Abstract
It was recently demonstrated that classification of posttransplantation lymphoproliferative disorders (PT-LPDs) into morphological and molecular categories is clinically relevant. It was also reported that PT-LPD not associated with Epstein-Barr virus (EBV) had a more aggressive course than most lesions associated with EBV. Because the cyclin-dependent kinase inhibitor p16/INK4a has been reported to be frequently inactivated in high-grade lymphomas, we evaluated 17 PT-LPD to determine whether p16/INK4a expression could be correlated to morphology, EBV detection, and a Ki-67 labeling index. We demonstrated that tumors with no p16/INK4a expression (n = 8) had a predominantly monomorphic appearance, and most were EBV negative (respectively, 7/8 and 5/8), whereas lesions with p16/INK4a expression (n = 9) were mostly polymorphic PT-LPD (6/9) (P = 0.049) and associated with EBV (9/9) (P = 0.015). In particular, strong p16/INK4a expression was observed in atypical immunoblasts and Reed-Sternberg-like cells. Furthermore, the proliferation index was significantly higher in tumors lacking p16/INK4a expression than in other lesions (P = 0.0008). In conclusion, down-regulation of p16/INK4a was mostly observed in PT-LPD lesions known to follow more aggressive courses: monomorphic tumors and EBV-negative PT-neoplasms. Conversely, overexpression of p16/INK4a was associated with EBV-positive PT-LPD. While p16/INK4a might play a role in the proliferative rate of LP-LPD, further investigations are needed to assess the clinical relevance of p16/INK4a expression in predicting the evolution of tumors and to explain how EBV could favor p16/INK4a protein accumulation in lesions.
Lymphoproliferative disorders (LPDs) are a major cause of morbidity in organ transplant recipients receiving immunosuppression, 1,2 varying between 1 and 12% of the graftees, for the most part according to the dose and duration of immunosuppressive therapy and the organ transplanted. 1,3-6 These tumors frequently involve extranodal sites and generally regress with reduction or withdrawal of immunosuppressive therapy, but some of them progress independently from the restoration of the immune system. It has been hypothesized that PT-LPDs arise from a polyclonal expansion of Epstein-Barr virus (EBV)-infected B cells and can evolve into a lymphoid tumor with the emergence of an oligoclonal or monoclonal dominant population. 7 PT-LPDs are usually classified into two subgroups, according to their appearance: polymorphic and monomorphic. 3 The latter closely resembles the diffuse large B-cell lymphoma (DLCL) of the Revised European American Lymphoma (REAL) classification. 8 Based on morphological and molecular analysis, Knowles et al 7 distinguished three main subgroups: 1) plasmacytic hyperplasia, 2) polymorphic B-cell hyperplasia and polymorphic B-cell lymphomas, and 3) malignant lymphoma or multiple myeloma, with the latter group being more frequently associated with genetic alterations, such as the inactivation of tumor suppressor gene p53 and the activation of c-myc and N-ras oncogenes. Moreover, Chadburn et al 9 recently showed the clinical relevance of this classification, as no response to aggressive clinical intervention was more frequently seen in patients with the lesions from the last group than those with the other tumors. Some patients with PT-LPDs that are not associated with EBV have a shorter survival time than those with EBV-positive lesions. 10
Expansion of B cells after organ transplantation should involve an excessive activation of the cell cycle, as in many tumors. The p16/INK4a gene is a major inhibitor of the cell cycle that is found to be inactivated in many cancers. 11 It prevents the association between cyclin D of the G1 phase and cyclin-dependent kinases (CDK)4–6 and thus the inactivation of the retinoblastoma protein (pRb) through its phosphorylation by these complexes. 12 Homozygous deletions, hypermethylation of 5′ CpG islands, and mutations are the most common mechanisms of p16/INK4a gene inactivation that have been demonstrated in cancers. 13,14 An alternative p16/INK4a transcript was recently described that encodes a structurally and functionally different protein, p14ARF, 15 which is able to inhibit the cell cycle in G1-S and G2-M phases via a p53-dependent pathway by interacting with murine double minute 2 (MDM2) protein. 16 Frequent modifications of the p16/INK4a gene have been reported in lymphoid malignancies, such as acute T-cell lymphoblastic leukemia 17,18 high-grade lymphomas, 19,20 and Burkitt’s lymphomas. 21 Furthermore, a high percentage of p16/INK4a knock-out mice develop B-cell lymphomas.
In in vitro experiments, EBV induced the phosphorylation of pRb in B cells through the up-regulation of cyclins D2 and E, and the down-regulation of p16/INK4a. 22,23 However, little is known about the regulation of p16/INK4a in EBV-driven lymphoid tumors.
Because inactivation of p16/INK4a is implicated in the transformation of indolent lymphomas, 19,20 it has been hypothesized that this gene could potentially play a role in the aggressiveness of PT-LPD. Our analysis of a series of PT-LPDs demonstrated that the expression of the p16/INK4a protein was associated with the morphological categories of tumors, EBV status, and Ki-67 proliferation index.
Materials and Methods
Materials
Formaldehyde-fixed PT-LPD biopsies from 17 adults were reviewed for the study. The following clinical data were collected for each patient: sex, age, tumor localization, and time since transplantation. Hematoxylin-eosin- and Giemsa-stained paraffin sections of all biopsies were reviewed by a panel of pathologists who classified them according to standard morphological criteria 3 as polymorphic or monomorphic PT-LPD. Immunophenotyping of the tumors by immunolabeling with B-cell and T-cell differentiation antigens, detection of EBV by in situ hybridization (ISH) and by Southern blotting, and immunoglobulin (Ig) gene heavy-chain and T-cell receptor rearrangements by Southern blotting had previously been conducted on these specimens. 10
Immunohistochemical Analysis
The streptavidin biotin-peroxidase labeling method was applied to 4-μm-thick paraffin sections with the LSAB kit (Dakopatts, Trappes, France) and monoclonal antibodies to p16/INK4a (G1175-405; PharMingen, San Diego, CA), p53 (DO7; Dakopatts), and Ki-67 (MIB-1; Immunotech, Marseille, France). Briefly, dewaxed sections were incubated overnight at 4°C with the anti-p16/INK4a antibodies diluted by 1:200 and 30 minutes at room temperature, after a microwave pretreatment of sections in citrate buffer (10 mmol/L, pH 6), for p53 and Ki-67 detection. Endogenous peroxidase blocking, inhibition of specific antibody binding, and visualization were carried out as previously described. 24 As previously reported, 20 a tumor was considered to be p16/INK4a negative when no nuclear labeling was observed in the tumor other than that of interspersed small reactive lymphocytes and endothelial cells used as an internal positive control. Ki-67-positive cells were counted on the digitized images (Visu Dis Light system; DIS, Blanc-Mesnil, France) of four high-power fields (magnification ×400) without any knowledge of the clinical information and morphological findings. p53 labeling was quantified as previously reported, 24 and only tumors with more than 10% labeled nuclei were considered positive. Normal preimmune mouse serum (Dakopatts) was used to assess nonspecific labeling.
To assess the cell-cycle status of the p16/INK4a-positive cells, double labeling with anti-p16/INK4a and anti-Ki-67 antibodies was performed using an EnVision system (Dakopatts). Briefly, after microwave treatment in citrate buffer for 15 minutes and blocking of endogenous peroxidase, the dewaxed sections were incubated with 1:200-diluted Ki-67 antibody for 30 minutes at room temperature and washed in Tris-buffered saline. Then the peroxidase-labeled polymer conjugated to goat anti-mouse Ig was applied for 30 minutes at room temperature. After revelation with 3,3′-diaminobenzidine and substrate chromogen solution, the rinsed sections were incubated with polyclonal antibodies to p16/INK4a (PharMingen) (diluted 1:400), overnight at 4°C, then with the phosphatase-labeled polymer conjugated to goat anti-mouse Ig, and were visualized with fast red and substrate-chromogen solution.
Southern Blot Analysis of the p16/INK4a Gene
High-molecular-weight DNA was extracted from frozen material by standard procedures. Southern blot analysis could be performed on seven samples that yielded sufficient DNA. EcoRI digests (15 μg) were analyzed as previously described. 18 The p16/INK4a probe (a gift from J.-M. Cayuela, Hôpital Saint-Louis, Paris, France) consisted of the exon 2 sequence cloned in PCR-Script SK(+) plasmid (Stratagene, La Jolla, CA), which was radiolabeled using a random DNA labeling kit (Boehringer-Mannheim, Mannheim, Germany) with [α-32P]dCTP. The hybridized membrane was exposed on autoradiographic film (Kodak, Rochester, NY).
Methylation-Specific Polymerase Chain Reaction
5′ CpG methylation was analyzed with the recently described methylation-specific polymerase chain reaction (PCR) (MSP), 25 using bisulfite-modified DNA, the unmethylated cytosines of which were converted into uracil. A 5′ CpG-rich site was amplified with pairs of primers distinguishing between methylated and unmethylated DNA in bisulfite-modified DNA. 25 DNA extracted from frozen samples was treated with 3 mol/L sodium bisulfite and purified as previously described. 25 Amplification was carried out in a Hybaid OmniGene thermocycler with 50 μl of PCR mix containing 1× PCR buffer (16.6 mmol/L ammonium sulfate, 67 mmol/L Tris (pH 8.8), 1.5 mmol/L MgCl2), deoxynucleoside triphosphates (each at 1.25 mmol/L), 300 ng of sense and antisense primers, and 50 ng of bisulfite-modified DNA. The p16 primers for methylated (p16-M) and unmethylated (p16-U) DNA were, respectively, 5′-TTATTAGAGGGTGGGGCGGATCGC (p16-M sense), 5′-GACCCCGAACCGCGACCGTAA (P16-M antisense), 5′-TTATTAGAGGGTGGGGTGGATTGT (p16-U sense), and 5′-CAACCCCAAACCACAACCATAA (p16-U antisense). Reactions without DNA or with untreated DNA from normal peripheral blood mononuclear cells and with treated DNA of the Jurkat lymphoblastic cell line with homozygous deletions of p16/INK4a gene were used as negative controls for MSP. A Burkitt cell line (Raji) served as the positive control for p16/INK4a methylation. PCR reaction mixture (10 μl) was electrophoresed in a nondenaturing 6% polyacrylamide gel and stained with ethidium bromide.
Statistical Analyses
Statistical analysis was performed with Stawiew software (Abacus Concepts, Berkeley, CA). A comparison between p16/INK4a status and the Ki-67 labeling index was made with the Mann-Whitney U-test, and the relationship between p16/INK4a expression and EBV detection or morphology was assessed using Fisher’s exact test. A P value less than 0.05 was considered significant.
Results
Clinical and Pathological Findings, Clonality, and EBV Detection
Clinical and demographic characteristics of the 17 patients are presented in Table 1 ▶ . Tumors involved extranodal sites in 15 of 17 cases. The time interval between organ allografting and tumor diagnosis ranged from 90 to 3450 days, with a mean of 658 ± 841 days and a median of 390 days.
Table 1.
Clinical and Demographic Characteristics, Pathologic Findings, Clonality, and EBV Detection for 17 Patients with Posttransplantation B-Cell Neoplasms
| Cases | Age | Sex | Organ transplanted | Graft-PT-LPD interval PT-LPDs | Morphology | Light chain | Clonality | EBV | |
|---|---|---|---|---|---|---|---|---|---|
| ISH | Southern | ||||||||
| 1 | 45 | M | Heart | 150 | Polymorphic | M | ND | + | ND |
| 2 | 50 | M | Heart | 90 | Polymorphic | M | ND | + | ND |
| 3 | 38 | F | Heart | 150 | Polymorphic | M | JHR | + | + |
| 4 | 68 | F | Heart | 210 | Polymorphic | M | JHR | + | + |
| 5 | 20 | M | Heart | 150 | Polymorphic | NM | ND | + | ND |
| 6 | 52 | M | Heart | 150 | Polymorphic | NM | GL | + | + |
| 7 | 51 | M | Lung | 180 | Polymorphic | NM | GL | + | + |
| 8 | 43 | M | Heart | 750 | Monomorphic | M | JHR | + | + |
| 9 | 56 | F | Kidney | 1200 | Monomorphic | M | ND | + | ND |
| 10 | 32 | M | Kidney | 450 | Monomorphic | M | JHR | + | + |
| 11 | 63 | M | Kidney | 680 | Monomorphic | M | JHR | + | + |
| 12 | 31 | M | Heart | 1660 | Monomorphic | M | JHR | + | + |
| 13 | 50 | F | Heart | 180 | Monomorphic | M | JHR | − | − |
| 14 | 43 | F | Liver | 900 | Monomorphic | M | JHR | − | − |
| 15 | 59 | M | Kidney | 3450 | Monomorphic | M | JHR | − | − |
| 16 | 57 | M | Kidney | 450 | Monomorphic | M | JHR | − | − |
| 17 | 56 | F | Kidney | 390 | Monomorphic | M | ND | − | − |
M, male; F, female; PT-LPD, posttransplantation lymphoproliferative disorders; M, monotypic light chain; NM, nonmonotypic; EBV, Epstein-Barr virus; ISH, in situ hybridization; ND, not done; Southern, Southern blot; JHR, rearrangement of Ig gene; GL, germline; +, positive; −, negative.
Among the 17 PT-LPDs examined, 41.2% were polymorphic and all were immunophenotype B. These diffuse lesions were composed of a mixture of small lymphocytes and large cells. The large cells varied in number but were generally numerous, consisting of centroblast-like cells, atypical immunoblasts, plasmablasts, Reed-Sternberg-like cells, and large areas of necrosis. The clonality of four of the polymorphic PT-LPDs was determined: two had Ig gene rearrangement and two exhibited a germline pattern. Among the three remaining samples, immunohistochemistry detected two monotypic light chains and one polytypic Ig. EBV genome and/or Epstein-Barr-encoded RNAs (EBERs) were detected in all of these polymorphic proliferations. The 10 remaining cases were monomorphic, met the REAL classification morphological criteria of diffuse large B-cell lymphomas, and were composed of uniform sheets of large noncleaved cells. All of these lesions exhibited Ig gene rearrangements and/or monotypic light chains. EBV was not detected in half of them.
Relationship between p16/INK4a Expression and Morphology
According to the level of p16/INK4a expression, two groups were distinguished: group 1 (n = 8), with no significant expression, and group 2 (n = 9), with nuclear labeling in 10–80% of the tumor cells. Most p16/INK4a-positive cells exhibited diffuse cytoplasmic labeling and were large B cells, corresponding to atypical immunoblasts or Reed-Sternberg-like cells (Figure 1A) ▶ . The majority of group 1 tumors had a monomorphic appearance (7/8). On the contrary, most in group 2 were polymorphic lesions (6/9) (P = 0.049). All tested tumors from group 1 (7/8) had monoclonal Ig gene rearrangement. In group 2, six tumors had Ig gene rearrangement and/or monotypic light-chain expression, and three lesions expressed the germline Ig gene pattern and/or had polytypic light chains (Tables 2 and 3) ▶ ▶ .
Figure 1.
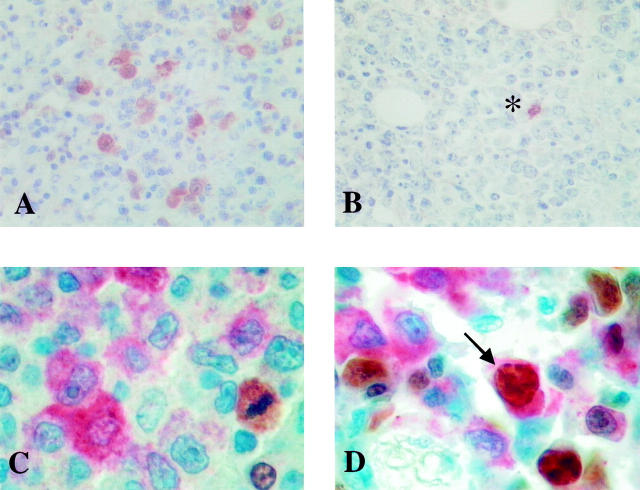
A and B: High-power (×400) field of p16/INK4a labeling in PT-LPD. A: Numerous atypical immunoblasts and Reed Sternberg-like cells in polymorphic lesion showing strong cytoplasmic and nuclear labeling. B: p16/INKa expression is detected in the tumor cells of a PT-LPD with the appearance of a large-cell lymphoma. In this case, a single reactive cell shows nuclear labeling (*). C and D: Double labeling for p16/INKa and Ki-67. C: The p16/INK4a-positive cells (red) are Ki-67 negative. Only one mitosis and one reactive cell are Ki-67 positive (brown). D: Note the double-labeled large cell (arrow).
Table 2.
Comparison between p16/INK4a Status and Ki-67 Labeling Index, p53 Expression, Morphology, EBV Detection, and Clonality
| Cases | Marker expression | Morphology | EBV | Clonality | Light chain | p16/INK4a | |||
|---|---|---|---|---|---|---|---|---|---|
| p16/INK4a | Ki-67 | p53 | Deletion | Methylation | |||||
| Group 1 | |||||||||
| 8 | − | 81% | − | Monomorphic | + | JHR | M | ND | ND |
| 13 | − | 61% | − | Monomorphic | − | JHR | M | + | − |
| 15 | − | 67% | − | Monomorphic | − | JHR | M | ND | ND |
| 14 | − | 56% | − | Monomorphic | − | JHR | M | − | + |
| 10 | − | 72% | − | Monomorphic | + | JHR | M | ND | − |
| 4 | − | 63% | − | Polymorphic | + | JHR | M | − | − |
| 16 | − | 54% | − | Monomorphic | − | JHR | M | ND | ND |
| 17 | − | 90% | − | Monomorphic | − | ND | M | ND | ND |
| Group 2 | |||||||||
| 11 | + | 40% | − | Monomorphic | + | JHR | M | ND | ND |
| 6 | + | 34% | − | Polymorphic | + | GL | NM | − | − |
| 1 | + | 33% | − | Polymorphic | + | ND | M | ND | ND |
| 5 | + | 28% | − | Polymorphic | + | ND | NM | ND | ND |
| 3 | + | 37% | − | Polymorphic | + | JHR | M | ND | ND |
| 2 | + | 29% | − | Polymorphic | + | ND | M | − | − |
| 12 | + | 55% | + (50%)* | Monomorphic | + | JHR | M | − | − |
| 7 | + | 27% | − | Polymorphic | + | GL | NM | − | − |
| 9 | + | 24% | − | Monomorphic | + | ND | M | ND | ND |
+, positive; −, negative. *50% of the tumor cells were p53 positive; for other abbreviations see footnote to Table 1 ▶ .
Table 3.
Statistical Comparisons between p16/INKK4a and Morphology, EBV Detection, and Ki-67 Proliferation Index
| Parameters | Group 1 (p16/INK4a−) (n = 8) | Group 2 (p16/INK4a+) (n = 9) | P values |
|---|---|---|---|
| Morphology | |||
| Polymorphic LP-PLDs | 1 | 6 | 0.049* |
| Monomorphic LP-PLDs | 7 | 3 | |
| EBV genome | |||
| Absent | 5 | 0 | 0.015* |
| Present | 3 | 9 | |
| Ki-67 labeling index | |||
| Mean± SD | 68 ± 12.4% | 34 ± 9.3% | 0.0008† |
SD: standard deviation.
*Fisher’s exact test.
†Mann-Whitney U-test.
Southern Blotting and MSP Analysis
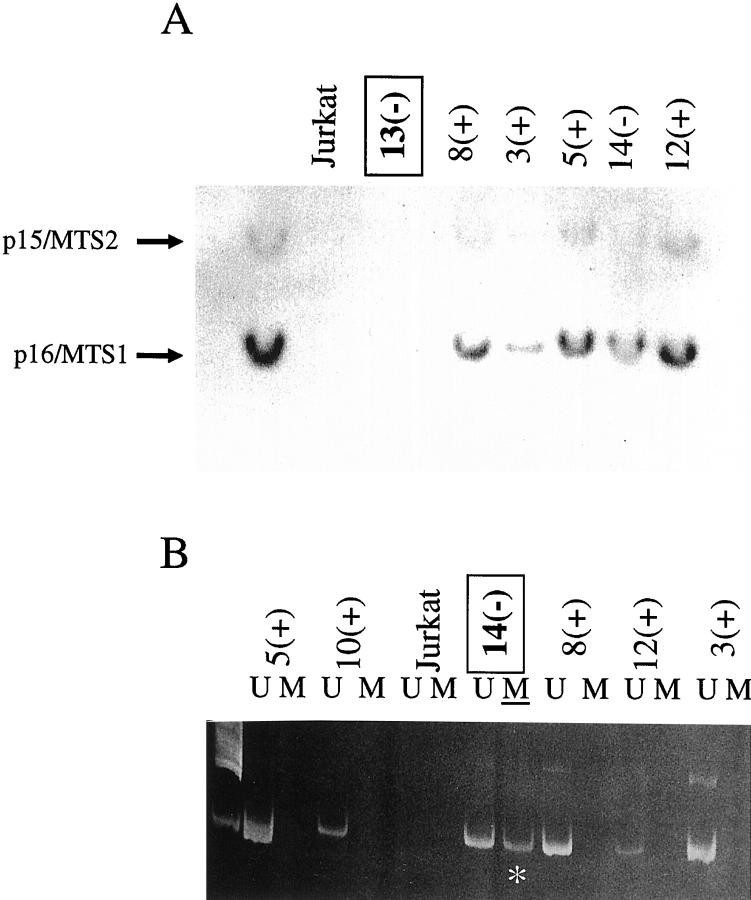
Homozygous deletion was detected in patient 13’s lesion, and methylation of a 5′ CpG was demonstrated in PT-LPD 14. p16/INK4a protein was not detected in either. The remaining tested cases, in which no deletion or methylation could be detected, always expressed p16/INK4a protein (Figure 2 ▶ and Table 2 ▶ ).
Figure 2.
A: Southern blot with a p16/MTS1-exon-2 probe that also recognized p15/INK4b-exon2. PT-LPD 13 showed a pattern similar to that of Jurkat cells, which are known to have homozygous deletion of the p16/MTS1-p15/MTS2 locus. −, 16/INK4a protein is absent; +, p16/INK4a protein is present. B: Methylation-specific PCR. Methylated (M) DNA (*) was detected in PT-LPD 14, from a patient belonging to group 1 (no detection of p16/INK4a protein). U, unmethylated.
Relationship between p16/INK4a Expression and EBV
EBV-genome sequences and/or nuclear EBER was detected in three of eight (37%) cases from group 1 and in nine of nine from group 2 (100%) (P = 0.015) (Tables 2 and 3) ▶ ▶ .
Ki-67 Labeling Index and Relationship with p16/INK4a Expression
The Ki-67 labeling was significantly higher in group 1 than in group 2 (P = 0.0008) (Tables 2 and 3) ▶ ▶ . Because of the polymorphic appearance of the majority of tumors with p16/INK4a expression, we wanted to assess the proliferative status of the p16/INK4a-positive cells. Taking advantage of the diffuse cytoplasmic labeling of p16/INK4a in the majority of positive cells, we performed double labeling with anti-p16/INK4a and anti-Ki-67 antibodies. Most of the p16/INK4a-positive cells were Ki-67 negative, except for patient 12, who had numerous double-labeled cells (Figure 1 ▶ , C and D). This case was the only one with p53 accumulation in more than 50% of tumor cells.
Discussion
In this series of PT-LPDs, we found a relationship between morphology and p16/INK4a expression, inasmuch as most lesions with no protein were monomorphic proliferations, whereas most lesions with p16/INK4a expression were polymorphic. Among the positive lesions, the labeling of the large atypical immunoblasts and Reed-Sternberg-like cells was particularly strong, whereas scattered lymphocytes and endothelial reactive cells for p16/INK4a, used as positive internal control, were weakly labeled. In the literature, some conflicting results have been reported concerning the reactivity of normal lymphocytes with anti-p16/INK4a antibodies. Betticher et al 26 did not observe labeling in normal lymphocytes in lung specimens, whereas Villuendas et al 20 found labeled lymphocytes in different compartments of reactive lymph nodes. Perhaps this discrepancy reflects differences between methods, as it is known that fixatives, duration of fixation, or use of older slides can influence the labeling of the p16/INK4a protein. To minimize false negative results, we used only sections freshly cut from paraffin blocks. In our experience we were able to detect weak labeling in some connective tissue cells and lymphocytes in different organs. Furthermore, Villuendas et al 20 showed by double labeling that most p16/INK4a-positive lymphocytes in reactive lymph nodes or in low-grade lymphomas were B cells, whereas T cells with p16/INK4a label were rare and scattered. Most of the reactive lymphocytes observed in PT-LPDs (as in Figure 1A ▶ ) were T cells, perhaps explaining why most small reactive cells were p16/INK4a negative.
The Ki-67 proliferation index was significantly higher in specimens with no p16/INK4a expression than in the others. However, it is necessary to exercise caution in interpreting the Ki-67 labeling index, because proliferating lymphoid cells can be dispersed among reactive T cells. Therefore, we verified that the tumor areas evaluated were mainly composed of B cells (data not shown), and we performed double labeling with anti-p16/INK4a and anti-Ki-67 antibodies. Interpretation of these slides was facilitated here because of the diffuse cytoplasmic labeling of p16/INK4a in large atypical cells. Although we cannot completely exclude the possibility that the number of double-labeled cells could be underestimated because of nucleus-localized p16/INK4a or poorly diffused cytoplasmic labeling, we showed in our series that most p16/INK4a-labeled large cells were Ki-67 negative, indicating that they were in G0. This observation is in agreement with the lower proliferation rate observed in PT-LPDs with p16/INK4a expression. Although diffuse cytoplasmic p16/INK4a labeling was previously reported for other tumors, 27,28 its significance remains uncertain. It might reflect artifacts or translocation of the protein into the cytoplasm by an unknown mechanism. p16/INK4a protein was reported to be present in quiescent B cells 22, and in a small percentage of peripheral blood lymphocytes. 20 Results of in vitro experiments are controversial. Cannell et al 22 showed a down-regulation of p16/INK4a in B cells after EBV infection, whereas for Villuendas et al, 20 its expression increased in circulating lymphocytes after mitogen stimulation and was more accentuated in S and G2/M phases. In reactive lymph nodes, p16/INK4a expression was observed in all compartments, but at higher levels in areas of proliferation. 20 However, it remains uncertain whether the positive cells in the proliferating compartments are effectively in cycle. In our specimens, p16/INK4a might have accumulated in cells to counteract excessive activation of the cell cycle. But the protein could be rendered ineffective by alterations of other genes implicated in the control of the cell cycle. Interestingly, one specimen with double labeling of numerous cells also had p53 protein accumulation, in contrast to the overwhelming majority of samples with only a few double-labeled cells.
A recent study showed that PT-LPDs with the large cell lymphoma pattern responded significantly less well to aggressive clinical intervention than did polymorphic lesions. 9 Furthermore, patients with EBV-negative PT-LPD seem to have a shorter survival time than patients with EBV-associated lesions. 10 Consequently, it is not surprising that a strong cell-cycle inhibitor like p16/INK4a was found to be down-regulated in lesions that are expected to have a more aggressive course.
Loss of p16/INK4a expression in tumors is strongly associated with gene deletions or 5′ CpG island methylation, and p16/INK4a inactivation is one of the most frequent genetic alterations seen in high-grade lymphomas and, particularly, in the progression of low-grade tumoral proliferations. 20 In PT-LPD, oncogene activation or tumor suppressor gene inactivation occurs in only a small percentage of lesions 7 and is associated with large-cell lymphomas or myelomas. Among the cases that we could test in this series, we found one case each of methylation and gene deletion in group 1 and no alterations in group 2.
We postulate that the integrity of p16/INK4a, in conjunction with cytotoxic cells and in the absence of genetic alterations of oncogenes or other tumor suppressor genes, could favor the rapid regression of PT-LPD frequently observed after the reduction of immunosuppressive therapy. Furthermore, it has been advanced that the loss of the p16/INK4a gene might play a role in the outgrowth of PT-LPD transplanted into mice with severe combined immunodeficiency. 29
It has been shown that in vitro EBV infection of B cells down-regulates p16/INK4a. 22 However, all of our p16/INK4a-positive PT-LPDs also expressed EBV genome. Furthermore, more than half of the p16/INK4a-negative specimens were also EBV negative. In EBV-positive lymphoblastoid cell lines that we tested previously, we did not detect p16/INK4a protein expression or gene deletion and methylation (data not shown). The discrepancy between p16/INK4a expression in in vitro cells and in vivo tumors remains to be elucidated. Finally, the relationship between p16/INK4a accumulation and morphological features of atypical immunoblasts or Reed-Sternberg-like cells infected by EBV raises some questions about the function of p16/INK4a in these cells. Indeed, p16/INK4a accumulation could reflect functions other than the regulation of the cell cycle. For example, it could favor the senescence of cells, because it is known that p16/INK4a accumulation induces senescence in in vitro cells. 30,31
Thus down-regulation of p16/INK4a is particularly observed in the lesions of PT-LPDs, which are known to have a more aggressive course, have a large-cell lymphoma appearance, and are more frequently EBV-negative than the other lesions. On the contrary, the p16/INK4a protein is often present in EBV-positive polymorphic lesions. We tried to correlate p16/INK4a expression with clinical response in our population. Unfortunately, no significant correlation with patient outcome could be established, but the number of studied cases was relatively small, and several patients died rapidly because of infections or conditions other than their LPDs (data not shown). Further investigations are necessary to determine the clinical relevance of p16/INK4a status and to specify the role of EBV in the expression of this cell cycle inhibitor.
Acknowledgments
We thank Florence Bouchard, Sandrine Stefanuto, and Karima Aliouat for their excellent technical assistance.
Footnotes
Address reprint requests to Dr. Antoine Martin, Service d’Anatomie et Cytologique Pathologiques, Hôpital Avicenne, 125 route de Stalingrad, 93009 Bobigny Cedex, France. E-mail: antoine.martin@avc.ap-hop-paris.fr.
Supported by grants from the Direction pour la Recherche Clinique de l’Assistance Publique des Hôpitaux de Paris (no. 97/147) and the Ligue contre le Cancer, Comité Départemental de Seine-Saint-Denis.
References
- 1.Penn I: Transmission of cancer with donor organs. Transplant Proc 1988, 20:739-740 [PubMed] [Google Scholar]
- 2.Penn I: Cancers complicating organ transplantation. N Engl J Med 1990, 323:1767-1769 [DOI] [PubMed] [Google Scholar]
- 3.Nalesnik MA, Jaffe R, Starzl TE, Demetris AJ, Porter K, Burnham JA, Makowka L, Ho M, Locker J: The pathology of posttransplant lymphoproliferative disorders occurring in the setting of cyclosporine A-prednisone immunosuppression. Am J Pathol 1988, 133:173-192 [PMC free article] [PubMed] [Google Scholar]
- 4.Ferry JA, Jacobson JO, Conti D, Delmonico F, Harris NL: Lymphoproliferative disorders and hematologic malignancies following organ transplantation. Mod Pathol 1989, 2:583-592 [PubMed] [Google Scholar]
- 5.Randhawa PS, Yousem SA, Paradis IL, Dauber JA, Griffith BP, Locker J: The clinical spectrum, pathology, and clonal analysis of Epstein Barr virus-associated lymphoproliferative disorders in heart-lung transplant recipients. Am J Clin Pathol 1989, 92:177-185 [DOI] [PubMed] [Google Scholar]
- 6.Opelz G, Henderson R: Incidence of non-Hodgkin’s lymphoma in kidney and heart transplant recipients. Lancet 1993, 342:1514-1516 [DOI] [PubMed] [Google Scholar]
- 7.Knowles DM, Cesarman E, Chadburn A, Frizzera G, Chen J, Rose EA, Michler RE: Correlative morphologic and molecular genetic analysis demonstrates three distinct categories of posttransplantation lymphoproliferative disorders. Blood 1995, 85:552-565 [PubMed] [Google Scholar]
- 8.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC: A revised European American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 9.Chadburn A, Chen JM, Hsu DT, Frizzera G, Cesarman E, Garrett TJ, Mears JG, Zangwill SD, Addonizio LJ, Michler RE, Knowles DM: The morphologic and molecular genetic categories of posttransplantation lymphoproliferative disorders are clinically relevant. Cancer 1998, 82:1978-1987 [PubMed] [Google Scholar]
- 10.Leblond V, Davi F, Charlotte F, Dorent R, Bitker MO, Sutton L, Gandjbakhch I, Binet JL, Raphaël M: Posttransplant lymphoproliferative disorders not associated with Epstein-Barr virus: a distinct entity? J Clin Oncol 1998, 16:2052-2059 [DOI] [PubMed] [Google Scholar]
- 11.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day R, Sr, Johnson BE, Skolnick MH: A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264:436-440 [DOI] [PubMed] [Google Scholar]
- 12.Serrano M, Hannon GJ, Beach D: A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366:704-707 [DOI] [PubMed] [Google Scholar]
- 13.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W: Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet 1995, 11:210-212 [DOI] [PubMed] [Google Scholar]
- 14.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D: 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995, 1:686-692 [DOI] [PubMed] [Google Scholar]
- 15.Quelle DE, Zindy F, Ashmun RA, Sherr CJ: Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 1995, 83:993-1000 [DOI] [PubMed] [Google Scholar]
- 16.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA: The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell 1998, 92:713-723 [DOI] [PubMed] [Google Scholar]
- 17.Hebert J, Cayuela JM, Berkeley J, Sigaux F: Candidate tumor-suppressor genes MTS1 (p16INK4a) and MTS2 (p15INK4b) display frequent homozygous deletions in primary cells from T- but not from B-cell lineage acute lymphoblastic leukemias. Blood 1994, 84:4038-4044 [PubMed] [Google Scholar]
- 18.Cayuela JM, Madani A, Sanhes L, Stern MH, Sigaux F: Multiple tumor-suppressor gene 1 inactivation is the most frequent genetic alteration in T-cell acute lymphoblastic leukemia. Blood 1996, 87:2180-2186 [PubMed] [Google Scholar]
- 19.Pinyol M, Cobo F, Bea S, Jares P, Nayach I, Fernandez PL, Montserrat E, Cardesa A, Campo E: p16(INK4a) gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin’s lymphomas. Blood 1998, 91:2977-2984 [PubMed] [Google Scholar]
- 20.Villuendas R, Sanchez-Beato M, Martinez JC, Saez AI, Martinez-Delgado B, Garcia JF, Mateo MS, Sanchez-Verde L, Benitez J, Martinez P, Piris MA: Loss of p16/INK4A protein expression in non-Hodgkin’s lymphomas is a frequent finding associated with tumor progression. Am J Pathol 1998, 153:887-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klangby U, Okan I, Magnusson KP, Wendland M, Lind P, Wiman KG: p16/Ink4a and p15/INK4b gene methylation and absence of p16/INK4a mRNA and protein expression in Burkitt’s lymphoma. Blood 1998, 91:1680-1687 [PubMed] [Google Scholar]
- 22.Cannell EJ, Farrell PJ, Sinclair AJ: Epstein Barr virus exploits the normal cell pathway to regulate Rb activity during the immortalisation of primary B-cells. Oncogene 1996, 13:1413-1421 [PubMed] [Google Scholar]
- 23.Arvanitakis L, Yaseen N, Sharma S: Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol 1995, 155:1047-1056 [PubMed] [Google Scholar]
- 24.Martin A, Flaman JM, Frebourg T, Davi F, El Mansouri S, Amouroux J, Raphaël M: Functional analysis of the p53 protein in AIDS-related non-Hodgkin’s lymphomas and polymorphic lymphoproliferations. Br J Haematol 1998, 101:311-317 [DOI] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betticher DC, White GR, Vonlanthen S, Liu X, Kappeler A, Altermatt HJ, Thatcher N, Heighway J: G1 control gene status is frequently altered in resectable non-small cell lung cancer. Int J Cancer 1997, 74:556-562 [DOI] [PubMed] [Google Scholar]
- 27.Geradts J, Kratzke RA, Niehans GA, Lincoln CE: Immunohistochemical detection of the cyclin-dependent kinase inhibitor 2/multiple tumor suppressor gene 1 (CDKN2/MTS1) product p16INK4a in archival human solid tumors: correlation with retinoblastoma protein expression. Cancer Res 1995, 55:6006-6011 [PubMed] [Google Scholar]
- 28.Fujita M, Enomoto T, Haba T, Nakashima R, Sasaki M, Yoshino K, Wada H, Buzard GS, Matsuzaki N, Wakasa K, Murata Y: Alteration of p16 and p15 genes in common epithelial ovarian tumors. Int J Cancer 1997, 74:148-155 [DOI] [PubMed] [Google Scholar]
- 29.Perera SM, Johannessen I, Thomas JA, Brooks LA, Jobe JN, Crawford DH, Radley-Smith R, Phillips M: Growth of Epstein Barr virus-associated B-lymphoproliferative disease tissue in a severe combined immunodeficient mouse. Blood 1996, 88:1123-1125 [PubMed] [Google Scholar]
- 30.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G: Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 1996, 16:859-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhrbom L, Nister M, Westermark B: Induction of senescence in human malignant glioma cells by p16INK4a. Oncogene 1997, 15:505-514 [DOI] [PubMed] [Google Scholar]