Abstract
Recent progress in molecular analysis of low-grade B cell lymphoma has revealed that API2 at 11q21 and a novel gene, MALT1 at 18q21, are involved in t(11;18)(q21;q21), a characteristic chromosome aberration for mucosa-associated lymphoid tissue (MALT) type lymphoma. We describe here the establishment of a reverse transcription-polymerase chain reaction (RT-PCR) assay that we used to analyze 22 cases of MALT lymphoma. All five cases that were shown to possess t(11;18)(q21;q21) showed the specific amplification of API2-MALT1 chimeric transcripts. Of the remaining 17 cases for which cytogenetic data were not available, three cases demonstrated the presence of fusion transcripts, indicating that a significant percentage of MALT lymphoma cases of the present series appeared to possess t(11;18). A single fragment was observed in each of these cases, but the size varied from case to case. Sequencing analysis revealed that there are two breakpoints in API2 and three in MALT1, and that all of the fusion transcripts are in-frame. On the basis of these results, four kinds of chimeric proteins can be predicted for the present series. Thus, the RT-PCR assay used here should serve as an effective molecular tool for understanding molecular pathogenesis and the clinical significance of API2-MALT1 for MALT lymphomas.
In B cell lymphoma, various oncogenes have been identified to be transcriptionally deregulated as a result of translocation with immunoglobulin genes, 1 including the c-myc gene in Burkitt’s lymphoma, 2 the cyclin D1 gene in mantle cell lymphoma, 3,4 the BCL2 gene in follicular lymphoma, 5 and the BCL6 gene in diffuse large cell lymphoma. 6 Recent advances in the research of these specific gene alterations have enabled us to investigate the pathogenesis of hematolymphoid malignancies, as well as to use these genetic techniques for clinical applications, for example, as an aid for diagnosis and for monitoring of minimal residual diseases.
Malignant lymphoma of mucosa-associated lymphoid tissue (MALT) was first described by Isaacson and Wright. 7 Later, this type of lymphoma was characterized by a representative histological appearance with lymphoepithelial lesions and follicular colonization, an indolent clinical course, and frequent multicentric and extranodal involvement including the gastrointestinal tract, lung, thyroid, and mammary, salivary, and lachrymal glands. 8 Now MALT lymphoma is recognized as constituting a distinct clinicopathological disease entity. On the basis of its supposed cell origin, this lymphoma has been categorized in extranodal marginal zone B cell lymphoma for the revised European-American lymphoma (REAL) classification. 9 MALT lymphomas are sometimes associated with chronic inflammation triggered by chronic infection or autoimmune disorders, such as Helicobacter pylori gastritis, Sjögren’s syndrome, and Hashimoto’s thyroiditis. 10-12 This suggests that the proliferation of the lymphoma cells may depend on the presence of activated, antigen-driven T cells. 13 The effectiveness of antibacterial therapy for gastric MALT lymphoma poses problems regarding oncogenesis of this type of lymphoma and whether MALT lymphoma is really neoplastic. 14
Despite its well-recognized clinical and pathological characteristics, the cytogenetic features of MALT lymphoma have not been thoroughly studied. This is probably because of difficulties caused by the low mitotic activity of the lymphoma cells and the relatively low percentage of tumor cells in the tissue specimens of MALT lymphoma, which include heterogeneous reactive small lymphocytes and plasma cells as well as epithelial cells. However, in two studies of relatively large series, the recurrent chromosomal translocation t(11;18)(q21;q21) was identified as characteristic of MALT lymphoma. 15,16 Recently, we and others have shown that c-IAP2/HIAP1/MIHC/API2 gene on chromosome 11 and a novel gene, MALT1/MLT, on chromosome 18 were fused as a result of this specific translocation. 17-19 (MALT1 is used for the gene name of MALT1/MLT in the present report, as it was so designated by the Genome Nomenclature Committee. 18 ) These analyses showed the presence of a chimeric API2-MALT1 transcript consisting of the N-terminal region of the API2 gene and the C-terminal region of the MALT1 gene in cases with t(11;18). The presence of different breakpoints on MALT1 cDNA was also demonstrated by these analyses. This prompted us to examine variations of the chimeric transcripts and the incidence of their involvement in a large number of MALT lymphoma cases. Detection of this chimeric product is important for exploration of the pathogenesis of MALT lymphoma as well as for clinical application, because it represents direct evidence of the clonal expansion of lymphoma cells.
Here we report the establishment of a reverse transcription-polymerase chain reaction (RT-PCR) assay that can detect a chimeric transcript in all of our five MALT lymphoma cases with t(11;18) translocation. Furthermore, the chimeric API2-MALT1 transcript was found in three cases for which karyotype data were not available, indicating that the RT-PCR analysis is useful for detecting clonally aberrant cells in MALT lymphoma cases.
Patients and Methods
Patient Samples
We retrieved cases with available mRNA that met the criteria of MALT lymphoma from those entered in the surgical pathology files of the Aichi Cancer Center Hospital between January 1990 and December 1998. A total of 22 cases with MALT lymphoma arising in the lung and gastrointestinal tract were enrolled in the present study. The clinicopathological profiles of the samples used for the present analysis are summarized in Table 1 ▶ . These cases included 16 cases of pure low-grade MALT lymphoma, five cases of low-grade with partially high-grade component, and one case of high-grade MALT lymphoma with residual low-grade component. The pathological diagnosis was established according to the REAL classification 9 in agreement with two independent pathologists (S. N. and T. S). The five cases with t(11;18)(q21;q21) were identical to those used for our previous analysis, which demonstrated aberrant transcripts in Northern blot analysis. 18 The karyotype data of the remaining 17 cases were not available.
Table 1.
Clinicopathological Features of MALT Lymphoma Patients
| Case | Age (years) | Sex | Origin | Histology | Stage | Other sites of involvement | Karyotype | RT-PCR product |
|---|---|---|---|---|---|---|---|---|
| 1* | 45 | F | Lung | LG | IVA | Hilar LN, Pleural effusion | 46,XX,t(11;18)(q21;q21) | + |
| 2* | 51 | M | Lung | LG | IA | — | 46,XY,t(11;18)(q21;q21) | + |
| 3* | 63 | F | Lung | LG | IA | — | 46,XX,t(11;18)(q21;q21), inv(16)(p13q22) | + |
| 4* | 45 | M | Colon | LG+H | IIA | Mesenteric LN | 46,XY,t(11;18)(q21;q21) | + |
| 5* | 56 | M | Colon | LG | IA | — | 46,XY,t(11;18)(q21;q21) | + |
| 6 | 61 | M | Lung | LG | IIA | Orbit | N.A. | − |
| 7 | 76 | M | Lung | LG | IA | — | N.A. | + |
| 8 | 63 | F | Lung | LG | IA | — | N.A. | + |
| 9 | 56 | M | Lung | LG | IIA | Salivary gland | N.A. | − |
| 10 | 74 | F | Lung | LG | IA | — | N.A. | − |
| 11 | 57 | F | Lung | LG | IA | — | N.A. | − |
| 12 | 72 | F | Stomach | LG+H | IIA | Gastric LN | N.A. | − |
| 13 | 39 | M | Stomach | LG | IA | — | N.A. | + |
| 14 | 38 | M | Stomach | LG | IA | — | N.A. | − |
| 15 | 67 | M | Stomach | LG+H | IIIA | Paratracheal LN Retroperitoneal LN | N.A. | − |
| 16 | 54 | F | Stomach | LG | IA | — | N.A. | − |
| 17 | 46 | M | Stomach | LG | IA | — | N.A. | − |
| 18 | 60 | F | Stomach | LG | IA | — | N.A. | − |
| 19 | 45 | F | Stomach | LG | IA | — | N.A. | − |
| 20 | 27 | M | Stomach | LG+H | IA | — | N.A. | − |
| 21 | 54 | F | Stomach | LG+H | IIA | Gastric LN | N.A. | − |
| 22 | 63 | M | Stomach | HG+L | IVA | Testis, Orbit Retroperitoneal LN | N.A. | − |
*Cases 1 through 5 are identical to the cases previously studied. 18
LG, low grade; LG+H, low grade with partially high grade component; HG+L, high grade with residual low grade component; LN, lymph node; N.A., not available.
RT-PCR Analysis
The representative RT-PCR results shown in this report are those conducted with sense primer API2/S897–916(5′-CTGGTGTGAATGACAAGGTC-3′) and antisense primer MALT1/AS1030–1049 (5′-CAAAGGCTGGTCAGTTGTTT-3′), and with sense primer API2/S1203–1222 (5′-GTTCCTACCACTGTGCAATG-3′) and antisense primer MALT1/AS1030–1049. The nucleotide numbers are based on the sequences registered in GenBank under the accession numbers NM 001165 for API2 and AB026118 for MALT1. To detect any possible breakpoints in the coding region of API2 and MALT1, various primers were used. These include API2/S (sense) 674–693, API2/S897–916, API2/S1203–1222, API2/S1505–1524, API2/S1810–1829, API2/S2110–2129, API2/S2403–2422, API2/S2758–2777, and MALT1/AS (antisense) 1030–1049, MALT1/AS1124–1143, MALT1/AS1363–1382, MALT1/AS1506–1525, MALT1/AS1713–1732 for detecting API2-MALT1 chimeric transcripts. Primers for MALT1-API2 detection are MALT1/S60–79, MALT1/S383–402, MALT1/S569–588, MALT1/S921–940, MALT1/S1189–1208, MALT1/S1403–1422, MALT1/S1644–1663, MALT1/S1981–2000, MALT1/S2269–2288, and API2/AS1430–1449, API2/AS2017–2036, API2/AS2603–2622, API2/AS3146–3165. All combinations were examined against lymphoma cases with t(11;18) (cases 1 to 5).
Five micrograms of total RNA prepared with the guanidium-isothiocyanate method were converted to cDNA by means of reverse transcriptase (Superscript II, GIBCO-BRL, Bethesda, MD) in a total volume of 40 μl, according to the manufacturer’s protocol. To test various primer combinations, 0.5 μl of the cDNA solution produced as described above was used for PCR amplification. The PCR was conducted in a total volume of 100 μl containing 2.5 units of TaKaRaTaq (Takara, Otsu, Japan), 10 mmol/L Tris (pH 8.3 at 25°C), 1.5 mmol/L MgCl2, 50 mmol/L KCl, 0.2 mmol/L of each of the dNTPs and 100 pmoles of each primer. The touchdown PCR protocol from 63°C to 58°C was used. 20 The PCR regimen was as follows: 95°C for 3 minutes followed by 10 cycles of 1.5 minutes at 95°C, 1.5 minutes at 63°C (−1°C per two cycles), 2.5 minutes at 72°C, and 25 cycles for which the annealing temperature was held constant at 58°C, followed by a final extension at 72°C for 10 minutes.
After the reaction, 10 μl of the PCR products was electrophoresed on a 1.2% gel and stained with ethidium bromide. For RNA quality control, a primer pair of 5′-GACTACCTCATGAAGATC-3′ and 5′-GATCCACATCTGCTGGAA-3′ based on β-actin was used under PCR conditions of 35 cycles of 1.5 minutes at 95°C, 1.5 minutes at 53°C, and 2.5 minutes at 72°C, followed by a final extension for 10 minutes at 72°C.
Nucleotide Sequence Analysis
The amplified fragments were separated on a low-melting-point gel electrophoresis and purified, after which 50 ng of the fragments were used for sequencing with the dideoxy chain termination method using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin Elmer, Foster City, CA).
Results
Amplified Fragments Vary in Size
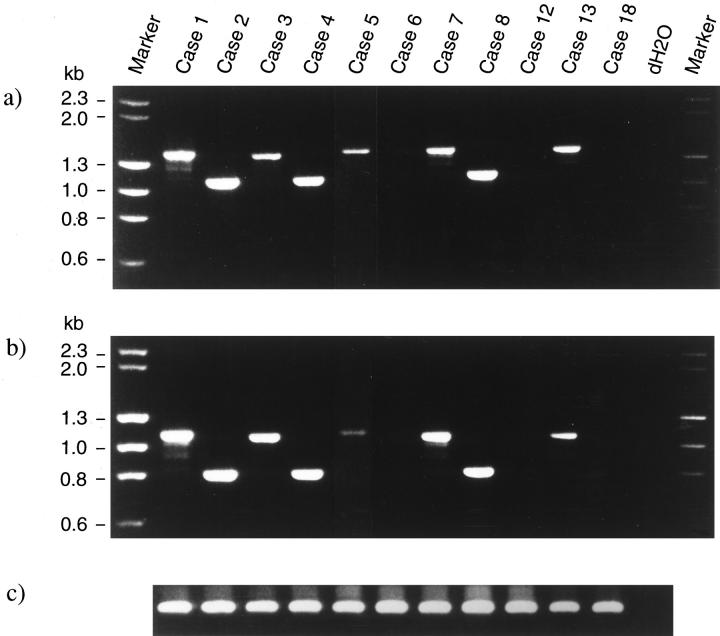
A previous study of ours revealed that the 5′ region of the MALT1 gene is frequently deleted, 18 indicating that the API2-MALT1 chimeric transcripts are more important for pathogenesis of MALT lymphoma than the reciprocal transcript, MALT1-API2. It was also shown by Northern blot analysis that the API2-MALT1 chimeric transcripts were heterogeneous. 18 Therefore, to detect API2-MALT1 transcripts, we first analyzed samples with various sense primers of API2 and antisense primers of MALT1. The primers were designed to cover any of the possible breakpoints in the coding region of each of the genes. Specific amplification was found in eight out of the 22 cases examined (Table 1) ▶ , and representative results obtained with the primer pair of API2/S897–916 and MALT1/AS1030–1049, and with that of API2/S1203–1222 and MALT1/AS1030–1049 are presented in Figure 1, a and b ▶ .
Figure 1.
Reverse transcription-polymerase chain reaction assay. a: RT-PCR with sense primer API2/S897–916 and antisense primer MALT1/AS1030–1049 demonstrated specific fragments in cases 1–5, 7, 8, and 13. The remaining cases did not show any amplified fragments. The size of the amplified fragments is 1487 bp for cases 1, 5, 7 and 13, 1184 bp for cases 2 and 4, 1466 bp for case 3, and 1211 bp for case 8. b: The same cases also showed amplification when analyzed with sense primer API2/S1203–1222 and antisense primer MALT1/AS1030–1049. The amplified fragments are 306 bp smaller than those shown in a. The size of the fragments is 1181 bp for cases 1, 5, 7, and 13, 878 bp for cases 2 and 4, 1160 bp for case 3, and 905 bp for case 8. c: RT-PCR with a β-actin primer pair that amplifies 512 bp is shown for RNA quality control. dH2O represents PCR without any first-strand cDNA. Markers are λHindIII and φHaeIII.
Figure 1a ▶ shows that amplified fragments varied in size from case to case ranging from 1487 to 1184 bp. When the primer pair of API2/S1203–1222 and MALT1/AS1030–1049 was used, the amplified fragments were reduced by 306 bp (Figure 1b) ▶ . It should be noted that all of the cases with t(11;18)(q21;q21) were found to produce these specific products (Figure 1, a and b ▶ , and Table 1 ▶ ).
For the reciprocal transcripts, MALT1-API2 chimeric products were examined for amplification with the sense primers of MALT1 and the antisense primers of API2, but none of the samples in the present series produced any specific products (data not shown), which is in accordance with our previously reported FISH analysis showing a deletion at the 18q21 breakpoint region 18,21 and with the Northern blot analysis result that a probe 5′ to the MALT1 breakpoints did not show any aberrant transcripts. 18
Nucleotide Sequence Analysis
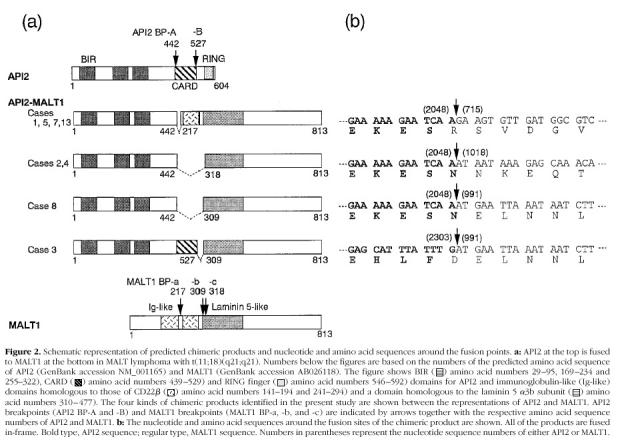
The nucleotide sequence revealed that all of the eight amplified fragments were fusion products of API2 and MALT1. An important finding was that all of the eight fragments were fused in-frame (Figure 2b) ▶ and were predicted to produce chimeric proteins between API2 and MALT1, as shown schematically in Figure 2a ▶ . It was demonstrated that there are two breakpoints (A and B) in API2, and three breakpoints (a, b, and c) in MALT1 cDNA (Figure 2, a and b) ▶ . The major breakpoint for API2 was breakpoint A, because seven out of the eight cases had their breakpoints at A. The MALT1 breakpoints, on the other hand, were dispersed: four of the eight cases had their breakpoint at MALT1 breakpoint a, two at b, and two at c. The resulting chimeric proteins were predicted to comprise four kinds as shown in Figure 2a ▶ , and the major chimeric protein is the one fused at API2 breakpoint A and MALT1 breakpoint a. It should be noted that all of these chimeric proteins retain three baculovirus inhibitor of apoptosis repeat (BIR) domains.
Figure 2.
Discussion
Recent molecular analyses by us and others of MALT lymphomas with t(11;18)(q21;q21) revealed that distinct regions of MALT1 gene were involved. 17,18 Indeed, the presence of various kinds of mRNA was demonstrated by our previous Northern blot analysis with MALT1, the size of which was found to range from 6.4 to 9.7 kb. 18 In the present study, we have demonstrated that there are three breakpoints in MALT1, although the difference in size observed in the Northern blot analysis cannot be explained simply by the difference in the fusion point. It was also found that there are two breakpoints in API2, a translocation partner gene of MALT1. Especially noteworthy is that all of the chimeric mRNAs were fused in-frame, leading to prediction of the chimeric API2-MALT1 products.
API2 was first identified as a molecule interacting with TRAF1 and TRAF2 and involved in the signal transduction of the anti-apoptotic pathway mediated by tumor necrosis factor receptor II. 22 API2 contains three BIR domains, one caspase recruitment domain (CARD), and one RING finger domain. 23 The common domain of the inhibitor of apoptosis family is the BIR motif, which has been shown to play an essential role for anti-apoptotic function. 24 It is therefore strongly suggested that the BIR motifs maintained in the API2-MALT1 chimeric protein may well be a key domain for the development of MALT lymphoma.
The CARD domain has also been demonstrated to be an important domain interacting with molecules involved in apoptosis. 25 The API2-MALT1 chimeric transcripts demonstrated by Dierlamm et al suggested that the truncation of the CARD domain from API2 is one of the characteristics of the API2-MALT/MLT fusion products. 17 Although most of the chimeric products identified in the present study have been shown to have lost the CARD domain, one case proved to have retained the CARD domain (case 3, Figure 2a ▶ ), suggesting that the removal of the CARD domain from the chimeric product is not essential.
MALT1, a novel gene at 18q21 and found in t(11;18)(q21;q21), shows homologies with the immunoglobulin-like domain of CD22β, the laminin-5 α3b subunit, and F22D3.6 of Caenorhabditis elegans. 18 The present study found the breakpoints of MALT1 to be relatively dispersed. Indeed, another breakpoint different from the three breakpoints of the present series has been reported by Dielamm et al, 17 suggesting that there are at least four different breakpoints in MALT1. Although the present analysis failed to identify important domain of MALT1 for the pathogenesis of MALT lymphoma, it should be noted that the C-terminal region may be important because of its consistent involvement in t(11;18) translocation. It is also conceivable that fusion with API2 might alter the function of API2 involved in the apoptotic pathway. Future studies focusing on API2, MALT1 and their chimeric protein API2-MALT1 are expected to shed further light on these issues.
In our current analysis, 8 of 22 (36%) MALT lymphoma cases were demonstrated, by means of the RT-PCR established in the present study, to possess chimeric products. Although the number of cases examined is limited, this assay is expected to be highly useful for clinical applications for diagnostic purposes and for the monitoring of clonal cells in MALT lymphoma patients. Recurrent detection of the API2-MALT1 chimeric transcripts suggests that low-grade lymphoma is in fact neoplastic (Table 1) ▶ . There are still some difficulties in defining the classification of low-grade B cell lymphomas. The identification of clonal cells containing these chimeric transcripts is especially important for MALT lymphoma because the heterogenous cell populations in the legion sometimes pose diagnostic problems. The present study suggested that MALT lymphomas with and without API2-MALT1 translocation might be subdivided into two groups. The pathogenesis of the latter can be expected to become the subject of intense investigation.
The incidence of the genetic alterations of API2-MALT1 may be somewhat related to their anatomical sites of origin. The 56% (5/9) detection of this genetic alteration in pulmonary lymphomas is much higher than the 9% (1/11) in gastric lymphoma. This point deserves further investigation with a larger-scale study.
In conclusion, we have established an RT-PCR assay for the detection of API2-MALT1 and found that a significant percentage of MALT lymphomas involves the API2-MALT1 gene alteration. This RT-PCR assay for detecting chimeric transcripts is expected to become an important tool for a better understanding of marginal zone lymphoma. The function of API2-MALT1 remains to be elucidated, and further studies, including those of the clinicopathological significance of the presence or absence of API2-MALT1, should provide new insights into the molecular pathogenesis of MALT lymphoma.
Acknowledgments
We thank Dr. T. Nishida at Japanese Red Cross Nagoya First Hospital for his valuable discussions, Ms. Y. Maeda for technical assistance, and Drs. I. Miura and A. Miura at the Akita University School of Medicine, Dr. M. Asaka at Hokkaido University, and Dr. S. Okabe at Kushiro-rousai Hospital for their encouragement and support throughout this study.
Footnotes
Address reprint requests to Masao Seto, M.D., Ph.D., Laboratory of Chemotherapy, Aichi Cancer Center Research Institute, 1–1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan. E-mail:mseto@aichi-cc.pref.aichi.jp.
Supported in part by grants-in-aid for the 2nd-Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health and Welfare, for science on primary areas (cancer research) and for the encouragement of young scientists from the Ministry of Education, Science and Culture of Japan, and from the Uehara Memorial Foundation; and by the Bristol-Myers Squibb Unrestricted Biomedical Research Grants Program.
References
- 1.Croce CM: Molecular biology of lymphomas. Semin Oncol 1993, 20:31-46 [PubMed] [Google Scholar]
- 2.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM: Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA 1982, 79:7824-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg CL, Wong E, Petty EM, Bale AE, Tsujimoto Y, Harris NL, Arnold A: PRAD1, a candidate BCL1 oncogene: mapping and expression in centrocytic lymphoma. Proc Natl Acad Sci USA 1991, 88:9638-9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seto M, Yamamoto K, Iida S, Akao Y, Utsumi KR, Kubonishi I, Miyoshi I, Ohtsuki T, Yawata Y, Namba M, Motokura T, Arnold A, Takahashi T, Ueda R: Gene rearrangement and overexpression of PRAD1 in lymphoid malignancy with t(11;14)(q13;q32) translocation. Oncogene 1992, 7:1401-1406 [PubMed] [Google Scholar]
- 5.Seto M, Jaeger U, Hockett RD, Graninger WB, Bennett S, Goldman P, Korsmeyer SJ: Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J 1988, 7:123-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R: Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993, 262:747–750 [DOI] [PubMed]
- 7.Isaacson PG, Wright DH: Malignant lymphoma of mucosa-associated lymphoid tissue: a distinctive type of B-cell lymphoma. Cancer 1983, 52:1410-1416 [DOI] [PubMed] [Google Scholar]
- 8.Isaacson PG, Spencer J: Malignant lymphoma of mucosa-associated lymphoid tissue. Histopathology 1987, 11:445-462 [DOI] [PubMed] [Google Scholar]
- 9.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JKC, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Müller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 10.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG: Helicobacter pylori-associated gastritis, and primary B-cell gastric lymphoma. Lancet 1991, 338:1175-1176 [DOI] [PubMed] [Google Scholar]
- 11.Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, Jellum E, Orentreich N, Vogelman JH, Friedman GD: Helicobacter pylori infection, and gastric lymphoma. N Engl J Med 1994, 330:1267-1271 [DOI] [PubMed] [Google Scholar]
- 12.Isaacson PG: Mucosa-associated lymphoid tissue lymphoma. Semin Hematol 1999, 36:139-147 [PubMed] [Google Scholar]
- 13.Greiner A, Knorr C, Qin Y, Sebald W, Schimpl A, Banchereau J, Müller-Hermelink HK: Low-grade B cell lymphomas of mucosa-associated lymphoid tissue (MALT-type) require CD40-mediated signaling, and Th2-type cytokines for in vitro growth and differentiation. Am J Pathol 1997, 150:1583-1593 [PMC free article] [PubMed] [Google Scholar]
- 14.Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG: Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 1993, 342:575-577 [DOI] [PubMed] [Google Scholar]
- 15.Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros LJ, Nathwani BN: Numerical cytogenetic abnormalities of chromosomes 3, 7, and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 16.Ott G, Katzenberger T, Greiner A, Kalla J, Rosenwald A, Heinrich U, Ott MM, Müller-Hermelink HK: The t(11;18)(q21;q21) chromosome translocation is a frequent and specific aberration in low-grade but not high-grade malignant non-Hodgkin’s lymphomas of the mucosa-associated lymphoid tissue (MALT-) type. Cancer Res 1997, 57:3944-3948 [PubMed] [Google Scholar]
- 17.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P: The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999, 93:3601-3609 [PubMed] [Google Scholar]
- 18.Akagi T, Motegi M, Tamura A, Suzuki R, Hosokawa Y, Suzuki H, Ota H, Nakamura S, Morishima M, Taniwaki M, Seto M: A novel gene, MALT1 at 18q21, is involved in t(11;18)(q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 1999, 18:5785-5794 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, Motegi M, Yonezumi M, Hosokawa Y, Seto M: API1-MALT1/MLT is involved in MALT lymphoma. Blood 1999, 94:3270-3271 [PubMed] [Google Scholar]
- 20.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS: ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 1991, 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akagi T, Tamura A, Motegi M, Suzuki R, Hosokawa Y, Nakamura S, Morishima Y, Seto M, Taniwaki M: Molecular cytogenetic delineation of the breakpoint at 18q21.1 in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Genes Chromosomes Cancer 1999, 24:315-321 [PubMed] [Google Scholar]
- 22.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV: The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995, 83:1243-1252 [DOI] [PubMed] [Google Scholar]
- 23.LaCasse EC, Baird S, Korneluk RG, MacKenzie AE: The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 1998, 17:3247-3259 [DOI] [PubMed] [Google Scholar]
- 24.Birnbaum MJ, Clem RJ, Miller LK: An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol 1994, 68:2521-2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunez G, Benedict MA, Hu Y, Inohara N: Caspases: the proteases of the apoptotic pathway. Oncogene 1998, 17:3237-3245 [DOI] [PubMed] [Google Scholar]