance gene in human cancers. This secreted semaphorin was found to be overexpressed in cis-platinum-resistant cell lines and was induced by diverse agents including X-ray and UV irradiation. Thus, the investigation of these molecules is likely to be very informative even outside the context of neurodevelopment.
Acknowledgments
We gratefully acknowledge C. Claraz and S. Veyrenc for technical help in immunochemistry, A. Cantereau for help in confocal microscopy, and J. Jacobsen for cell culture.
Footnotes
Address reprint requests to Prof. Joëlle Roche, IBMIG, Université de Poitiers, 40 avenue du Recteur Pineau, 86022 Poitiers Cédex, France. E-mail: joelle.roche@campus.univ-poitiers.fr.
Supported by La Ligue Nationale Contre le Cancer (E. B.), CNRS UMR 6558 (B. C.), grant P30 CA 46934 and University of Colorado Lung Cancer SPORE CA5187–07 (H. D.), the generous equipment support from the Lulu Frankel Foundation (H. D.), the Association pour la Recherche sur le Cancer (E. B., J. R.), La Ligue Nationale Contre le Cancer, Comité de la Vienne (J. R.) and NATO (J. R., H. D.).
References
- 1.: Semaphorin Nomenclature Committee: Unified nomenclature for the semaphorins/collapsins. Cell 1999, 97:551-552 [DOI] [PubMed] [Google Scholar]
- 2.Roche J, Boldog F, Robinson M, Robinson L, Varella-Garcia M, Swanton M, Waggoner B, Fishel R, Franklin W, Gemmill R, Drabkin H: Distinct 3p21.3 deletions in lung cancer, analysis of deleted genes, and identification of a new human semaphorin. Oncogene 1996, 12:1289-1297 [PubMed] [Google Scholar]
- 3.Xiang R, Hensel C, Garcia D, Carlson H, Kok K, Daly M, Kerbacher K, Van Den Berg A, Veldhuis P, Buys C, Naylor S: Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics 1996, 32:39-48 [DOI] [PubMed] [Google Scholar]
- 4.Sekido Y, Bader S, Latif F, Chen JY, Duh FM, Wei MH, Albanes JP, Lee CC, Lerman MI, Minna JD: Human semaphorins A (V) and (IV) reside in the 3p21.3 small cell lung cancer deletion region and demonstrate distinct expression patterns. Proc Natl Acad Sci USA 1996, 93:4120-4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Y, Raible D, Raper A: Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 1993, 75:217-227 [DOI] [PubMed] [Google Scholar]
- 6.Kolodkin A: Semaphorins: mediators of repulsive growth cone guidance. Trends Cell Biol 1996, 6:15-22 [DOI] [PubMed] [Google Scholar]
- 7.Püschel A: The semaphorins: a family of axonal guidance molecule. Eur J Neurosci 1996, 8:1317-1321 [DOI] [PubMed] [Google Scholar]
- 8.Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M, Goodman CS: Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 1998, 95:903-916 [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Ng S, Wu Z, Nguyen D, Homburger S, Seidel-Dugan C, Ebens A, Luo Y: Human semaphorin K1 is glycosylphosphatidylinositol-linked and defines a new subfamily of viral-related semaphorins. J Biol Chem 1998, 273:22428-22434 [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Kolodkin A: Semaphorin signaling: a little less per-plexin. Neuron 1999, 22:11-14 [DOI] [PubMed] [Google Scholar]
- 11.Kolodkin A, Matthes D, Goodman C: The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 1993, 75:1389-1399 [DOI] [PubMed] [Google Scholar]
- 12.Wong J, Yu W, O’Connor T: Transmembrane grasshopper semaphorin I promotes axon outgrowth in vivo. Development 1997, 124:3597-3607 [DOI] [PubMed] [Google Scholar]
- 13.Bagnard D, Lohrum M, Uziel D, Puschel AW, Bolz J: Semaphorins act as attractive and repulsive guidance signals during the development of cortical projections. Development 1998, 125:5043-5053 [DOI] [PubMed] [Google Scholar]
- 14.Song H, Ming G, He Z, Lehmann M, McKerracher L, Tessier-Lavigne M, Poo M: Conversion of neuronal growth cone responses from repulsion to attraction by cyclic nucleotides. Science 1998, 281:1515-1518 [DOI] [PubMed] [Google Scholar]
- 15.Chen H, He Z, Tessier-Lavigne M: Axon guidance mechanisms: semaphorins as simultaneous repellents and antirepellents. Nature Neurosci 1998, 1:436-439 [DOI] [PubMed] [Google Scholar]
- 16.de Castro F, Hu L, Drabkin H, Sotelo C, Chédotal A: Chemoattraction and chemorepulsion of olfactory bulb axons by different secreted semaphorins. J Neurosci 1999, 19:4428-4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC: Semaphorin III is needed for normal patterning, and growth of nerves, bones, and heart. Nature 1996, 383:525-528 [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi M, Yuasa S, Fujisawa H, Naruse I, Saga S, Mishina M, Yagi T: Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron 1997, 19:519-530 [DOI] [PubMed] [Google Scholar]
- 19.Fujisawa H, Kitsukawa T: Receptors for collapsin/semaphorins. Curr Opin Neurobiol 1998, 8:587-592 [DOI] [PubMed] [Google Scholar]
- 20.Kolodkin A, Ginty D: Steering clear of semaphorins: neuropilins sound the retreat. Neuron 1997, 19:1159-1162 [DOI] [PubMed] [Google Scholar]
- 21.He Z, Tessier-Lavigne M: Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell 1997, 90:739-751 [DOI] [PubMed] [Google Scholar]
- 22.Kolodkin A, Levengood D, Rowe E, Tai Y, Giger R, Ginty D: Neuropilin is a semaphorin III receptor. Cell 1997, 90:753-762 [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Chédotal A, He Z, Goodman CS, Tessier-Lavigne M: Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron 1997, 19:547-559 [DOI] [PubMed] [Google Scholar]
- 24.Soker S, Takashima S, Miao H, Neufeld G, Klagsbrun M: Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92:735-745 [DOI] [PubMed] [Google Scholar]
- 25.Wise LM, Veikkola T, Mercer AA, Savory LJ, Fleming SB, Caesar C, Vitali A, Makinen T, Alitalo K, Stacker SA: Vascular endothelial growth factor (VEGF)-like protein from orf virus NZ2 binds to VEGFR2 and neuropilin-1. Proc Natl Acad Sci USA 1999, 96:3071-3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migdal M, Huppertz B, Tessler S, Comforti A, Shibuya M, Reich R, Baumann H, Neufeld G: Neuropilin-1 is a placenta growth factor-2 receptor. J Biol Chem 1998, 273:22272-22278 [DOI] [PubMed] [Google Scholar]
- 27.Takagi S, Kasuya Y, Shimizu M, Matsuura T, Tsuboi M, Kawakami A, Fujisawa H: Expression of a cell adhesion molecule, neuropilin, in the developing chick nervous system. Dev Biol 1995, 170:207-222 [DOI] [PubMed] [Google Scholar]
- 28.Giger RJ, Urquhart ER, Gillespie SK, Levengood DV, Ginty DD, Kolodkin AL: Neuropilin-2 is a receptor for semaphorin IV: insight into the structural basis of receptor function and specificity. Neuron 1998, 21:1079-1092 [DOI] [PubMed] [Google Scholar]
- 29.Kok K, Naylor S, Buys C: Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res 1997, 71:27-92 [DOI] [PubMed] [Google Scholar]
- 30.Todd M, Xiang R, Garcia D, Kerbacher K, Moore S, Hensel C, Liu P, Siciliano M, Kok K, van den Berg A, Veldhuis P, Buys C, Killary A, Naylor S: An 80 Kb P1 clone from chromosome 3p21.3 suppresses tumor growth in vivo. Oncogene 1996, 13:2387–2396 [PubMed]
- 31.Daly M, Xiang R, Buchhagen D, Hensel C, Garcia D, Killary A, Minna J, Naylor S: A homozygous deletion on chromosome 3 in a small cell lung cancer cell line correlates with a region of tumor suppressor activity. Oncogene 1993, 8:1721-1729 [PubMed] [Google Scholar]
- 32.Hirsch E, Hu L-J, Prigent A, Constantin B, Agid Y, Drabkin H, Roche J: Distribution of semaphorin IV in adult human brain. Brain Res 1999, 823:67-79 [DOI] [PubMed] [Google Scholar]
- 33.Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M: Neuropilin-1 mediates collapsin-1/semaphorinIII inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol 1999, 146:233-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E, and collaborators from 14 countries: Histological typing of lung and pleural tumors. International histological classification of tumors, 3rd ed. Edited by Springer. World Health Organization Pathology Panel: World Health Organization, 1999
- 35.Kok K, van den Berg A, Veldhuis P, van der Veen A, Franke M, Schoenmakers E, Hulsbeek M, van der Hout A, de Leij L, van de Ven W, Buys C: A homozygous deletion in a small cell lung cancer cell line involving a 3p21 region with a marked instability in yeast artificial chromosomes. Cancer Res 1994, 54:4183-4187 [PubMed] [Google Scholar]
- 36.Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N: Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood 1991, 77:2031-2036 [PubMed] [Google Scholar]
- 37.Travis WD, Rush W, Flieder DB, Falk R, Fleming MV, Gal AA, Koss MN: Survival analysis of 200 pulmonary neuroendocrine tumors with clarification of criteria for atypical carcinoid and its separation from typical carcinoid. Am J Surg Pathol 1998, 22:934-944 [DOI] [PubMed] [Google Scholar]
- 38.Eckhardt F, Meyerhans A: Cloning and expression pattern of a murine semaphorin homologous to H-sema IV. Neuroreport 1998, 9:3975-3979 [DOI] [PubMed] [Google Scholar]
- 39.Wilson CA, Ramos L, Villasenor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D, Zuch RH, Kanter MH, Cohen S, Calzone FJ, Slamon DJ: Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet 1999, 21:236-240 [DOI] [PubMed] [Google Scholar]
- 40.Lantuejoul S, Moro D, Michalides RJ, Brambilla C, Brambilla E: Neural cell adhesion molecules (NCAM) and NCAM-PSA expression in neuroendocrine lung tumors. Am J Surg Pathol 1998, 22:1267-1276 [DOI] [PubMed] [Google Scholar]
- 41.Weaver V, Petersen O, Wang F, Larabell C, Briand P, Damsky C, Bissell M: Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 1997, 137:231-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams R, Lohrum M, Klostermann A, Betz H, Püschel A: The chemorepulsive activity of secreted semaphorins is regulated by furin-dependent proteolytic processing. EMBO J 1997, 16:6077-6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klostermann A, Lohrum M, Adams RH, Puschel AW: The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. J Biol Chem 1998, 273:7326-7331 [DOI] [PubMed] [Google Scholar]
- 44.Koppel A, Raper J: Collapsin-1 covalently dimerizes, and dimerization is necessary for collapsing activity. J Biol Chem 1998, 273:15708-15713 [DOI] [PubMed] [Google Scholar]
- 45.Chen H, He Z, Bagri A, Tessier-Lavigne M: Semaphorin-neuropilin interactions underlying sympathetic axon responses to class III semaphorins. Neuron 1998, 21:1283-1290 [DOI] [PubMed] [Google Scholar]
- 46.Roskies AL: Dissecting semaphorin signaling. Neuron 1998, 21:935-936 [DOI] [PubMed] [Google Scholar]
- 47.Renzi MJ, Feiner L, Koppel AM, Raper JA: A dominant negative receptor for specific secreted semaphorins is generated by deleting an extracellular domain from neuropilin-1. J Neurosci 1999, 19:7870-7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi T, Fournier A, Nakamura F, Wang L-H, Murakami Y, Kalb RG, Fujisawa H, Strittmatter SM: Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999, 99:59-69 [DOI] [PubMed] [Google Scholar]
- 49.Tamagnone L, Artigiani S, Chen H, He Z, Ming G-L, Song H-J, Chedotal A, Winberg ML, Goodman CS, Poo M-M, Tessier-Lavigne M, Comoglio PM: Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 1999, 99:71-80 [DOI] [PubMed] [Google Scholar]
- 50.Takeichi M: Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol 1993, 5:806-811 [DOI] [PubMed] [Google Scholar]
- 51.Nawrocki B, Polette M, Van Hengel J, Tournier JM, Van Roy F, Birembault P: Cytoplasmic redistribution of E-cadherin-catenin adhesion complex is associated with down-regulated tyrosine phosphorylation of E-cadherin in human bronchopulmonary carcinomas. Am J Pathol 1998, 153:1521-1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ballestrem C, Wehrle-Haller B, Imhof BA: Actin dynamics in living mammalian cells. J Cell Sci 1998, 111:1649-1658 [DOI] [PubMed] [Google Scholar]
- 53.Fan J, Mansfield SG, Redmond T, Gorgon-Weeks PR, Raper RA: The organization of F-actin and microtubules in growth cones exposed to a brain-derived collapsing factor. J Cell Biol 1993, 121:867-878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan J, Raper R: Localized collapsing cues can steer growth cones without inducing their full collapse. Neuron 1995, 14:263-274 [DOI] [PubMed] [Google Scholar]
- 55.Eickholt BJ, Mackenzie SL, Graham A, Walsh FS, Doherty P: Evidence for collapsin-1 functioning in the control of neural crest migration in both trunk and hindbrain regions. Development 1999, 126:2181-2189 [DOI] [PubMed] [Google Scholar]
- 56.Giraudo E, Primo L, Audero E, Gerber H, Koolwijk P, Soker S, Klagsbrun M, Ferrara N, Bussolino F: Tumor necrosis factor-alpha regulates expression of vascular endothelial growth factor receptor-2 and of its co-receptor neuropilin-1 in human vascular endothelial cells. J Biol Chem 1998, 273:22128-22135 [DOI] [PubMed] [Google Scholar]
- 57.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z: Vascular endothelial growth factor (VEGF) and its receptors. FASEB J 1999, 13:9-22 [PubMed] [Google Scholar]
- 58.Delaire S, Elhabazi A, Bensussan A, Boumsell L: CD100 is a leukocyte semaphorin. Cell Mol Life Sci 1998, 54:1265-1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall K, Boumsell L, Schultze J, Boussiotis V, Dorfman D, Cardoso A, Bensussan A, Nadler L, Freeman G: Human CD100, a novel leukocyte semaphorin that promotes B-cell aggregation, and differentiation. Proc Natl Acad Sci USA 1996, 93:11780-11785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dorfman D, Shahsafaei A, Nadler L, Freeman G: The leukocyte semaphorin CD100 is expressed in most T-cell, but few B-cell, non-Hodgkin’s lymphomas. Am J Pathol 1998, 153:255-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueno K, Kumagai T, Kijima T, Kishimoto T, Hosoe S: Cloning and tissue expression of cDNAs from chromosome 5q21–22 which is frequently deleted in advanced lung cancer. Hum Genet 1998, 102:63-68 [DOI] [PubMed] [Google Scholar]
- 62.Christensen CR, Klingelhofer J, Tarabykina S, Hulgaard EF, Kramerov D, Lukanidin E: Transcription of a novel mouse semaphorin gene, M-semaH, correlates with the metastatic ability of mouse tumor cell lines. Cancer Res 1998, 58:1238-1244 [PubMed] [Google Scholar]
- 63.Yamada T, Endo R, Gotoh M, Hirohashi S: Identification of semaphorin E as non-MDR drug resistance gene of human cancers. Proc Natl Acad Sci USA 1997, 94:14713-14718 [DOI] [PMC free article] [PubMed] [Google Scholar]