Abstract
Presence of microdomains has been postulated in the cell membrane, but two-dimensional distribution of lipid molecules has been difficult to determine in the submicrometer scale. In the present paper, we examined the distribution of gangliosides GM1 and GM3, putative raft molecules in the cell membrane, by immunoelectron microscopy using quick-frozen and freeze-fractured specimens. This method physically immobilized molecules in situ and thus minimized the possibility of artifactual perturbation. By point pattern analysis of immunogold labeling, GM1 was shown to make clusters of <100 nm in diameter in normal mouse fibroblasts. GM1-null fibroblasts were not labeled, but developed a similar clustered pattern when GM1 was administered. On cholesterol depletion or chilling, the clustering of both endogenous and exogenously-loaded GM1 decreased significantly, but the distribution showed marked regional heterogeneity in the cells. GM3 also showed cholesterol-dependent clustering, and although clusters of GM1 and GM3 were found to occasionally coincide, these aggregates were separated in most cases, suggesting the presence of heterogeneous microdomains. The present method enabled to capture the molecular distribution of lipids in the cell membrane, and demonstrated that GM1 and GM3 form clusters that are susceptible to cholesterol depletion and chilling.
INTRODUCTION
Microdomains enriched with cholesterol and sphingolipids, or rafts, have been postulated to exist in the cell membrane (Simons and Ikonen, 1997). Domains showing a liquid-ordered state have been visualized in model membranes (Korlach et al., 1999; Dietrich et al., 2001a), but whether similar domains exist in the biological membranes of living cells, and what their basic properties would be, including size, life span and dynamics, are still under debate (Simons and Ikonen, 1997; Edidin, 2003; Munro, 2003; Kusumi et al., 2004; Mayor and Rao, 2004; Mukherjee and Maxfield, 2004). Recent results, obtained by single-particle tracking and fluorescent resonance energy transfer experiments, have suggested that rafts in normal unstimulated cells are extremely small and may last for <1 ms (Kenworthy et al., 2004; Kusumi et al., 2004; Sharma et al., 2004). Detergent-resistant membranes have often been regarded as an in vitro correlate of rafts, but detergents themselves have been found to cause domain formation artificially (Heerklotz, 2002). These results have thus posed questions regarding the true existence of rafts in living, nonstimulated cells. In addition, although microscopic identification of rafts has been attempted in a number of studies, putative raft molecules generally show diffuse distribution in the cell membrane without any concentration at the resolution of light microscopy. This result has been interpreted in several different ways, i.e., that rafts do not in fact exist, rafts are too small to be resolved by light microscopy, rafts occupy the majority of the membrane, or rafts do exist but may be disrupted by experimental procedures.
GM1 has been generally regarded as an authentic raft molecule, and cholera toxin B-subunit (CtxB) has been used to probe its distribution in many microscopic studies including FRET (Kenworthy et al., 2000; Nichols, 2003). However, because CtxB is a pentameric molecule that can bind to five GM1 molecules, binding of CtxB itself is likely to change the distribution of GM1 in the membrane as shown for membrane proteins that were cross-linked with antibodies (Mayor et al., 1994; Fujimoto, 1996). Some studies used specimens that were fixed before the CtxB labeling (Parton, 1994), but chemical fixatives are unlikely to preserve the in situ localization of membrane molecules, particularly lipids, and may even cause artifactual results (Jost et al., 1973; Chandler, 1984).
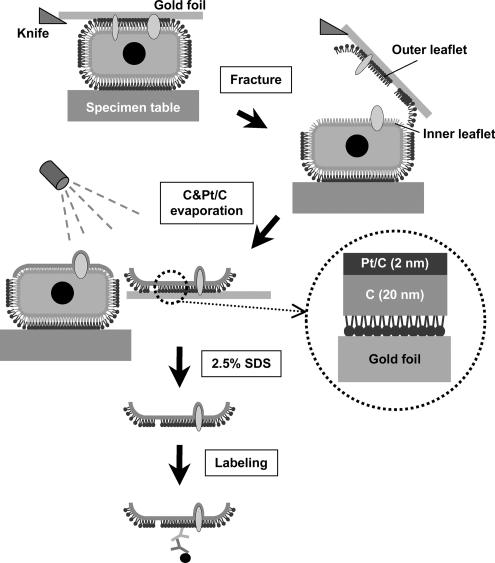
To avoid the possible artifacts caused by probe binding and chemical fixation, and also to take advantage of the high resolving power of electron microscopy (EM), we turned to immuno-EM using SDS-treated freeze-fracture replicas (SDS- FRL) (Figure 1). In combination with rapid freezing, SDS-FRL can immobilize membrane molecules physically and determine their localization in the nanometer range (Fujimoto, 1995). By use of this method, we wanted to examine whether putative raft lipids are distributed in clusters in the living cell membrane as postulated in the raft hypothesis.
Figure 1.
Outline of the SDS-FRL technique used in the present study. Cells cultured on thin gold foils were quick-frozen and freeze-fractured. Replicas were made by evaporation of carbon followed by platinum/carbon in most experiments. The replicas were treated with SDS and labeled with antibodies.
In the present study, we labeled gangliosides GM1 and GM3 in the mouse fibroblast membrane, and we analyzed the result by spatial statistical methods in wide membrane areas. As a result, both GM1 and GM3 were found to form clusters in normal mouse fibroblasts, and their clustering became less obvious after cholesterol depletion. Contrary to the simplest raft hypothesis, dissolution of the cluster upon cholesterol depletion was not complete, and it showed regional heterogeneity; and, unexpectedly, a similar result was observed when cells were cooled on ice before rapid freezing. Additionally, clusters of GM1 and GM3 overlapped only partially, suggesting the presence of heterogeneous microdomains. These observations were discussed with reference to the findings obtained using other methodologies.
MATERIALS AND METHODS
Antibodies and Probes
Rabbit anti-GM1 antibodies were raised and affinity-purified as described previously (Kusunoki et al., 1996). Two different batches of antibodies were used in the present study, and they gave equivalent results. Mouse anti-GM3 antibody (clone GMR6; Seikagaku-Kogyo, Tokyo, Japan) and biotin-conjugated cholera toxin B-subunit (b-CtxB; Invitrogen, Carlsbad, CA) were purchased. Mouse anti-phosphatidylcholine (PC) antibody (JE-1) (Nam et al., 1990) was kindly donated by Dr. Masato Umeda (Kyoto University). Secondary antibodies conjugated with fluorochromes (Invitrogen), horseradish peroxidase (Pierce Chemical, Rockford, IL), and colloidal gold (BioCell, Cardiff, United Kingdom), and protein A coupled to colloidal gold (University Medical Center Utrecht, Utrecht, The Netherlands) were obtained from respective suppliers.
Cells
Fibroblasts were explanted from the dermis of wild-type mice and mice lacking the β1,4-N-acetylgalactosamine-transferase gene (Takamiya et al., 1996). They were maintained in DMEM supplemented with 10% fetal calf serum, 50 U/ml penicillin, and 0.05 mg/ml streptomycin at 37°C under 5% CO2, 95% air. The latter cells were referred to as GM1-null fibroblasts. To load GM1-null cells with exogenous GM1 (Sigma-Aldrich, St. Louis, MO), the cells were incubated in serum-free DMEM containing GM1 (0.3, 3, or 30 μM) for 1 h at 37°C in the CO2 incubator. After incubation, the cells were washed twice with phosphate-buffered saline (PBS) and incubated with PBS containing 0.2% fatty acid-free bovine serum albumin (BSA) (Wako Pure Chemicals, Osaka, Japan) to remove GM1 loosely bound to the cell surface (Schwarzmann et al., 1983). For cholesterol depletion, cells were treated with 5 mM methyl-β-cyclodextrin (MβCD) in DMEM for 60 min.
Thin-Layer Chromatography (TLC) Immunoblotting and Dot Blotting
Total lipids were extracted from cells by using chloroform/methanol, and glycosphingolipids were obtained by reverse phase chromatography with Sep-Pak C18 cartridge (Waters, Milford, MA) (Williams and McCluer, 1980). TLC was performed with high-performance TLC plates (Merck, Darmstadt, Germany) with a solvent system of chloroform/methanol/0.25% CaCl2 (60:35:8). For staining by resorcinol, 0.5 μg of gangliosides was loaded. For TLC immunoblotting, 0.25 μg of gangliosides was developed on TLC plates, transferred to polyvinylidene difluoride membranes, and immunolabeled as described previously (Taki and Ishikawa, 1997). For dot blotting, glycosphingolipids were blotted on the nitrocellulose membrane. The signals of blotting were detected using the SuperSignal West Dura Extended Substrate (Pierce Chemical) according to the manufacturer's instruction.
Immunofluorescence Microscopy
Cells cultured on glass coverslips were fixed with buffered 4% formaldehyde, pretreated with 3% BSA, and incubated with antibodies or b-CtxB, followed by the fluorochrome-conjugated secondary reagents. In some experiments, unfixed cells were incubated with the primary probes on ice, fixed, and then labeled by the secondary reagents. The labeled samples were observed by an Axiophot2 microscope (Carl Zeiss, Jena, Germany) equipped with an AxioCam charge-coupled device camera using an Apochromat oil immersion 63× objective lens with a numerical aperture of 1.4. For quantification of filipin labeling intensity, focus was adjusted by rhodamine-phalloidin in the same specimen, and filipin fluorescence was pictured without observation by an identical microscope setting, and analyzed by ImageJ (National Institute of Health, Bethesda, MD).
Quick-Freezing and Freeze-Fracture
Cells grown on a small gold foil (∼4 mm2 in area; 20 μm in thickness) were inverted upon prewarmed 10% gelatin on a gold-plated copper specimen table with the cell side down according to the metal sandwich method (Fujimoto and Fujimoto, 1997). The cell sandwich was slammed onto the copper block precooled to the liquid helium temperature (−269°C) by using the rapid freezing apparatus HIF-4K (Hitachi High-Technologies, Tokyo, Japan).
For freeze-fracturing, the specimens were transferred to a cold stage of a Balzers BAF400 apparatus, and they were fractured at −95°C and ∼2 × 10−6 mbar. Replicas of the fractured membrane were made by electron-beam evaporation of platinum/carbon (Pt/C) and carbon (C), and the replica thickness was controlled by a crystal thickness monitor. Three different evaporation protocols were tested: 1) Pt/C (2 nm) followed by C (20 nm), 2) C (20 nm) followed by Pt/C (2 nm), 3) C (2 nm) followed by Pt/C (2 nm), and then by C (20 nm). After thawing, the replicas were immediately treated for 5 min in 2.5% SDS in PBS at 70°C. They were adjusted to 50% glycerol and kept at −20°C until labeling. Immunogold labeling was done as described previously (Fujimoto et al., 1996), and the specimens were observed with a JEOL 1200EX electron microscope operated at 100 kV.
Statistical Analysis of Immunogold Labeling
Electron micrographs were digitized with an image scanner. The x-y coordinates of gold particles were obtained by Image Processing Tool Kit version 5 plug-in (Reindeer Graphics, Asheville, NC) for Adobe Photoshop version 6 (Adobe system, Mountain View, CA), and areas of 1 μm × 1 μm chosen randomly were analyzed by Ripley's K-function (Ripley, 1979) by using a program provided by John Hancock (Prior et al., 2003). For significance tests, 99% confidence envelopes for complete spatial randomness (CSR) were generated from 100 Monte Carlo simulations. The frequency distribution of the nearest neighbor distance distribution was obtained by using the Image Processing Tool Kit.
Measurement of Phospholipids Bound to Replicas
The amount of phospholipids retained in SDS-treated C-Pt/C replicas was measured as described previously. Briefly, a suspension of small unilamellar PC liposomes was sandwiched between two thin copper foils, quickly frozen, freeze-fractured, and shadowed. One of the complementary replicas was washed in distilled water, and the other replica was treated with 2.5% SDS and washed with distilled water. The replicas were treated with 70% perchloric acid at 200°C for 2 h, and the released phosphorus was measured (Zhou and Arthur, 1992).
Biochemical Assay of Cholesterol
The total lipids were extracted from cells by a mixture of hexane and isopropanol (3:2), and the amount of free cholesterol was measured by enzymatic fluorometric method as described previously (Heider and Boyett, 1978).
On-Grid Model Experiment
Colloidal gold (2 nm; BioCell) conjugated with recombinant glutathione S-transferase (GST) was applied to Formvar-coated EM grids, blocked, and labeled with rabbit anti-GST followed by GAR-Fab5 or by PAG5. In other experiments, a dilute solution of recombinant GST was put on Formvar-coated grids and labeled by the same protocol.
RESULTS
Antibody Specificity
In enzyme-linked immunosorbent assay analysis with purified gangliosides, one of the two rabbit anti-GM1 antibodies used in our present study reacted with GM1 alone, whereas the other one also bound to GD1b and GA1. However, neither of these GM1 antibodies recognized GM2, GM3, GD1a, GD3, or GT1b (data not shown). In addition, by TLC blotting of gangliosides extracted from mouse fibroblasts, both of these antibodies showed positive reactivity only at the GM1 position and not GD1b or GA1 (Supplemental Figure 1). b-CtxB also reacted at the same position as the anti-GM1 antibodies. The monoclonal anti-GM3 antibody produced a band in cell extracts at the correct GM3 position and not GM1 (Supplemental Figure 1). The bands recognized by anti-GM1 and anti-GM3 antibodies were broader in the cell samples than that were for purified GM1 and GM3 molecules, but this is probably due to the presence of heterogeneous fatty acids, and/or the presence of glycolyl and acetylated forms of sialic acids in fibroblasts.
Immunogold Labeling of Freeze-Fracture Replicas
We previously showed that phospholipids are retained on SDS-treated freeze-fracture replicas and that they can be labeled by antibodies, but the labeling density was relatively low (Fujimoto et al., 1996). To explore different conditions that may facilitate the efficient labeling of GM1, we compared replicas prepared in three different ways. In replicas produced by the conventional evaporation method, e.g., Pt/C (2 nm) followed by C (20 nm), labeling was found to be extremely low either by anti-GM1 antibody or by b-CtxB. The labeling intensity was improved, however, when the replicas were prepared by evaporating C before Pt/C. C (20 nm)-Pt/C (2 nm) replicas and C (2 nm)-Pt/C (2 nm)-C (20 nm) replicas gave equivalent results (Supplemental Figure 2). The retention of phospholipids in the SDS-treated replicas was not compromised by the change of the evaporation method: 82.6 ± 13.0% (n = 3) phosphatidylcholine was retained in the C (20 nm)-Pt/C (2 nm) replicas, which was even better than the retention ratio obtained for the conventional Pt/C (2 nm)-C (20 nm) replicas (Fujimoto et al., 1996). Although the structural details became somewhat less defined in the C (20 nm)-Pt/C (2 nm) replica, we adopted this method in the present study due to its simplicity. Regardless of the procedure that was used for replica preparation, the labeling of GM1 was observed only in the E face, which represents the outer leaflet, and not in the P face, which represents the inner leaflet, or in the cytoplasm (Figure 3A, inset). Because the rabbit anti-GM1 antibodies gave significantly better labeling than b-CtxB, we used these antibodies in all of the subsequent EM experiments. The labeling efficiency of GM1 by the current method was calculated as described in Supplemental Table 1, and it was estimated that no less than 19.4% of GM1 in the original membrane was captured by immunogold labeling on the replicas.
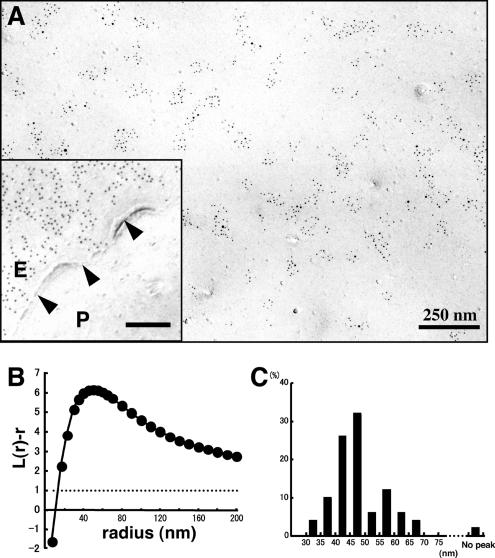
Figure 3.
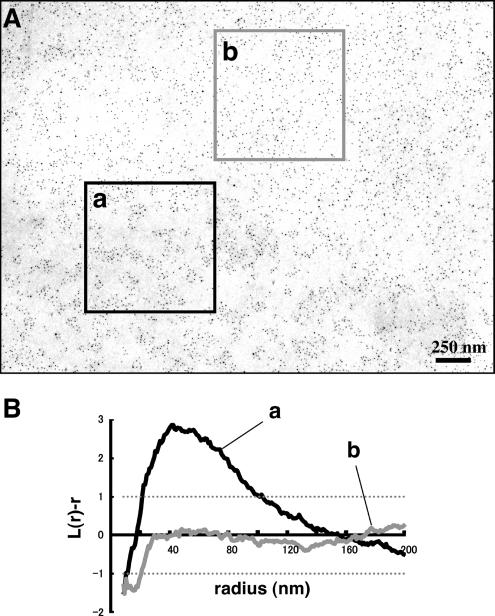
Labeling of freeze-fracture replicas of normal mouse fibroblasts by rabbit anti-GM1 antibody and colloidal gold (5-nm)-conjugated anti-rabbit IgG F(ab′)2 fragment (GAR-Fab5). (A) GM1 labeling by 5-nm colloidal gold particles was detectable as clusters in the E face of the freeze-fractured plasma membrane. Inset, the P face was unlabeled. The cell boundary is marked by arrowheads. Ten-nanometer colloidal gold was used for labeling in this sample. (B) Fifty areas (1 × 1 μm) were randomly photographed, and the gold point patterns were analyzed by Ripley's K-function. The mean L(r) − r curve showed maximal deflection from CSR (99% CI is shown by a dotted line) at a 47.0-nm radius. (C) Radii of maximal deflection for 50 sample areas ranging from 32 to 68 nm, with only one area showing no apparent peak.
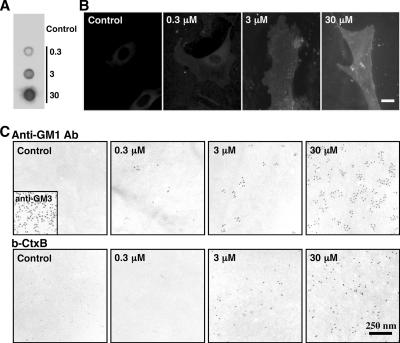
The specificity of labeling in the replicas was confirmed using GM1-null cells (Takamiya et al., 1996). Replicas of those cells were devoid of labeling either by the anti-GM1 antibodies or by b-CtxB (Figure 2C). Significantly, however, positive labeling by both probes was observed when exogenous GM1 was added to the culture medium before freezing. The quantitative loading of GM1 was confirmed by dot blotting and immunofluorescence microscopy (Figure 2, A and B), and the density of replica labeling increased as the GM1 concentration in the medium was raised from 0.3 to 30 μM (Figure 2C). The distribution of the labeling in the GM1-loaded cells was indistinguishable from that in normal mouse fibroblasts. Detailed analysis of the labeling is described below. Because the current methodology precludes the observation of molecules outside the lipid bilayer, the aforementioned results also demonstrate that exogenous GM1 molecules are incorporated into the cell membrane and that they adopt a similar disposition to endogenous GM1.
Figure 2.
Fibroblasts obtained from GM1-null mice were cultured with or without 0.3, 3, or 30 μM GM1 for 60 min. (A) Dot blot labeling by b-CtxB. (B) Fluorescence microscopy using b-CtxB, followed by fluorescein isothiocyanate-avidin. The addition of GM1 to the culture medium increases the GM1 content in GM1-null mouse fibroblasts. Scale bar, 10 μm. (C) Freeze-fracture replicas were labeled by anti-GM1 antibody (top row), or by b-CtxB (bottom row). With either probe, labeling was not observed without GM1 loading, and labeling increased according to the amount of loading. Labeling of GM3 was observed without GM1 loading (inset). Ten-nanometer colloidal gold was used for labeling.
Distribution of GM1 in the Cell Membrane
By use of the aforementioned technique, GM1 distribution in normal mouse fibroblasts was examined. Immunogold labeling gave apparently clustered distribution (Figure 3A). To obtain objective data, 50 areas of 1 × 1 μm were randomly chosen from samples obtained in more than three independent experiments, and the distribution patterns were assessed by point pattern analysis using Ripley's K-function (Ripley, 1977, 1979; Prior et al., 2003). When all the samples were compiled, the L(r) − r curve was found to deviate most from the 99% confidence interval (CI) at a radius of 47.0 nm (Figure 3B). When individual samples were analyzed, the L(r) − r curve showed a prominent peak except in a few cases (Supplemental Figure 3), and the peak size ranged from 32 to 68 nm (Figure 3C). We assumed that the basic cluster is in this size range (the size includes the arm length of the antibodies, which will be discussed later), and in subsequent experiments we classified the GM1 distribution patterns as “clustered” when the K-function was above the 99% CI at more than one point below a 100-nm radius. By this criterion, the GM1 labeling was clustered in all of the randomly chosen areas (50/50). The density of immunogold particles per unit area was found to be quite variable (Figure 4F), but GM1 clustering was observed irrespective of the labeling density.
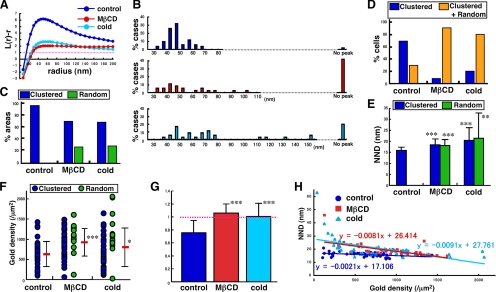
Figure 4.
Analysis of GM1 distribution in normal mouse fibroblasts under three different conditions: control, cholesterol depletion, and incubation on ice for 30 min. (A) Mean L(r) − r curves. The pooled data show clustering even after cholesterol depletion or chilling, but deviation from CSR was considerably smaller than the control. (B) Radii of maximal deflection in 50 areas. L(r) − r curves without any peak below r = 200 nm increased after either treatment. (C) Classification based on K-function analysis. Areas showing more than one point above the 99% CI below r = 100 nm were regarded as clustered. (D) Classification of 10 randomly chosen cells. The entire area in each cell was analyzed. Cells were classified by whether they showed clustered areas only or both clustered and random areas. (E) NND analysis. NND values increased significantly after cholesterol depletion or chilling, compared with control cells (Student's t test; **p < 0.005, ***p < 0.001). (F) The average labeling density showed a wide range for each sample and increased significantly after cholesterol depletion or chilling (Student's t test; *p < 0.05, ***p < 0.001). (G) NND normalized to the value expected for random distribution. Only the control sample showed a value significantly <1 (***p < 0.001). (H) Correlation of NND and the average labeling density. The dependence of NND upon the labeling density was far less in the control than in the treated samples.
The aforementioned result was obtained using rabbit anti-GM1 as the primary antibody, and colloidal gold (5 nm)-conjugated anti-rabbit IgG F(ab′)2 fragment (GAR-Fab5) as the secondary probe. Because of the small size of the GM1 head group and the highly selective binding characteristics of the anti-GM1 antibody, it is unlikely that more than two primary antibodies bound to a GM1 molecule. In contrast, more than two GAR-Fab5 particles could bind to a primary antibody. However, we concluded that the clustering of GM1 labeling was not due to multivalency based on the following results. First, a model experiment showed that two or three GAR-Fab5 particles could bind to an IgG molecule in 15.7 and 3.6% of the cases, respectively. When random point patterns were generated and the above-mentioned proportions of points were duplicated or triplicated, however, the resultant patterns did not show clustering as analyzed by Ripley's K-function (Supplemental Figure 4). Second, a very similar clustering was obtained when using colloidal gold (5 nm)-conjugated protein A (PAG5) as the secondary probe (Supplemental Figure 5, A and B). Only one PAG5 particle should bind to an IgG molecule, and the result of the model experiment was consistent with this principle (Supplemental Figure 4).
We next analyzed the entire area of randomly chosen cells to examine possible local heterogeneity within a single cell. Replicas were often disrupted within the cellular boundary, but areas of 145 μm2 (ranging from 55 to 327 μm2) could be observed on average for each cell. From this analysis, 70% of the cells showed only a clustered distribution throughout their surface, but the remainder showed small areas of random distribution (Figure 4D). An example of a whole cell profile and local L(r) − r curves is shown in Supplemental Figure 6.
Effects of Cholesterol Depletion and Low Temperatures on GM1 Clustering
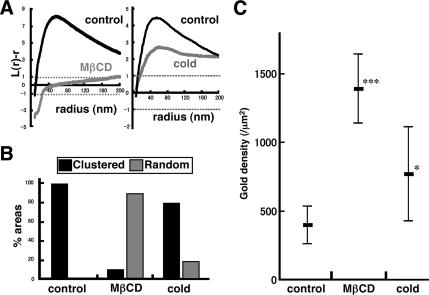
We next examined distribution of GM1 in mouse fibroblasts after depleting cholesterol to disrupt rafts. In cells treated with 5 mM MβCD for 60 min, the free cholesterol content was reduced considerably (Supplemental Figure 7). In these cells, 28% (14/50) of the areas showed random GM1 distribution, but 72% (36/50) still showed clustering (Figure 4C). Essentially, the same result was obtained when PAG5 was used for labeling (Supplemental Figure 5, B and C). Analysis of the compiled data also showed clustering (Figure 4A), but when individual samples were assessed, the peak of the L(r) − r curves was clearly lower, broader, and less distinct compared with the control sample (Supplemental Figure 3). In cholesterol-depleted wild-type mouse fibroblasts, the clustered and random distribution patterns of GM1 coexisted in >90% of the cells (Figure 4D). As shown in representative cases (Figure 5 and Supplemental Figure 6), the two different patterns were often observed side by side in narrow areas of the membrane. The result confirmed that the clustering of GM1 was not caused artifactually by the experimental procedures after freezing.
Figure 5.
GM1 labeling in mouse fibroblasts treated with 5 mM MβCD for 60 min. (A) The labeling shows a marked regional heterogeneity in most cells. (B) The gold point patterns of adjacent areas in A show clustered (a) and random (b) distributions, respectively.
With the assumption that a low temperature would increase the raft area, and thus induce a further GM1 clustering, we kept mouse fibroblasts on ice for 30 min before rapid freezing. In contrast to the assumption, however, the distribution of the GM1 labeling in those cells showed changes that were very similar to those observed in cholesterol-depleted cells. The areas showing both clustered and random GM1 patterns were 70% (35/50) and 30% (15/50), respectively (Figure 4C). Individual samples also revealed a similar tendency, and the L(r) − r curves generally became less distinct and broader than the control (Supplemental Figure 3). The coexistence of both clustered and random distribution patterns of GM1 in these cells was also observed in 80% of cases (Figure 4D). The changes caused by the cold were not likely to be caused by energy depletion because treatment with 10 mM sodium azide and 10 mM 2-deoxyglucose for 30 min, which inhibited endocytosis of CtxB completely, did not affect the GM1 distribution (data not shown).
Cholesterol depletion and low temperatures caused similar changes in two other indices: both the nearest neighbor distance (NND) and the average labeling density values increased significantly (Figure 4, E and F). The average NND, normalized to the value expected for random distribution, was significantly <1 in the control cells, but it became close to 1 after either cholesterol depletion or chilling (Figure 4G). Most significantly, the NND was virtually independent of the labeling density in control cells, whereas the NND decreased as the labeling density increased in cells after cholesterol depletion or chilling (Figure 4H). These data strongly indicate that GM1 in control mouse fibroblasts is clustered and that it becomes less so after cholesterol depletion or chilling.
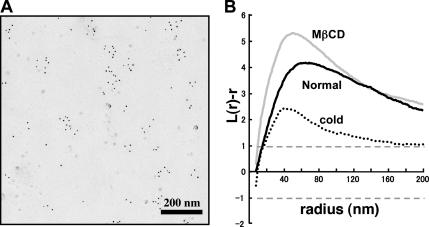
The changes caused by cholesterol depletion and the cold were observed similarly in GM1-null cells preloaded with 10–15 μM GM1. The peak of the L(r) − r curves became lower (Figure 6A), the frequency of random areas increased (Figure 6B), and the average labeling density increased (Figure 6C). Cholesterol depletion caused a more drastic change in the GM1-loaded cells than in the normal mouse fibroblasts. The reason is not clear, but some difference in the fatty acid composition between exogenous GM1 derived from bovine brain and GM1 expressed endogenously in mouse fibroblasts may be involved.
Figure 6.
Changes observed after cholesterol depletion and chilling in normal mouse fibroblasts were also observed in GM1-null fibroblasts loaded with exogenous GM1. (A) The mean L(r) − r curve shows a decrease of clustering after cholesterol depletion by MβCD or after chilling. The change was more drastic by cholesterol depletion than by chilling. (B and C) The proportion of areas showing random distribution as well as the average labeling density also increased significantly by cholesterol depletion or chilling.
As a control, PC was labeled by SDS-FRL. Probably because the epitope of the anti-PC antibody includes a hydrophobic portion of PC that is located deeper in the lipid bilayer (Nam et al., 1990), the antibody can only bind to a certain population of PC molecules, and the labeling does not occur homogenously even on liposome replicas (see figure 2A of Fujimoto et al., 1996). A similar patchy labeling pattern was observed in the mouse fibroblast (Figure 7A). In contrast to GM1 (Figure 3) and GM3 (see below), the distribution of the PC labeling was not affected by cholesterol depletion, whereas chilling caused reduction of clustering (Figure 7B). Because of the unidentified binding characteristics of the anti-PC antibody, definite interpretation of the result is not possible, but the result showed that the distributional changes do not occur in the same manner in membrane lipids.
Figure 7.
Distribution of PC labeled by mouse anti-PC antibody (IgM) and colloidal gold (5-nm)-conjugated goat anti-mouse Ig antibody. (A) The labeling of PC in the normal mouse fibroblast was observed as clusters in the E face, which was similar to that seen in liposomes and in other cell types (Fujimoto et al., 1996). (B) The K-function analysis of 10 areas (1 × 1 μm) showed that the labeling was clustered in the normal mouse fibroblast. The clustering was observed similarly in cells depleted of cholesterol, but it decreased in chilled cells.
Distribution of GM3 in Relation to GM1
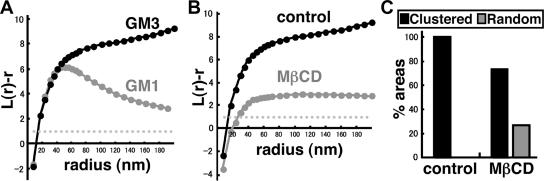
We next examined distribution of another ganglioside, GM3, by the same technique. GM3 also showed a clustered pattern in normal mouse fibroblasts (Figure 8). In comparison with GM1, the L(r) − r curve of the GM3 labeling did not show an evident peak (Figure 8A), indicating that the size of the GM3 cluster was highly variable and generally larger than that of the GM1 cluster. We confirmed that the indirect immunolabeling protocol, i.e., mouse immunoglobulin M (IgM) followed by colloidal gold-conjugated anti-mouse Ig antibody, did not cause prominent clustering by a model experiment similar to Supplemental Figure 4 (data not shown). In cells depleted of cholesterol by MβCD, the clustering of GM3 decreased significantly (Figure 8B). According to the criterion used for GM1, 26.7% (8/30) of the area showed random distribution, and in the rest of the cases, the L(r) − r curve was out of the CI range (Figure 8C). But as in the case of GM1, analysis of the individual areas showed that the L(r) − r curves became generally lower in cholesterol-depleted cells even when they were classified as “clustered” by the criterion used for GM1 (Supplemental Figure 8). These results indicated that the clustering of GM3 is also related to the cholesterol content of the plasma membrane.
Figure 8.
GM3 labeling in normal mouse fibroblasts. (A) Thirty randomly chosen areas (1 × 1 μm) were analyzed by Ripley's K-function. In contrast to GM1, the mean L(r) − r curve of GM3 did not show any apparent deflection below r = 200 nm. (B) The mean L(r) − r curve became lower after cholesterol depletion. (C) Classification of the 30 areas according to K-function analysis. Cholesterol depletion caused an increase in the number of areas with random distribution.
The relationship between the GM1 and GM3 clusters in normal mouse fibroblasts was examined by double labeling by using colloidal gold particles of two different diameters. By use of the bivariate K-function, two different patterns were observed. In a relatively limited number of areas (13.3%), GM1 and GM3 were observed to make coclusters, whereas in most areas (86.7%), the GM1 and GM3 clusters were segregated and formed independently (Figure 9, A and B). When coclustering was observed, GM1 clustering was less discrete than that observed by single labeling, which was evident in the L(r) − r curve of the univariate K-function analysis (data not shown). This difference was likely caused by steric hindrance between the anti-GM1 and anti-GM3 antibodies binding to overlapping areas.
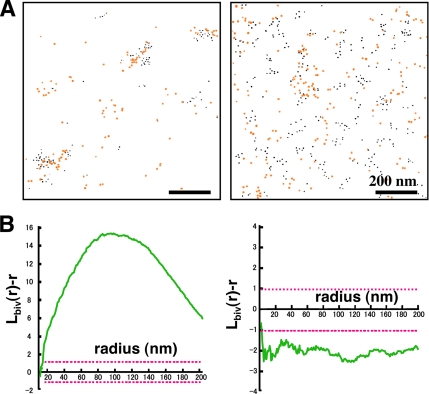
Figure 9.
Double labeling of GM1 and GM3 in normal mouse fibroblasts. (A) GM1 and GM3 were marked with colloidal gold particles of different sizes, which were colored artificially: GM1, black; GM3, orange. (B) Analysis of the two areas shown in A by using a bivariate K-function. GM1 and GM3 were found to be coclustered in the left sample, but they segregated from each other in the right sample. Coclustering as shown in the left graph was seen only in 13.3% of the cases.
DISCUSSION
Methodological Considerations
SDS-FRL has several advantages for analyzing the distribution of membrane molecules in the submicrometer range. By freezing live cells quickly to the liquid helium temperature, the molecular motion in the cell should stop immediately. Phase separation may occur in membranes that were cooled slowly, but it was prevented by this quick-freeze method (Thompson et al., 1985). In their pioneering article, Heuser and his colleagues showed that the surface of a muscle piece became frozen within 1 ms after the impact with the liquid helium-cooled copper block (Heuser et al., 1979). They later found that the freezing rate is at least 10 times faster than that (Heuser, personal communication). Although details in the present experiment were different from their experiment, the cell membrane of thin culture cells should be frozen at a better cooling rate than the muscle piece. Further sophistication in the measurement of the freezing rate is required to assess the time and spatial resolution of this method precisely.
During the freeze-fracture procedure, the specimens were kept frozen below −100°C, and the fractured membrane half was coated by evaporation of C and Pt/C to immobilize membrane molecules in situ. The physical fixation provided by the combination of quick-freezing and freeze-fracturing contrasts with chemical fixation by aldehydes in many respects. Aldehydes have only limited reactivity with membrane lipids and could even cause redistribution of membrane proteins by a cross-linking effect (Kusumi and Suzuki, 2005). More than several seconds are required for the aldehyde fixation to complete, and even after fixation, membrane molecules retain their two-dimensional mobility (Jost et al., 1973; Chandler, 1984). Thus, in specimens fixed by formaldehydes, binding of probes, especially multivalent probes, could cause redistribution of membrane molecules (Mayor et al., 1994). Although glutaraldehyde was shown to prevent gross redistribution of membrane proteins (Mayor et al., 1994), it is unlikely to stabilize membrane lipids in the nanometer range. The membranes stabilized in replicas preclude the possibility of such artifacts caused by probes.
Compared with the conventional method that evaporates Pt/C before C, the labeling of gangliosides was improved drastically by evaporating C before Pt/C. A similar effect has been reported for some membrane proteins (Hagiwara et al., 2005). The reason for this improvement is not fully clear, but compared with Pt/C, pure C casting may allow the molecules to retain some flexibility, which may facilitate their interaction with probes. Additionally, the use of antibodies improved the GM1 labeling in SDS-FRL. CtxB worked well for immunofluorescence microscopy, but it labeled the replica to a much lesser extent than the anti-GM1 antibodies. For high-affinity binding of pentameric CtxB molecules, GM1 molecules and surrounding lipids may need to be rearranged properly, and this probably occurs frequently in the fluid membrane. The relatively low labeling by SDS-FRL may have resulted because molecules in the replica are immobilized, and they do not take an appropriate arrangement very often.
GM1 Clustering in the Cell Membrane
The GM1 labeling formed clusters with an average radius of 47.0 nm in control mouse fibroblasts. The average labeling density ranged from <100 to >1400 particles/μm2, but clusters of similar sizes were observed irrespective of the GM1 density. In our present method, two layers of probes, either a whole IgG molecule and a F(ab′)2 portion (GAR-Fab5) or a whole IgG and protein A (PAG5), were intercalated between colloidal gold (5 nm) and the antigen. Using a model experiment, the spacer distances were estimated as 16.3 ± 4.1 nm (GAR-Fab5) and 16.1 ± 5.3 nm (PAG5) (Supplemental Figure 10). This lead to a calculated size of the GM1 cluster of ∼61 nm in diameter, which roughly corresponds to that which was estimated by several studies for glycosylphosphatidylinositol (GPI)-anchored protein clusters (Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998; Pralle et al., 2000), but it is larger than the estimate obtained in more recent studies by using sophisticated biophysical techniques (Kusumi et al., 2004; Sharma et al., 2004). Ras proteins were also shown to make smaller clusters by immuno-EM of mechanically detached membranes (Plowman et al., 2005). The size difference of the clusters is not surprising, however, because GPI-anchored proteins were shown to behave differently from GM1 in several instances (Schnitzer et al., 1995; Simons et al., 1999; Dietrich et al., 2001b). It is also likely that the Ras cluster in the inner leaflet does not coincide with that of GM1 in the outer leaflet. SDS-FRL should be able to study the distribution of those proteins and to compare the cluster sizes directly.
After cholesterol depletion or incubation of cells on ice, GM1 labeling became less clustered, and areas showing random distribution increased significantly. In addition, cholesterol depletion and chilling increased the average labeling density as well as the NND. The increase of the labeling density could reflect a real increase of GM1 molecules in the plasma membrane, but the change of the aforementioned three parameters together, and above all, the virtual independence of NND values from the labeling density in control cells (Figure 4H), is more likely to suggest a dispersion of GM1 clusters by cholesterol depletion or by chilling. That is, GM1 molecules densely packed in control cells are probably not labeled efficiently due to steric hindrance, but after cholesterol depletion or chilling, they may be dispersed and more GM1 molecules may become accessible to antibodies. This possibility also suggests that the density of GM1 molecules in the cluster is underestimated in immunogold labeling.
An important question that arises from our current data is the very nature of these GM1 clusters. The clusters may be explained by the presence of rafts, or liquid-ordered (lo) domains, and by preferential partitioning of GM1 in the domains. The results of our chilling experiments would not seem consistent with this supposition, because low temperatures are expected to increase more ordered domains, whereas cholesterol depletion should disrupt lo domains (Simons and Ikonen, 1997; Gaus et al., 2003). However, as the temperature is decreased, the lo domain should become the dominant or percolating phase, and GM1 and other raft-philic molecules that are confined to small nonpercolating lo domains at the ambient temperature may adopt a dispersed distribution in the expanded lo domains (Meder et al., 2006). Hence, the dispersed GM1 distribution after chilling can be interpreted to reflect the larger lo domain, or an increase of raft areas. This contrasts with the similarly dispersed GM1 distribution after cholesterol depletion, which may be caused by an increase of the liquid-disordered domains, or disruption of rafts.
However, the incomplete dissolution of the GM1 cluster and the marked regional heterogeneity upon cholesterol depletion suggest that preferential partitioning in the ordered membrane may not explain the whole phenomenon. In this context, it is notable that several studies showed formation of GM1-rich microdomains within the ordered phase of model membranes (Vie et al., 1998; Yuan and Johnston, 2000; Yuan et al., 2002), suggesting that glycolipids are capable of self-organization (but please note that other studies failed to observe the clustering of charged gangliosides; Thompson et al., 1985; Wang and Silvius, 2003). Furthermore, the extracellular matrix, the cytoskeleton, and membrane proteins may influence distribution of membrane molecules in various ways (Edidin, 2003; Ritchie et al., 2003; Mukherjee and Maxfield, 2004). These results raised a possibility that those nonraft factors are sufficient to generate the GM1 cluster in mouse fibroblasts and that rafts are not involved. Under this conjecture, the result of cholesterol depletion might be attributed to its effect on the actin cytoskeleton (Kwik et al., 2003). However, we observed that manipulation of actin filaments did not affect the GM1 distribution to the extent that can explain the result of cholesterol depletion (Fujita, Cheng, and Fujimoto, unpublished observation). Moreover, chilling is not likely to affect the actin cytoskeleton in the same manner. We thus inferred that preferential partitioning in the ordered membrane is involved at least partially for the generation of GM1 clusters.
Irrespective of the mechanism that facilitates GM1 clustering in living cells, the present result has several implications. First, it has not been easy to understand why cholesterol depletion and chilling could both activate mitogen-activated protein kinase signaling (Furuchi and Anderson, 1998; Kabouridis et al., 2000; Chen and Resh, 2002; Gousset et al., 2002; Magee et al., 2005), because the two manipulations are thought to exert opposite effects on rafts, but the present result on GM1 suggests that the two manipulations may affect the distribution of individual raft-philic molecules in the same manner and could cause a similar outcome. This tenet may be tested directly by probing fine distribution of signaling proteins that reside in the inner leaflet of the cell membrane. Second, cooling on ice has been often used to label cells for microscopy with the tacit supposition that the molecular distribution at an ambient temperature is retained. However, the present results indicated that cooling could change the distribution of some membrane molecules, so data that have been obtained from chilled cells need to be interpreted with caution.
Heterogeneous Clusters of GM1 and GM3
We observed that both GM1 and GM3 formed clusters susceptible to cholesterol depletion in normal mouse fibroblasts but that the respective clusters were segregated from each other in most cases. In migrating T cells, a distinct segregation of GM1 and GM3, i.e., GM1 in the uropod and GM3 in the leading edge, was observed (Gomez-Mouton et al., 2001). Our result suggests that GM1 and GM3 also distribute differentially in other cell types, although on a much more minuscule scale. The result also implies that the high distribution density of the two gangliosides has precluded the observation of their segregation by light microscopy in the past.
Biochemical fractions enriched with GM1 and GM3 have been shown to contain different sets of signaling molecules (Iwabuchi et al., 1998; Chigorno et al., 2000). Those fractions are likely to correspond to the GM1 and GM3 domains that we observed, and this would raise two immediate questions. One question is whether the spatial relationship of the two domains would modulate signaling by altering molecular interactions among the signaling proteins. We observed occasional coclustering of GM1 and GM3, which suggests that the two domains could coalesce under certain conditions. It would be interesting to study how coalescence and segregation of those domains are regulated and what is brought about as a result of the domain interactions. The second question is how signaling proteins in the inner leaflet partition to one of the ganglioside domains preferentially. This may be related to the mechanism that generates separate clusters of GM1 and GM3. An intriguing possibility is that GM1 and GM3 are diversified in the ceramide portion and thus favor homologous interactions (Sonnino and Chigorno, 2000; Hakomori Si, 2002). This is not an unrealistic speculation, because glycosyltransferases for the late ganglioside biosynthetic pathways are in the distal Golgi membranes (Lannert et al., 1998), which harbor cholesterol-dependent microdomains, and the probability that a GM3 molecule is processed to become GM1 may depend on the affinity of its ceramide portion to the microdomain where the enzymes exist. As a result, gangliosides remaining as GM3, and those that became GM1, are likely to make homologous clusters more often than heterogeneous gangliosides. Analysis of the ceramide composition of gangliosides is awaited to test the possibility.
Although the underlying mechanism needs to be explored further, our result showed that GM1 and GM3 exist as distinct clusters in the native cell membrane. But the GM1 and GM3 clusters may not be the only microdomains that are affected by cholesterol depletion, and additional microdomains with variable contents are likely to coexist (Marwali et al., 2003; Nagatsuka et al., 2003; Brugger et al., 2004; Kiyokawa et al., 2005). How all those heterogeneous microdomains are formed and how their mutual relationship is controlled warrant further investigation. Hopefully, the present method would help to address these unsolved questions in the near future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Atsuyuki Okabe and Ken-ichiro Shimatani for advice on point pattern analysis, Dr. John Hancock for providing the K-function program, Dr. Masato Umeda for anti-PC antibody, Drs. Akihiro Kusumi and John E. Heuser for helpful discussions, and Kumi Tauchi-Sato and Tetsuo Okumura for assistance. This work was supported by grants-in-aid for scientific research and the 21st Century Center of Excellence Program “Integrated Molecular Medicine for Neuronal and Neoplastic Disorders” of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
Abbreviations used:
- b-CtxB
biotinylated cholera toxin B
- CI
confidence interval
- CSR
complete spatial randomness
- EM
electron microscopy
- GAR-Fab5
colloidal gold (5-nm)-conjugated anti-rabbit IgG F(ab′)2 fragment
- PAG5
colloidal gold (5-nm)-conjugated protein A
- MβCD
methyl-β-cyclodextrin
- NND
nearest neighbor distance
- PC
phosphatidylcholine
- SDS-FRL
SDS-treated freeze-fracture replicas.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0071) on March 28, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Brugger B., Graham C., Leibrecht I., Mombelli E., Jen A., Wieland F., Morris R. The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J. Biol. Chem. 2004;279:7530–7536. doi: 10.1074/jbc.M310207200. [DOI] [PubMed] [Google Scholar]
- Chandler D. E. Comparison of quick-frozen and chemically fixed sea-urchin eggs: structural evidence that cortical granule exocytosis is preceded by a local increase in membrane mobility. J. Cell Sci. 1984;72:23–36. doi: 10.1242/jcs.72.1.23. [DOI] [PubMed] [Google Scholar]
- Chen X., Resh M. D. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J. Biol. Chem. 2002;277:49631–49637. doi: 10.1074/jbc.M208327200. [DOI] [PubMed] [Google Scholar]
- Chigorno V., Palestini P., Sciannamblo M., Dolo V., Pavan A., Tettamanti G., Sonnino S. Evidence that ganglioside enriched domains are distinct from caveolae in MDCK II and human fibroblast cells in culture. Eur. J. Biochem. 2000;267:4187–4197. doi: 10.1046/j.1432-1327.2000.01454.x. [DOI] [PubMed] [Google Scholar]
- Dietrich C., Bagatolli L. A., Volovyk Z. N., Thompson N. L., Levi M., Jacobson K., Gratton E. Lipid rafts reconstituted in model membranes. Biophys. J. 2001a;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C., Volovyk Z. N., Levi M., Thompson N. L., Jacobson K. Partitioning of Thy-1, GM1, and cross-linked phospholipid analogs into lipid rafts reconstituted in supported model membrane monolayers. Proc. Natl. Acad. Sci. USA. 2001b;98:10642–10647. doi: 10.1073/pnas.191168698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- Friedrichson T., Kurzchalia T. V. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Fujimoto K., Umeda M., Fujimoto T. Transmembrane phospholipid distribution revealed by freeze-fracture replica labeling. J. Cell Sci. 1996;109:2453–2460. doi: 10.1242/jcs.109.10.2453. [DOI] [PubMed] [Google Scholar]
- Fujimoto T. GPI-anchored proteins, glycosphingolipids, and sphingomyelin are sequestered to caveolae only after crosslinking. J. Histochem. Cytochem. 1996;44:929–941. doi: 10.1177/44.8.8756764. [DOI] [PubMed] [Google Scholar]
- Fujimoto T., Fujimoto K. Metal sandwich method to quick-freeze monolayer cultured cells for freeze-fracture. J. Histochem. Cytochem. 1997;45:595–598. doi: 10.1177/002215549704500411. [DOI] [PubMed] [Google Scholar]
- Furuchi T., Anderson R. G. Cholesterol depletion of caveolae causes hyperactivation of extracellular signal-related kinase (ERK) J. Biol. Chem. 1998;273:21099–21104. doi: 10.1074/jbc.273.33.21099. [DOI] [PubMed] [Google Scholar]
- Gaus K., Gratton E., Kable E. P., Jones A. S., Gelissen I., Kritharides L., Jessup W. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mouton C., Abad J. L., Mira E., Lacalle R. A., Gallardo E., Jimenez-Baranda S., Illa I., Bernad A., Manes S., Martinez A. C. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc. Natl. Acad. Sci. USA. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousset K., Wolkers W. F., Tsvetkova N. M., Oliver A. E., Field C. L., Walker N. J., Crowe J. H., Tablin F. Evidence for a physiological role for membrane rafts in human platelets. J. Cell. Physiol. 2002;190:117–128. doi: 10.1002/jcp.10039. [DOI] [PubMed] [Google Scholar]
- Hagiwara A., Fukazawa Y., Deguchi-Tawarada M., Ohtsuka T., Shigemoto R. Differential distribution of release-related proteins in the hippocampal CA3 area as revealed by freeze-fracture replica labeling. J. Comp. Neurol. 2005;489:195–216. doi: 10.1002/cne.20633. [DOI] [PubMed] [Google Scholar]
- Hakomori Si S. I. Inaugural article: the glycosynapse. Proc. Natl. Acad. Sci. USA. 2002;99:225–232. doi: 10.1073/pnas.012540899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophys. J. 2002;83:2693–2701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider J. G., Boyett R. L. The picomole determination of free and total cholesterol in cells in culture. J. Lipid Res. 1978;19:514–518. [PubMed] [Google Scholar]
- Heuser J. E., Reese T. S., Dennis M. J., Jan Y., Jan L., Evans L. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell Biol. 1979;81:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K., Handa K., Hakomori S. Separation of “glycosphingolipid signaling domain” from caveolin-containing membrane fraction in mouse melanoma B16 cells and its role in cell adhesion coupled with signaling. J. Biol. Chem. 1998;273:33766–33773. doi: 10.1074/jbc.273.50.33766. [DOI] [PubMed] [Google Scholar]
- Jost P., Brooks U. J., Griffith O. H. Fluidity of phospholipid bilayers and membranes after exposure to osmium tetroxide and gluteraldehyde. J. Mol. Biol. 1973;76:313–318. doi: 10.1016/0022-2836(73)90394-x. [DOI] [PubMed] [Google Scholar]
- Kabouridis P. S., Janzen J., Magee A. L., Ley S. C. Cholesterol depletion disrupts lipid rafts and modulates the activity of multiple signaling pathways in T lymphocytes. Eur. J. Immunol. 2000;30:954–963. doi: 10.1002/1521-4141(200003)30:3<954::AID-IMMU954>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Kenworthy A. K., Nichols B. J., Remmert C. L., Hendrix G. M., Kumar M., Zimmerberg J., Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy A. K., Petranova N., Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyokawa E., Baba T., Otsuka N., Makino A., Ohno S., Kobayashi T. Spatial and functional heterogeneity of sphingolipid-rich membrane domains. J. Biol. Chem. 2005;280:24072–24084. doi: 10.1074/jbc.M502244200. [DOI] [PubMed] [Google Scholar]
- Korlach J., Schwille P., Webb W. W., Feigenson G. W. Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 1999;96:8461–8466. doi: 10.1073/pnas.96.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A., Koyama-Honda I., Suzuki K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Suzuki K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta. 2005;1746:234–251. doi: 10.1016/j.bbamcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Kusunoki S., Shimizu J., Chiba A., Ugawa Y., Hitoshi S., Kanazawa I. Experimental sensory neuropathy induced by sensitization with ganglioside GD1b. Ann. Neurol. 1996;39:424–431. doi: 10.1002/ana.410390404. [DOI] [PubMed] [Google Scholar]
- Kwik J., Boyle S., Fooksman D., Margolis L., Sheetz M. P., Edidin M. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 2003;100:13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannert H., Gorgas K., Meissner I., Wieland F. T., Jeckel D. Functional organization of the Golgi apparatus in glycosphingolipid biosynthesis. Lactosylceramide and subsequent glycosphingolipids are formed in the lumen of the late Golgi. J. Biol. Chem. 1998;273:2939–2946. doi: 10.1074/jbc.273.5.2939. [DOI] [PubMed] [Google Scholar]
- Magee A. I., Adler J., Parmryd I. Cold-induced coalescence of T-cell plasma membrane microdomains activates signalling pathways. J. Cell Sci. 2005;118:3141–3151. doi: 10.1242/jcs.02442. [DOI] [PubMed] [Google Scholar]
- Marwali M. R., Rey-Ladino J., Dreolini L., Shaw D., Takei F. Membrane cholesterol regulates LFA-1 function and lipid raft heterogeneity. Blood. 2003;102:215–222. doi: 10.1182/blood-2002-10-3195. [DOI] [PubMed] [Google Scholar]
- Mayor S., Rao M. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 2004;5:231–240. doi: 10.1111/j.1600-0854.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- Mayor S., Rothberg K. G., Maxfield F. R. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264:1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- Meder D., Moreno M. J., Verkade P., Vaz W. L., Simons K. Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc. Natl. Acad. Sci. USA. 2006;103:329–334. doi: 10.1073/pnas.0509885103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Maxfield F. R. Membrane domains. Annu. Rev. Cell Dev. Biol. 2004;20:839–866. doi: 10.1146/annurev.cellbio.20.010403.095451. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Nagatsuka Y., et al. Carbohydrate-dependent signaling from the phosphatidylglucoside-based microdomain induces granulocytic differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA. 2003;100:7454–7459. doi: 10.1073/pnas.1232503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K. S., Igarashi K., Umeda M., Inoue K. Production and characterization of monoclonal antibodies that specifically bind to phosphatidylcholine. Biochim. Biophys. Acta. 1990;1046:89–96. doi: 10.1016/0005-2760(90)90098-i. [DOI] [PubMed] [Google Scholar]
- Nichols B. J. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 2003;13:686–690. doi: 10.1016/s0960-9822(03)00209-4. [DOI] [PubMed] [Google Scholar]
- Parton R. G. Ultrastructural localization of gangliosides; GM1 is concentrated in caveolae. J. Histochem. Cytochem. 1994;42:155–166. doi: 10.1177/42.2.8288861. [DOI] [PubMed] [Google Scholar]
- Plowman S. J., Muncke C., Parton R. G., Hancock J. F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A., Keller P., Florin E. L., Simons K., Horber J. K. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I. A., Muncke C., Parton R. G., Hancock J. F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley B. D. Modeling spatial patterns. J. R. Stat. Soc. Ser. B. 1977;39:172–212. [Google Scholar]
- Ripley B. D. Tests of randomness for spatial point patterns. J. R. Stat. Soc. Ser. B. 1979;41:368–374. [Google Scholar]
- Ritchie K., Iino R., Fujiwara T., Murase K., Kusumi A. The fence and picket structure of the plasma membrane of live cells as revealed by single molecule techniques. Mol. Membr. Biol. 2003;20:13–18. doi: 10.1080/0968768021000055698. [DOI] [PubMed] [Google Scholar]
- Schnitzer J. E., McIntosh D. P., Dvorak A. M., Liu J., Oh P. Separation of caveolae from associated microdomains of GPI-anchored proteins. Science. 1995;269:1435–1439. doi: 10.1126/science.7660128. [DOI] [PubMed] [Google Scholar]
- Schwarzmann G., Hoffmann-Bleihauer P., Schubert J., Sandhoff K., Marsh D. Incorporation of ganglioside analogues into fibroblast cell membranes. A spin-label study. Biochemistry. 1983;22:5041–5048. doi: 10.1021/bi00290a025. [DOI] [PubMed] [Google Scholar]
- Sharma P., Varma R., Sarasij R. C., Ira, Gousset K., Krishnamoorthy G., Rao M., Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons M., Friedrichson T., Schulz J. B., Pitto M., Masserini M., Kurzchalia T. V. Exogenous administration of gangliosides displaces GPI-anchored proteins from lipid microdomains in living cells. Mol. Biol. Cell. 1999;10:3187–3196. doi: 10.1091/mbc.10.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnino S., Chigorno V. Ganglioside molecular species containing C18- and C20-sphingosine in mammalian nervous tissues and neuronal cell cultures. Biochim. Biophys. Acta. 2000;1469:63–77. doi: 10.1016/s0005-2736(00)00210-8. [DOI] [PubMed] [Google Scholar]
- Takamiya K., et al. Mice with disrupted GM2/GD2 synthase gene lack complex gangliosides but exhibit only subtle defects in their nervous system. Proc. Natl. Acad. Sci. USA. 1996;93:10662–10667. doi: 10.1073/pnas.93.20.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki T., Ishikawa D. TLC blotting: application to microscale analysis of lipids and as a new approach to lipid-protein interaction. Anal. Biochem. 1997;251:135–143. doi: 10.1006/abio.1997.2247. [DOI] [PubMed] [Google Scholar]
- Thompson T. E., Allietta M., Brown R. E., Johnson M. L., Tillack T. W. Organization of ganglioside GM1 in phosphatidylcholine bilayers. Biochim. Biophys. Acta. 1985;817:229–237. doi: 10.1016/0005-2736(85)90024-0. [DOI] [PubMed] [Google Scholar]
- Varma R., Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Vie V., Van Mau N., Lesniewska E., Goudonnet J. P., Heitz F., Le Grimelle C. Distribution of ganglioside GM1 between two-component, two-phase phosphatidylcholine monolayers. Langmuir. 1998;14:4754–4783. [Google Scholar]
- Wang T. Y., Silvius J. R. Sphingolipid partitioning into ordered domains in cholesterol-free and cholesterol-containing lipid bilayers. Biophys. J. 2003;84:367–378. doi: 10.1016/S0006-3495(03)74857-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. A., McCluer R. H. The use of Sep-Pak C18 cartridges during the isolation of gangliosides. J. Neurochem. 1980;35:266–269. doi: 10.1111/j.1471-4159.1980.tb12515.x. [DOI] [PubMed] [Google Scholar]
- Yuan C., Furlong J., Burgos P., Johnston L. J. The size of lipid rafts: an atomic force microscopy study of ganglioside GM1 domains in sphingomyelin/DOPC/cholesterol membranes. Biophys. J. 2002;82:2526–2535. doi: 10.1016/S0006-3495(02)75596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Johnston L. J. Distribution of ganglioside GM1 in L-alpha-dipalmitoylphosphatidylcholine/cholesterol monolayers: a model for lipid rafts. Biophys. J. 2000;79:2768–2781. doi: 10.1016/S0006-3495(00)76516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Arthur G. Improved procedures for the determination of lipid phosphorus by malachite green. J. Lipid Res. 1992;33:1233–1236. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.