Abstract
The t(11;18)(q21;q21) is thought to represent an important primary event in the development of marginal zone lymphomas, although an accurate estimation of the frequency and distribution of this genetic alteration among nodal, splenic, and extranodal marginal zone lymphoma types has yet to be determined. Recently, molecular genetic studies have shown that this translocation results in the fusion of the API2 gene on chromosome 11 and a novel gene termed MALT1 on chromosome 18. To investigate the incidence of API2-MALT1 fusion transcripts among marginal zone lymphomas and to determine possible marginal zone lymphoma subtype associations, we used reverse transcriptase-polymerase chain reaction to analyze RNAs extracted from frozen tissue samples of 99 marginal zone lymphomas. Fifty-seven involved diverse extranodal sites including 14 stomach, 11 lung, 7 orbit, 7 parotid, 5 thyroid, 5 lacrimal gland, 3 small intestine, 2 large intestine, 1 kidney, 1 paraspinal region and 1 skin. Twenty-one primary splenic and twenty-one primary nodal marginal zone lymphomas were also studied. API2-MALT1 fusion transcripts were detected in 12 of 57 extranodal marginal zone lymphomas (21%), but in none of the nodal or splenic cases. The cDNA sequences of the fusion transcripts were determined, revealing variation in the coding sequence fusion point for both API2 and MALT1. The findings suggest that t(11;18)(q21;q21) is restricted to extranodal marginal zone lymphomas and that these tumors have distinct genetic etiologies in comparison with their splenic and nodal counterparts.
Although they share a common CD5 and CD10 negative B cell phenotype 1,2 and are thought to arise from post follicular memory B cells, 3-5 marginal zone lymphomas are separated into three distinct disorders by the Revised European Lymphoma Classification: primary nodal marginal zone lymphoma, primary splenic marginal zone lymphoma, and extranodal marginal zone lymphoma of MALT type. 2 These lymphoma types have subtle morphological differences 6-9 and distinct clinical behaviors. 10,11 In addition, the few cases that have been studied by routine cytogenetics or by fluorescence in situ hybridization have displayed differences in the incidence of chromosome 3 abnormalities. 12-15 Thus, it is likely that marginal zone lymphomas arising in lymph nodes, spleen, and extranodal sites are also pathophysiologically distinct from one another.
The t(11;18)(q21;q21) has been identified as a recurring cytogenetic abnormality in marginal zone lymphomas, particularly in those that arise in extranodal sites. 16-20 Because balanced translocations are thought to play a pathogenetic role in the development of B cell lineage lymphomas such as Burkitt’s lymphomas, 21,22 follicular lymphomas, 23,24 and mantle cell lymphomas, 25 the t(11;18)(q21;q21) may provide clues to the pathogenesis of extranodal marginal zone lymphomas. Recently, in a detailed molecular genetic analysis of two cases of marginal zone lymphoma with the t(11;18)(q21;q21), Dierlamm and colleagues identified the fusion of two genes at the 11q21 and 18q21 breakpoints: API2 and MLT (MALT lymphoma-associated translocation), respectively. 1 API2 codes for an inhibitor of apoptosis termed c-IAP2 (also called HIAP1 or MIHC), suggesting that cells harboring the API2-MLT fusion might exhibit enhanced resistance to apoptosis. No function has yet been assigned to MLT, but this gene has also been identified by Akagi and colleagues 26 and termed MALT1. By consensus MLT is now referred to as MALT1 (Marynen P, unpublished communication), which will be its designation for the remainder of this manuscript.
Many questions regarding the frequency, clinical significance, and functional role of the API2-MALT1 gene fusion in marginal zone lymphomas remain to be addressed. For example, if t(11;18)(q21;q21) plays a consistent role in the pathogenesis of marginal zone lymphoma, then the frequency of its occurrence in such tumors should be high. Because the cytogenetic features of only a small number of marginal zone lymphomas have been reported, however, the incidence of the t(11;18)(q21;q21) is unknown at present. In addition, the number of well characterized cases of nodal and splenic marginal zone lymphomas studied by cytogenetics has been insufficient to determine whether or not these related lymphoma types also harbor the t(11;18)(q21;q21). Finally, it is not known whether the t(11;18)(q21;q21) is disproportionately represented in extranodal marginal zone lymphomas involving different anatomical sites.
Given the practical limitations of performing routine cytogenetic studies on a large number of marginal zone lymphoma cases, we undertook this study to determine the incidence of t(11;18)(q21;q21) by using reverse transcriptase-polymerase chain reaction (RT-PCR) to detect API2-MALT1 fusion transcripts. We applied this form of analysis to RNAs from a large number of marginal zone lymphomas to address the frequency of the API2-MALT1 fusion in examples of extranodal, nodal, and splenic marginal zone lymphomas. We then sequenced cDNAs from all cases in which reaction products were formed to determine the extent of variation of API2-MALT1 fusion sites.
Materials and Methods
Patient Samples
Ninety-nine marginal zone lymphoma cases were selected from files of the Division of Anatomic Pathology of the Mayo Clinic according to three criteria: diagnosis of marginal zone lymphoma based on morphology and immunophenotype (frozen section immunohistochemistry or flow cytometry in all cases) using the criteria of the Revised European American Lymphoma Classification; availability of frozen tissue for extraction of RNA; and consent of the patients for research use of their tissue. Of the 99 cases, 57 involved diverse extranodal sites including: 14 stomach, 11 lung, 7 orbit, 7 parotid, 5 thyroid, 5 lacrimal gland, 3 small bowel, 2 colon, 1 each from kidney, skin of scalp, and paravertebral region. Twenty-one cases were primary splenic marginal zone lymphomas and 21 represented nodal marginal zone lymphomas. From one patient, contemporaneous samples from intestine and spleen were studied. From a second patient 3 gastric specimens were studied: 1 at the time of diagnosis and 2 others representing recurrences at 6 months and 19 months after the date of the original biopsy. One sample of an extranodal marginal zone lymphoma involving the lung and known to contain t(11;18)(q21;q21) by standard cytogenetic analysis was used as a positive control. In addition, frozen samples of 7 cases representing reactive lymphoid hyperplasia (3 lymph nodes, 1 spleen, 1 nasopharynx, 1 tonsil, and 1 rectum with follicular lymphoid hyperplasia) were also studied as negative controls. Clinical data were collected in all cases. The study was approved by the Mayo Clinic Institutional Review Board.
RNA Extractions
Sections of frozen tumor tissue approximately 2 mm 3 were homogenized in a guanidinium isothiocyanate-phenol solution (Gibco BRL, Grand Island, NY). Following the addition of one-fifth volume of chloroform to the homogenates, organic and aqueous phases were separated by microcentrifugation at 4°C. The aqueous phases were isolated and the RNAs they contained were precipitated by the addition of an equal volume of isopropanol followed by sample incubation at −20°C for 1 hour. RNA precipitates were pelleted by microcentrifugation and the pellets were dissolved in DEPC-treated H2O. RNA samples were stored at −80°C until they were used for RT-PCR reactions (below).
cDNA Synthesis and Sequence Analysis
One-microgram samples of total RNA were reverse transcribed at 37°C for 1 hour in 10-μl reaction volumes containing random hexamer primers, MMLV reverse transcriptase, and buffer supplied by the manufacturer (GibcoBRL). Following a 2-minute, 95°C heat denaturation step, cDNA amplifications were performed in 50 μl reaction volumes containing 1 μl of product from the reverse transcription reaction, 20 pmoles of forward and reverse primers in various combinations, 200 μmol/L dNTPs (Perkin-Elmer, Foster City, CA), 1.25 U of Taq polymerase (AmpliTaq Gold, Perkin-Elmer), 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, and 0.001% gelatin, and using a reaction profile consisting of 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute (final extension at 72°C for 10 minutes). The sequence identities for the sense primers used for cDNA synthesis are according to National Center for Biotechnology Information (NCBI) accession number L49432 (API2): S1 = 4–23, S2 = 123–143, and S3 = 1098–1118. Sequences for antisense primers were based on NCBI entry AF130356 (MALT1): AS1 = 822–842, AS2 = 1452–1469, AS3 = 1908–1931, and AS4 = 2672–2693. After electrophoresis, cDNA products were excised from 1.2% agarose gels, purified (gel extraction kit, Qiagen, Valencia, CA), incubated with 40 U exonuclease I and 8 U shrimp alkaline phosphatase (PCR Product Pre-Sequencing kit, Amersham, Piscataway, NJ) at 37°C for 15 minutes, and then incubated at 80°C for 15 minutes. Approximately 100 ng quantities of purified and treated cDNA products were sequenced using 2 pmoles of forward or reverse primer (API2 sense primers 1138–1158, 1368–1391, or 1652–1673, and MALT1 antisense primers 901–921 or 1223–1243) and reagents from Amersham’s Thermosequenase kit. All fusion cDNA sites were determined and corroborated by sequencing in the sense and antisense directions. Cycling parameters were 20 seconds at 95°C, 30 seconds at 58°C, and 1 minute at 72°C for 30 cycles. Stop solution was added to each reaction, and these were subsequently denatured at 95°C for 2 minutes, quenched on ice, and electrophoresed through a 6% polyacrylamide gel with 15% formamide and 7 mol/L urea. Gels were dried for 1 hour and exposed to film overnight.
Results
Detection of API2-MALT1 Fusion Transcripts
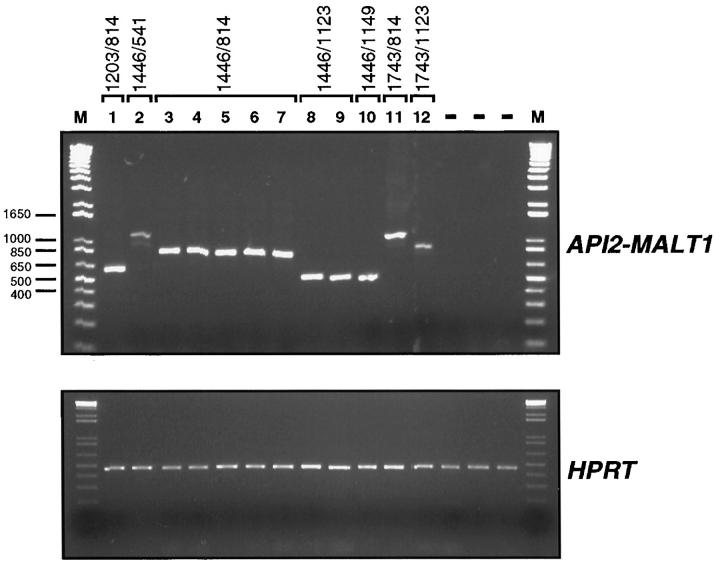
Total RNA was isolated from each biopsy specimen and analyzed by RT-PCR for API2-MALT1 fusion transcripts. Each RNA sample was examined with a minimum of four primer pairs. Results, including all of those from the marginal zone lymphoma specimens containing amplifiable API2-MALT1 fusion transcripts, are shown in Figure 1 ▶ . All 99 lymphoma specimens and all 7 lymphoid hyperplasia specimens contained detectable HPRT mRNA. API2-MALT1 fusion transcripts were identified in 12 of the 57 (21%) extranodal marginal zone lymphoma cases (6 lung, 2 colon, 2 small intestine, 1 stomach and 1 parotid). The single primer combination S2 and AS3 (Materials and Methods) produced fusion cDNAs from all 12 cases. When only those extranodal marginal zone lymphoma samples that represented the initial diagnostic specimens were considered (n = 35) the site specific frequencies of API2-MALT1 fusion were as follows: lung 4 of 9, stomach 1 of 5, colon 1 of 1, orbit 0 of 7, thyroid 0 of 4, parotid 0 of 4, lacrimal gland 0 of 3, paravertebral region 0 of 1 and kidney 0 of 1. API2-MALT1 fusion transcripts were not identified from any of the 21 primary nodal or 21 splenic marginal zone lymphoma specimens, or from any of the samples of reactive lymphoid hyperplasia. When the frequencies of detection of API2-MALT1 fusion transcripts was compared between extranodal marginal zone lymphomas and either nodal or splenic marginal zone lymphoma the differences were statistically significantly different (P < 0.05; Fisher’s exact test). The pulmonary extranodal marginal zone lymphoma case that displayed t(11;18)(q21;q21) by standard cytogenetic techniques contained an API2-MALT1 fusion transcript and its corresponding cDNA is shown in lane 1 of Figure 1 ▶ .
Figure 1.
RT-PCR detection of API2-MALT1 fusion transcripts. Total RNA, extracted from tumor specimens as described in the Materials and Methods, was reverse transcribed and RT products were used for the amplification of fusion cDNAs with primers S3 and AS2; approximate primer locations are shown in Figure 2 ▶ . Control reactions using primers for the HPRT gene were run in parallel to document that sample RNA was of adequate quality to yield RT-PCR product in all cases. Reaction products were resolved in a 1.2% agarose gel that was stained with ethidium bromide and photographed. Marker (M) sizes (in bp) are indicated to the left and sample identities are indicated at the top.
Sequencing API2-MALT1 Fusion Transcripts
As suggested from the variation in electrophoretic mobility of the fusion cDNAs and demonstrated by direct sequencing of the RT-PCR products, several different API2-MALT1 fusion sites were determined. Portions of the sequencing gels corresponding to API2-MALT1 fusion sites and schematic representations of the corresponding chimeric protein for each of the variant sequences identified are displayed in Figure 2 ▶ . In each case, the portion of the API2 gene that encodes the BIR sequences and the VDJ4-like domain from the MALT1 gene were retained in the fusion transcript. In cases 11 and 12, the CARD domain of the API2 gene was contained in the cDNA product, and in cases 1 through 7 and case 11, one or two C2-like domain sequences from MALT1 were present. Finally, the contemporaneously obtained samples of spleen and small intestine involved by extranodal marginal zone lymphoma had transcripts with identical API2-MALT1 fusion sites, as did the serially obtained specimens from the stomach.
Figure 2.
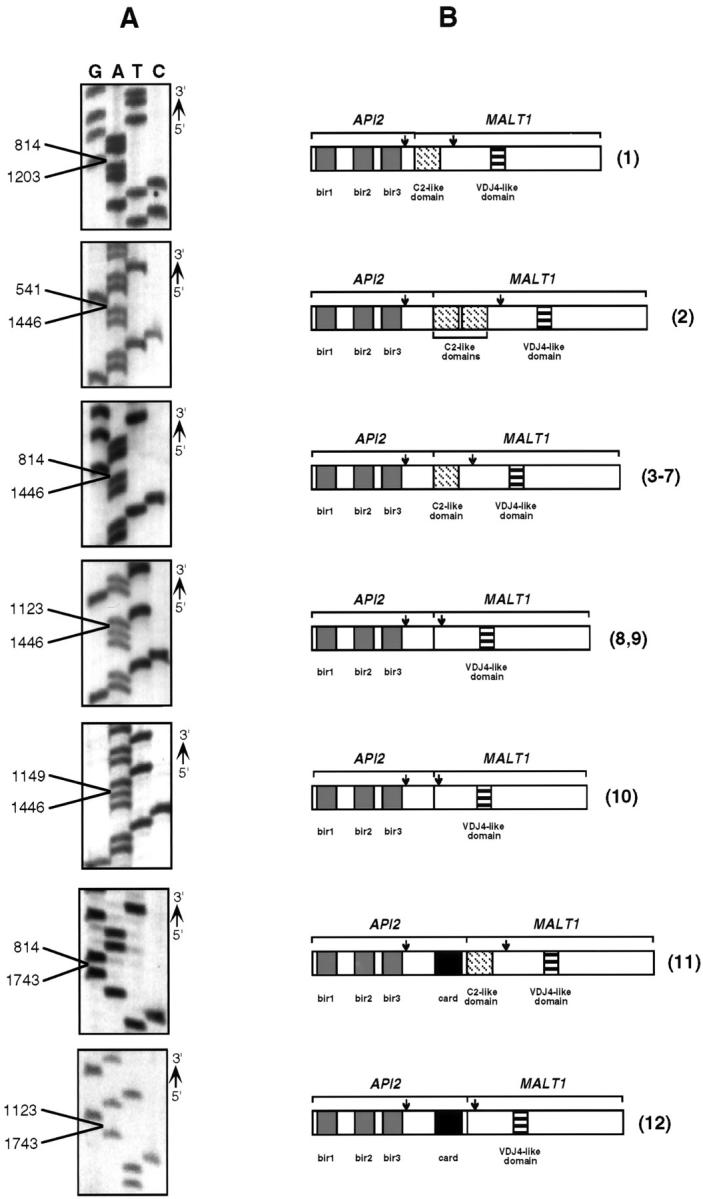
Detection of API2-MALT1 transcript fusion sites. A: Portions of sequencing gels showing examples of each type of fusion that was identified in this study. Points of fusion are indicated for each, with the lower number representing the API2 fusion site (accession no. L49432) and the upper number the fusion site for MALT1 (accession no. AF130356). B: Corresponding predicted fusion protein structures. The arrows show the locations of the RT-PCR primers that were used to produce the cDNAs shown in Figure 1 ▶ . The numbers in parentheses to the right of each fusion protein correspond to the case numbers from the top of Figure 1 ▶ .
Clinical Data
Clinical data in this study were available for 76 patients, 46 women and 30 men (F:M = 1.5:1), with a median age of 67 years (range, 29–95 years). Fifty-seven had extranodal marginal zone lymphomas, 8 had primary nodal marginal zone lymphomas, and 11 had primary splenic marginal zone lymphomas. Fifty patients, including 6 in whose specimens the API2-MALT1 fusion was demonstrated, had no prior history of lymphoma. Twenty-five patients, including 5 in whose specimens the API2-MALT1 fusion was demonstrated, had a prior diagnosis of malignant lymphoma. Twenty-one of these were extranodal marginal zone lymphomas, and the prior malignant lymphoma involved either the same (n = 13) or different (n = 5) extranodal sites, lymph nodes (n = 2), or bone marrow (n = 1). Three were nodal marginal zone lymphomas and 1 was a splenic marginal zone lymphoma. In 1 case, the patient’s prior history was unknown. Follow-up data were available for 69 patients; the median duration of follow-up was 30 months (range, 1–105 months). In 8 cases the tumor underwent transformation to large cell lymphoma; 6 of these patients, including both whose tumors were API2-MALT1 fusion-positive, died. There were no significant differences in the general clinical features or survivals between patients whose tumors contained API2-MALT1 transcripts and those whose tumors did not contain API2-MALT1 transcripts. However, API2-MALT1 transcripts were disproportionately detected in marginal zone lymphomas involving the lung.
Discussion
In 1989, Levine and colleagues first reported that t(11;18)(q21;q21) was a recurring chromosomal abnormality in malignant lymphoma. 20 Subsequent studies linked this translocation with low grade lymphomas that involved extranodal sites 19 and that had a CD5- and CD10-negative B cell phenotype. 17 As extranodal marginal zone lymphomas became recognized as a unique lymphoma type, t(11;18)(q21;q21) became specifically associated with these tumors. 16,18 When found, t(11;18)(q21;q21) almost always existed as the sole genetic anomaly, suggesting that it was a primary chromosomal abnormality that involved genes that were important in lymphomagenesis or in lymphoma progression. 16
Using fluorescence in situ hybridization with yeast, bacterial, and P1 artificial chromosome (YAC, BAC, and PAC, respectively) probes to metaphases containing t(11;18)(q21;q21), Stoffel et al 27 and Akagi et al 28 identified an 18q21 breakpoint distal to DCC1and proximal to BCL-2. These studies were followed by a detailed mapping of two cases with this type of translocation, which led to the identification of two genes, API2 at chromosome 11q21 and MALT1 at chromosome 18q21, that had been rearranged and fused. 1
The API2 gene is one member of a five-gene family in humans that codes for proteins involved in regulation of apoptosis. Common features in all but one of the gene family members include one to three copies of a motif termed the baculovirus inhibitor of apoptosis (IAP) repeat (BIR) sequence, a caspase recruitment domain (CARD), and a RING finger domain. 29 The protein coded for by API2 has been shown to be highly expressed in lymphoid cells of the spleen and thymus 29 and to suppress apoptosis by binding to and inhibiting caspase-3 as well as caspase-7 activity, and by inhibiting cytochrome C activation of procaspase-9. 30,31 Caspase binding and inhibition are maintained by functional mutant proteins containing only the BIR domains. 30 The BIR domains of c-IAP2 can also associate with TRAF1 and TRAF2, and they are thus likely to be involved in signal transduction through the 75 kd tumor necrosis factor receptor. 29 The function of the protein encoded by MALT1 is unknown. It is homologous to a hypothetical Caenorhabditis elegans gene and contains two immunoglobulin-like C2 domains and a domain similar to the mouse immunoglobulin γ chain VDJ4 sequence. 1
The cDNA sequences of the API2-MALT1 fusion transcripts identified in this study showed seven variations, including the two variants identified by Dierlamm et al. 1 In all cases the 5′ end of the fusion transcripts contained all three BIR domains encoded by API2, suggesting that if a chimeric protein is produced by the API2-MALT1 fusion transcripts, it would have an antiapoptotic function. In each of the 12 cases the RING finger domain of c-IAP2 was eliminated from the API2-MALT1 fusion transcript. It is possible that the API2-MALT1 fusion liberates the BIR motifs from regulatory control by the RING finger domain, and thereby produces an enhanced anti-apoptotic effect in the cells harboring the API2-MALT1 fusion. In 2 cases reported here, but in neither of the previously reported cases, the CARD domain of API2 was present in the API2-MALT1 transcript. The pathogenetic significance of its inclusion is unknown, but the CARD domain could conceivably alter the caspase-binding affinity of a resulting chimeric protein. The function of the MALT1 gene product in the context of the API2-MALT1 fusion transcript is unknown, but as shown (Figure 2) ▶ , the amount of MALT1 sequence in the fusion gene is variable.
As applied to extranodal marginal zone lymphomas, inhibition of apoptosis due to the presence of API2-MALT1 fusion transcripts would confer a survival advantage on B-lymphocytes, releasing them from the usual controls regulating antigen dependent proliferation. This may be particularly important for examples of gastric MALT type lymphomas related to Helicobacter pylori infection 32,33 or to extranodal lymphomas related to autoimmune disorders 34-36 where B cell recruitment to and proliferation at the anatomical sites of disease are hallmarks of the milieu in which these lymphomas develop. Enhanced inhibition of apoptosis might also favor the development of additional genetic abnormalities that cause transformation of low grade extranodal marginal zone lymphomas to aggressive large cell lymphomas. 37-40 In this series, the marginal zone lymphomas harboring the API2-MALT1 fusion did not seem to show a greater likelihood of transforming to large cell lymphoma, but the number of cases with transformation was small.
Because they arise from postfollicular memory B cells, nodal, extranodal, and splenic marginal zone lymphomas are often considered to be related neoplasms. However, mounting evidence suggests that they are pathophysiologically distinct. Extranodal marginal zone lymphomas arise in lymphoid tissue recruited to sites such as the stomach, thyroid, salivary glands, and lung in the setting of chronic antigenic stimulation. 32,34-36,41,42 They typically remain localized to the initial site of disease for long periods of time and disseminate preferentially to other extranodal sites, recapitulating the homing patterns of the normal lymphocytes of mucosa-associated lymphoid tissues. 43-46 By contrast, splenic marginal zone lymphomas preferentially involve the spleen. 7,47-49 They infrequently involve extranodal sites other than the bone marrow and blood and can produce a leukemic phase sometimes termed “splenic lymphoma with villous lymphocytes.” 9,50-52 Primary nodal marginal zone lymphomas have clinical features similar to other low grade lymphomas, with disease based predominantly in the lymph nodes. 10,11 An additional point of distinction between the marginal zone lymphoma subtypes has been suggested by studies showing differences in the frequency of trisomy 3 and in translocations involving chromosome 3q in extranodal, splenic, and nodal marginal zone lymphomas. 12-15 Furthermore, instances of t(11;18)(q21;q21) have been limited to low grade lymphomas involving extranodal sites. 16-20 The latter finding is consistent with the results of this study in which API2-MALT1 fusion transcripts were found exclusively in extranodal marginal zone lymphomas. Finally, specimens from primary marginal zone lymphomas involving the lung had a disproportionately higher rate of API2-MALT1 fusion compared to primary extranodal marginal zone lymphomas involving other sites, suggesting a site-specific molecular pathogenesis of this tumor type.
Technical factors such as the necessity for tumor cell viability and the low proliferative rate of marginal zone lymphoma cells 53 have limited the ability of routine cytogenetic testing to provide an accurate estimate of the incidence of t(11;18)(q21;q21) in marginal zone lymphomas. In addition, the studies that report on the t(11;18)(q21;q21) in marginal zone lymphomas were not specifically designed to determine the incidence of the translocation in large numbers of cases. In this study based on the RT-PCR technique, the API2-MALT1 fusion transcript was detected in 21% of 57 extranodal marginal zone lymphoma specimens. Because the primers that were used in the assay covered nearly the entire coding sequences of both API2 and MALT1, it is unlikely that cases having a fusion translocation of these genes would have escaped detection. The RT-PCR assay used here, in combination with the development of probes for fluorescence in situ hybridization detection of t(11;18)(q21;q21) in fixed, paraffin-embedded tissue, should facilitate further investigation of the clinical significance of this gene alteration.
Footnotes
Address reprint requests to Paul J. Kurtin, M.D., Hilton Building, Room 1156, Mayo Clinic, 200 First Street S.W., Rochester, MN 55905. E-mail: kurtin.paul@mayo.edu.
References
- 1.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez J, Hossfeld D, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P: The apoptosis inhibitor gene API2 and a novel 18q gene, MLT are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999, 93:3601-3609 [PubMed] [Google Scholar]
- 2.Harris N, Jaffe E, Stein H, Banks P, Chan J, Cleary M, Delsol G, De Wolf-Peeters C, Falini B, Gatter K, Grogan T, Isaacson P, Knowles D, Mason D, Muller-Hermelink H-K, Pileri S, Piris M, Ralfkiaer E, Warnke R: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 3.Miranda R, Cousar J, Hammer R, Collins R, Vnencak-Jones C: Somatic mutation analysis of IgH variable regions reveals that tumor cells of most parafollicular (monocytoid) B-cell lymphomas, splenic marginal zone B-cell lymphoma, and some hairy cell leukemia are composed of memory B lymphocytes. Hum Pathol 1999, 30:306-312 [DOI] [PubMed] [Google Scholar]
- 4.Qin Y, Greiner A, Trunk M, Schmausser B, Ott M, Müller-Hermelink H: Somatic hypermutation in low-grade mucosa-associated lymphoid tissue-type B-cell lymphoma. Blood 1995, 1995:3528-3534 [PubMed] [Google Scholar]
- 5.Zhu D, Oscier D, Stevenson F: Splenic lymphoma with villous lymphocytes involves B cells with extensively mutated Ig heavy chain variable region genes. Blood 1995, 85:1603-1607 [PubMed] [Google Scholar]
- 6.Campo E, Miquel R, Krenacs L, Sorbara L, Raffeld M, Jaffe E: Primary nodal marginal zone lymphomas of splenic and MALT types. Am J Surg Pathol 1999, 23:59-68 [DOI] [PubMed] [Google Scholar]
- 7.Mollejo M, Menárguez J, Lloret E, Sánchez A, Campo E, Algara P, Christóbal E, Sánchez E, Piris M: Splenic marginal zone lymphoma: a distinctive type of low-grade B-cell lymphoma: a clinicopathological study of 13 cases. Am J Surg Pathol 1995, 19:1146-1157 [PubMed] [Google Scholar]
- 8.Oritz-Hidalgo C, Wright D: The morphological spectrum of monocytoid B-cell lymphoma and its relationship to lymphomas of mucosa-associated lymphoid tissue. Histopathology 1992, 21:555-561 [DOI] [PubMed] [Google Scholar]
- 9.Isaacson P, Matutes E, Burke M, Catovsky D: The histopathology of splenic lymphoma with villous lymphocytes. Blood 1994, 84:3828-3834 [PubMed] [Google Scholar]
- 10.Nathwani B, Anderson J, Armitage J, Cavalli F, Diebold J, Drachenberg M, Harris N, MacLennan K, Müller-Hermelink H, Ullrich F, Weisenburger D: Marginal zone B-cell lymphoma: a clinical comparison of nodal and mucosa-associated lymphoid tissue types. J Clin Oncol 1999, 17:2486-2492 [DOI] [PubMed] [Google Scholar]
- 11.Armitage J, Weisenburger D: New approach to classifying non-Hodgkin’s lymphomas: clinical features of the major histologic subtypes. J Clin Oncol 1998, 16:2780-2795 [DOI] [PubMed] [Google Scholar]
- 12.Brynes R, Almaguer P, Leathery K, McCourty A, Arber D, Medeiros J, Nathwani B: Numerical cytogenetic abnormalities of chromosomes 3, 7 and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 13.Dierlamm J, Michauzx L, Wlodarska I, Pittaluga S, Zeller W, Stul M, Criel A, Thomas J, Boogaerts M, Delaere P, Cassiman J-J, De Wolf-Peeters C, Mecucci C, Van den Berghe H: Trisomy 3 in marginal zone B-cell lymphoma: a study based on cytogenetic analysis and fluorescence in situ hybridization. Br J Haematol 1996, 93:242-249 [DOI] [PubMed] [Google Scholar]
- 14.Wotherspoon A, Finn T, Isaacson P: Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995, 85:2000-2004 [PubMed] [Google Scholar]
- 15.Gruszka-Westwood A, Matutes E, Coignet J, Wotherspoon A, Catovsky D: The incidence of trisomy in splenic lymphoma with villous lymphocytes: a study by FISH. Br J Haematol 1999, 104:600-604 [DOI] [PubMed] [Google Scholar]
- 16.Auer I, Gascoyne R, Connors J, Cotter F, Greiner T, Sanger W, Horsman D: t(11;18)(q21;q21) is the most common translocation in MALT lymphomas. Ann Oncol 1997, 8:979-985 [DOI] [PubMed] [Google Scholar]
- 17.Griffin C, Zehnbauer B, Beschorner W, Ambinder R, Mann R: t(11;18)(q21;q21) is a recurrent chromosome abnormality in small lymphocytic lymphoma. Genes Chromosomes Cancer 1992, 4:153-157 [DOI] [PubMed] [Google Scholar]
- 18.Horsman D, Gascoyne R, Klasa R, Coupland R: t(11;18)(q21;q21): a recurring translocation in lymphomas of mucosa-associated lymphoid tissue (MALT)? Genes Chromosomes Cancer 1992, 4:183-187 [DOI] [PubMed] [Google Scholar]
- 19.Leroux D, Sieite P, Hillion J, LeMarc’hadour F, Pegourie-Bandelier B, Jacob M, Larsen C, Sotto J: t(11;18)(q21;q21) may delineate a spectrum of diffuse small B-cell lymphoma with extranodal involvement. Genes Chromosomes Cancer 1993, 7:54-56 [DOI] [PubMed] [Google Scholar]
- 20.Levine E, Arthur D, Machnicke J, Frizzera G, Hurd D, Peterson B, Gajl-Peczalska K, Bloomfield C: Four new recurring translocations in non-Hodgkin lymphoma. Blood 1989, 74:1796-1800 [PubMed] [Google Scholar]
- 21.Zech L, Haglund U, Nilsson K, Klein G: Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int J Cancer 1976, 17:47-56 [DOI] [PubMed] [Google Scholar]
- 22.Dalla-Favera R, Bregni M, Erickson J, Patterson D, Gallo R, Croce C: Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA 1982, 79:7824-7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss L, Warnke R, Sklar J, Cleary M: Molecular analysis of the t(14;18) chromosomal translocation in malignant lymphomas. N Engl J Med 1987, 317:1185-1189 [DOI] [PubMed] [Google Scholar]
- 24.Tsujimoto Y, Finger L, Yunis J, Nowell P, Croce C: Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984, 226:1097-1099 [DOI] [PubMed] [Google Scholar]
- 25.Williams M, Swerdlow S, Meeker T: Chromosome t(11;14)(q13;q32) breakpoints in centrocytic lymphoma are highly localized at the bcl-1 major translocation cluster. Leukemia 1993, 7:1437-1440 [PubMed] [Google Scholar]
- 26.Akagi T, Motegi T, Tamura A, Suzuki R, Hosokawa Y, Suzuki H, Ota H, Nakamura S, Morishima Y, Taniwaki M, Seto M: A novel gene, MALT1 at 18q21, is involved in t(11;18)(q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 1999, 18:5785-5794 [DOI] [PubMed] [Google Scholar]
- 27.Stoffel A, Rao P, Louie D, Krauter K, Leibowitz D, Koeppen H, LeBeau M, Chaganti R: Chromosome 18 breakpoint in t(11;18)(q21;q21) translocation associated with MALT lymphoma is proximal to BCL2, and distal to DCC. Genes Chromosomes Cancer 1999, 24:156-159 [PubMed] [Google Scholar]
- 28.Akagi T, Tamura A, Motegi M, Suzuki R, Hosokawa Y, Nakamura S, Morishima Y, Seto M, Taniwaki M: Molecular cytogenetic delineation of the breakpoint at 18q21.1 in low-grade lymphoma of mucosa-associated lymphoid tissue. Genes Chromosomes Cancer 1999, 24:315-321 [PubMed] [Google Scholar]
- 29.Rothe M, Pan M-G, Henzel W, Ayers T, Doeddel D: The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995, 83:1243-1252 [DOI] [PubMed] [Google Scholar]
- 30.Roy N, Deveraux Q, Takahashi R, Salvesen G, Reed J: The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J 1997, 23:6914-6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deveraux Q, Roy N, Stennicke H, Van Arsdale T, Zhou Q, Srinivasula S, Alnemri E, Salvesen G, Reed J: IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 1998, 17:2215-2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsonnet J, Hansen S, Rodriguez L, Gelb A, Warnke R, Jellum E, Orentreich N, Vogelman J, Friedman G: Helicobacter pylori infection and gastric lymphoma. N Engl J Med 1994, 330:1267-1271 [DOI] [PubMed] [Google Scholar]
- 33.Hussell T, Isaacson P, Crabtree J, Spencer J: The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 1993, 342:571-574 [DOI] [PubMed] [Google Scholar]
- 34.Hyjek E, Isaacson P: Primary B cell lymphoma of the thyroid, and its relationship to Hashimoto’s thyroiditis. Hum Pathol 1988, 19:1315-1326 [DOI] [PubMed] [Google Scholar]
- 35.Hyjek E, Smith W, Isaacson P: Primary B-cell lymphoma of salivary glands, and its relationship to myoepithelial sialadenitis. Hum Pathol 1988, 19:766-776 [DOI] [PubMed] [Google Scholar]
- 36.Royer B, Cazals-Hatem D, Sibila J, Agbalika F, Cayuela J-M, Soussim T, Maloisel F, Clauvel J-P, Brouet J-C, Mariette X: Lymphomas in patients with Sjögren’s syndrome are marginal zone B-cell neoplasms, arise in diverse extranodal and nodal sites, and are not associated with viruses. Blood 1997, 90:766-775 [PubMed] [Google Scholar]
- 37.Baldini L, Fracchiolla N, Cro L, Trecca D, Romitti L, Polli E, Maiolo A, Neri A: Frequent p53 gene involvement in splenic B-cell leukemia/lymphomas of possible marginal zone origin. Blood 1994, 84:270-278 [PubMed] [Google Scholar]
- 38.Du M, Peng H, Singh N, Isaacson P, Pan L: The accumulation of p53 abnormalities is associated with progression of mucosa-associated lymphoid tissue lymphoma. Blood 1995, 86:4587-4593 [PubMed] [Google Scholar]
- 39.Peng H, Chen G, Du M, Singh N, Isaacson P, Pan L: Replication error phenotype and p53 gene mutation in lymphomas of mucosa-associated lymphoid tissue. Am J Pathol 1996, 148:643-648 [PMC free article] [PubMed] [Google Scholar]
- 40.Gaidano G, Volpe G, Pastore C, Chiarle R, Capello D, Gloghini A, Perissinotto E, Savinelli F, Bosco M, Mazza U, Pileri S, Palestro G, Carbone A, Saglio G: Detection of BCL-6 rearrangements and p53 mutations in malt-lymphomas. Am J Hematol 1997, 56:206-213 [DOI] [PubMed] [Google Scholar]
- 41.Li G, Hansmann M-L, Zwingers T, Lennert K: Primary lymphomas of the lung: morphological, immunohistochemical and clinical features. Histopathology 1990, 16:519-531 [DOI] [PubMed] [Google Scholar]
- 42.Addis B, Hyjek E, Isaacson P: Primary pulmonary lymphoma: a re-appraisal of its histogenesis and its relationship to pseudolymphoma and lymphoid interstitial pneumonia. Histopathology 1988, 13:1-17 [DOI] [PubMed] [Google Scholar]
- 43.Isaacson P, Spencer J: Malignant lymphoma of mucosa-associated lymphoid tissue. Histopathology 1987, 11:445-462 [DOI] [PubMed] [Google Scholar]
- 44.Isaacson P: B cell lymphomas of mucosa-associated lymphoid tissue. Bull Cancer (Paris) 1991, 78:203-205 [PubMed] [Google Scholar]
- 45.Zucca E, Roggero E: Biology and treatment of MALT lymphoma: the state-of-the-art in 1996. A workshop at the 6th International Conference on Malignant Lymphoma. Ann Oncol 1996, 7:787-792 [DOI] [PubMed] [Google Scholar]
- 46.Du M-Q, Xu C-F, Diss T, Peng H-Z, Wotherspoon A, Isaacson P, Pan L-X: Intestinal dissemination of gastric mucosa-associated lymphoid tissue lymphoma. Blood 1996, 89:4445–4451 [PubMed]
- 47.Hammer R, Glick A, Greer J, Collins R, Cousar J: Splenic marginal zone lymphoma: a distinct B-cell neoplasm. Am J Surg Pathol 1996, 20:613-626 [DOI] [PubMed] [Google Scholar]
- 48.Pittaluga S, Verhoef G, Criel A, Wlodarska I, Dierlamm J, Mecucci C, Van den Berghe H, De Wolf-Peeters C: “Small” B-cell non-Hodgkin’s lymphomas with splenomegaly at presentation are either mantle cell lymphoma or marginal zone cell lymphoma: a study based on histology, cytology, immunohistochemistry and cytogenetic analsysis. Am J Surg Pathol 1996, 20:211-223 [DOI] [PubMed] [Google Scholar]
- 49.Hollema H, Visser L, Poppema S: Small lymphocytic lymphomas with predominant splenomegaly: a comparison of immunophenotypes with cases of predominant lymphadenopathy. Mod Pathol 1991, 4:712-717 [PubMed] [Google Scholar]
- 50.Melo J, Robinson D, Gregory C, Catovsky D: Splenic B cell lymphoma with “villous” lymphocytes in the peripheral blood: a disorder distinct from hairy cell leukemia. Leukemia 1987, 1:294-299 [PubMed] [Google Scholar]
- 51.Melo J, Hegde U, Parreira A, Thompson I, Lampert I, Catovsky D: Splenic B cell lymphoma with circulating villous lymphocytes: differential diagnosis of B cell leukaemias with large spleens. J Clin Pathol 1987, 40:642-651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulligan S, Matutes E, Dearden C, Catovsky D: Splenic lymphoma with villous lymphocytes: natural history and response to therapy in 50 cases. Br J Haematol 1991, 78:206-209 [DOI] [PubMed] [Google Scholar]
- 53.Du M, Singh N, Husseuin A, Isaacson P, Pan L: Positive correlation between apoptotic and proliferative indices in gastrointestinal lymphomas of mucosa-associated lymphoid tissue (MALT). J Pathol 1996, 178:379-384 [DOI] [PubMed] [Google Scholar]