Abstract
Nopp140 associates with small nucleolar RNPs to chaperone pre-rRNA processing and ribosome assembly. Alternative splicing yields two isoforms in Drosophila: Nopp140-True is homologous to vertebrate Nopp140 particularly in its carboxy terminus, whereas Nopp140-RGG contains a glycine and arginine-rich (RGG) carboxy terminus typically found in vertebrate nucleolin. Loss of ribosome function or production at critical points in development leads to Minute phenotypes in Drosophila or the Treacher Collins syndrome (TCS) in humans. To ascertain the functional significance of Nopp140 in Drosophila development, we expressed interfering RNA using the GAL4/UAS system. Reverse transcription-PCR showed variable losses of Nopp140 mRNA in larvae from separate RNAi-expressing transgenic lines, whereas immunofluorescence microscopy with isoform-specific antibodies showed losses of Nopp140 in imaginal and polyploid tissues. Phenotypic expression correlated with the percent loss of Nopp140 transcripts: a ≥50% loss correlated with larval and pupal lethality, disrupted nuclear structures, and in some cases melanotic tumors, whereas a 30% loss correlated with adult wing, leg, and tergite deformities. We consider these adult phenotypes to be Minute-like and reminiscent of human craniofacial malformations associated with TCS. Similarly, overexpression of either isoform caused embryonic and larval lethality, thus indicating proper expression of Nopp140 is critical for normal development.
INTRODUCTION
Nucleolar phosphoprotein 140 (Nopp140, Mr = 140–180 kDa) has several purported functions in ribosome assembly (Meier, 2005). The greatest steady state concentrations of vertebrate Nopp140 occur within the dense fibrillar component of nucleoli and within extra-nucleolar Cajal bodies. Nopp140 associates with box C/D small nucleolar ribonucleoprotein (snoRNPs) that guide site-specific 2′-O-methylation of the pre-rRNA and with box H/ACA snoRNPs that guide site-specific pseudouridylation of pre-rRNA and snRNAs (Yang et al., 2000; Wang et al., 2002; reviewed by Meier, 2005). Nopp140 likely acts as an assembly factor for these snoRNPs or as a chaperone in their transport between Cajal bodies and nucleoli (Isaac et al., 1998; Lemm et al., 2006). Nopp140 also interacts with RNA Pol I to regulate rRNA transcription within mammalian nucleoli (Chen et al., 1999) and with Pol II transcription factors in the protein kinase A–mediated activation of the α1-acid glycoprotein gene in liver cells during acute phase response (Miau et al., 1997; Chiu et al., 2002). Nopp140 shuttles rapidly between the nucleus and the cytoplasm (Meier and Blobel, 1992), perhaps to facilitate the import of nucleolar ribosome assembly factors or the export of nucleolar products (Meier and Blobel, 1990).
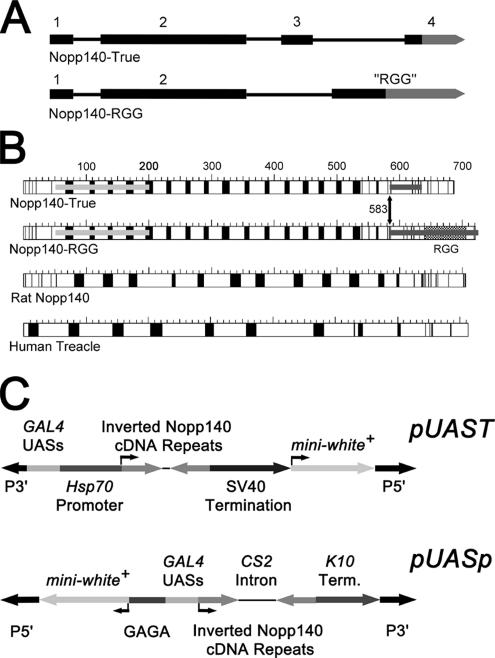
In Drosophila melanogaster, the Nopp140 gene maps to the proximal left arm of chromosome 3 in cytological region 78F4 (http://flybase.bio.indiana.edu/.bin/fbidq.html? FBgn0037137). Two isoforms of the protein exist: Nopp140-True contains 686 amino acid residues and Nopp140-RGG contains 720 residues. Their mRNAs share the first two exons that together encode amino acids 1–583. Alternative splicing generates distinctively different carboxy termini: in one pathway, exons 3 and 4 encode a carboxy terminus that is 65% identical to the carboxy terminus of human Nopp140 over a 94-amino acid stretch. We considered this isoform to be the true ortholog of vertebrate Nopp140 in Drosophila (Waggener and DiMario, 2002). In the second splicing pathway, an alternative acceptor site within the intron separating exons 3 and 4 of the first pathway links the common exon 2 to a single longer “RGG” (glycine and arginine-rich) exon that encodes the carboxy tail of Nopp140-RGG (Figure 1A). What were intronic sequences in the first pathway now encode the entire carboxy terminus of Nopp140-RGG. Clustered Arg-Gly-Gly motifs within this terminus also exist in several other RNA-associated proteins (Burd and Dreyfuss, 1994), including the nucleolar proteins nucleolin (Lapeyre et al., 1987) and fibrillarin (Aris and Blobel, 1991). Both Nopp140-True and Nopp140-RGG localize to Drosophila nucleoli when exogenously expressed as green fluorescent protein (GFP) fusions in transgenic embryos, larvae, and adults (McCain et al., 2006). Here we report that isoform-specific antibodies label the nucleoli in all cells examined to date.
Figure 1.
(A) Alternative splicing of the Nopp140 pre-mRNA (3943 nt) produces Nopp140-True and Nopp140-RGG. Exons 1 and 2 are common for both mature mRNAs. Nopp140-True is encoded by exons 3 and 4, whereas Nopp140-RGG is encoded by the RGG exon. Gray segments represent the 3′ untranslated regions (UTRs). The coding sequence for the carboxy terminus of Nopp140-RGG is totally contained within the intron separating exons 3 and 4 in the first pathway. Notice that exon 4 in the first pathway is expressed as a part of the 3′ UTR in the RGG exon. (B) Primary structures of the two Drosophila Nopp140 isoforms (Nopp140-True and Nopp140-RGG) compared with prototypical rat Nopp140 (Meier, 1996) and human Treacle. Acidic regions are in black and basic or neutral regions are in white. The carboxy terminus of the Drosophila Nopp140-True is 64% identical to the carboxy terminus of rat Nopp140. The light and dark gray bars spanning the amino and carboxy terminal regions of the Drosophila proteins denote the corresponding cDNA sequences that were used to express RNAi. (C) pUAST (Brand and Perrimon, 1993) harbors a transposable P-element, five tandem yeast GAL4 UASs, the minimal Hsp70 gene promoter, the common 450-base pair Nopp140 cDNA repeats ligated in inverted manner with an intervening polylinker spacer sequence of 25 base pairs, followed by the SV40 small t termination sequence. The other plasmid used in this work was pUASp-Nba-CS2-BgX (Zhu and Stein, 2004; simply designated pUASp), which contains upstream GAGA sites, 14 GAL4 UASs, the P-element transposase gene promoter with first intron, and the fs(1)K10 gene 3′ UTR. The mini-white+ genes with their promoters in both plasmids allowed selection of transformed (red-eyed) adults after microinjecting the recombinant plasmids into homozygous and hemizygous w1118 embryos, along with a helper plasmid that encoded transposase.
We also describe our efforts to deplete Nopp140 using interfering RNA (RNAi) induced by the GAL4/UAS system in transgenic flies. Phenotypes resulting from the knockdown of Nopp140 fall within the Minute syndrome of Drosophila. Classic Minute phenotypes include shortened and thin thoracic bristles, rough eyes, missing or deformed antennae, abnormal wings, defective abdominal cuticle segmentation (i.e., fused tergites), reduced viability and fertility, prolonged development, and recessive lethality (Schultz, 1929; Lambertsson, 1998; Marygold et al., 2005). Many of the Minute genes encode ribosomal proteins, and Minute phenotypes caused by mutations in these genes likely arise from insufficient ribosome function in larval imaginal discs, pupal histoblasts, and adult germ cells, all of which proliferate and differentiate within defined developmental time frames.
The large central region of all Nopp140 orthologues consists of alternating acidic and basic motifs (Meier, 1996; Figure 1B of this report). This central region is structurally related to Treacle (see Figure 1), another nucleolar protein of 144 kDa identified thus far only in vertebrates (Wise et al., 1997; Isaac et al., 2000; Valdez et al., 2004; Gonzales et al., 2005). Treacle has unique amino and carboxy terminal domains that contain nuclear localization signals, whereas its large central domain contains 10 alternating acidic and basic domains. Isaac et al. (2000) showed that both Treacle and Nopp140 are phosphorylated by casein kinase type II (CKII) enzymes and that both proteins localize to the dense fibrillar components of HeLa cell nucleoli; but of the two, only Nopp140 localizes to extranucleolar Cajal bodies.
With similar structural features and nucleolar colocalizations, Treacle and Nopp140 likely share related functions in ribosome assembly, but then diverge in function with respect to Cajal bodies. Treacle interacts with the nucleolar protein, Nop56, a component of the ribonucleoprotein complex that methylates specific nucleotides (2′-O-ribose) in the pre-rRNA (Hayano et al., 2003). Knockdown of Treacle in Xenopus oocytes inhibits site-directed methylation of the pre-rRNA. Besides Nop56, Valdez et al. (2004) showed that Treacle also interacts with the upstream binding factor (UBF), an RNA polymerase I transcription factor specific for rRNA genes within vertebrate nucleoli. Treacle, therefore, seems to have two important roles in the biosynthesis of ribosomes; first by regulating transcription of pre-rRNA and then by chemically modifying (processing) pre-rRNA as ribosomal proteins assemble with the rRNA to form ribosomal subunits.
Mutations in human Treacle lead to the human Treacher Collins syndrome (TCS), an autosomal dominant disorder afflicting 1 in 50,000 live births. TCS patients display craniofacial malformations that include hypoplasia of facial bones, specifically the mandible and the zygomatic complex (cheek bones), and underdevelopment of the outer ear, external ear canal, and middle ear, thus causing conductive hearing loss. Other defects include a downward slant of the palpebral fissure (eye lids) causing an anti-mongoloid eye slant, hypoplasia of the lower eye lids, including a coloboma (an eyelid defect) in the outer third of the lower eye lid, with a loss of cilia medial to the coloboma, and finally, cleft palate with or without cleft lip in ∼35% of TCS cases. These various deformities are thought to arise from a loss of ribosome production or function in progenitor neural crest cells (Dixon et al., 2006) before their migration into branchial arches I and II by day 24 ± 1 of human embryogenesis (reviewed by Dixon, 1996; Marszalek et al., 2002). Human Treacle is encoded by TCOF1 in 5q32-q33.1 (Treacher Collins Syndrome Collaboration Group, 1996). Mutations in TCOF1 are haplo-insufficient, but significant variability in the penetrance and expressivity of TCS in humans is likely due to differences in genetic background (Dixon and Dixon, 2004), although these genetic factors remain unknown.
BLAST searches of the Drosophila genome indicate that Nopp140-True is the closest homologue to human Treacle. Similar to the purported loss of ribosome function for Minute mutations in Drosophila, a loss of ribosome production in embryonic neural crest cells is believed to be responsible for the TCS in humans. Because of these apparent similarities, we hope to advance further the Drosophila Minute-like phenotypes resulting from the loss of ribosome assembly factors (e.g., Nopp140) as molecular-genetic models for the human TCS, especially as defined Drosophila genetic backgrounds or environmental conditions may affect the penetrance and expressivity of these phenotypes.
MATERIALS AND METHODS
Fly Stocks
Unless otherwise stated, fly lines were obtained from the Bloomington Drosophila Stock Center at Indiana University. Strains used in this study included the w1118 line (stock #3605), the second chromosome balancer stock w*/w*; Sp1/CyO originally from W. M. Saxton (Indiana University), and the third chromosome balancer stock w−/w−; ScmEt50 e/TM3 Sb originally from J. A. Simon (University of Minnesota). The third chromosome daughterless-GAL4 driver line was stock no. 8461. The salivary gland GAL4 third chromosome driver was Sgs3-GAL4 (stock no. 6870). Details of the various strains, chromosomes, and marker genes are available at Flybase (Grumbling et al., 2006).
Plasmid Construction
GAL4-induced expression in transgenic flies (Brand and Perrimon, 1993; Duffy, 2002) used pUAST and a derivative of pUASp (Rorth, 1998) called pUASp-Nba-CS2-BgX (Zhu and Stein, 2004) that contains the CS-2 gene intron flanked by restriction enzyme sites. Both pUAST and pUASp contain GAL4 upstream activation sequences (UASs); pUAST also contains the Hsp70 promoter (Figure 1C).
To express RNAi specific for both Nopp140-True and Nopp140-RGG mRNAs, we amplified a 450-base pair cDNA fragment encoding Thr52-Ser201 (light gray bar Figure 1B) using primers Common-F (5′-CCAAGAATTCATAGGTACCACCAAAAGCCCCAAAAAGATC-3′) and Common-R (5′-CATTCTAGATATACTCGAGCTTTCTTCGCTGCTGGAGTC-3′). The resulting PCR product contained flanking EcoRI and KpnI sites on the 5′ end, and XbaI and XhoI sites on the 3′ end. The fragment was propagated in pBluescript using the EcoRI and XbaI sites. The forward fragment was ligated into pUAST using the EcoRI and XbaI sites, whereas the reverse fragment was ligated using the KpnI and XhoI sites. A 25-base pair poly-linker sequence separated the inverted repeats in the final pUAST construct.
Similar strategies were used to introduce inverted repeats into pUASp-Nba-CS2-BgX. We used the primers Common-F (5′-CCAACGAGATCTACCAAAAGCCCCAAAAAGATC-3′) and Common-R (5′-CAACATTCTAGACTTTCTTCGCTGCTGGAGTC-3′) to amplify the same 450-base pair fragment common to both cDNAs. The resulting PCR product was trimmed at its 5′ and 3′ ends with BglII and XbaI, respectively, and ligated into pUASp-Nba-CS2-BgX in the forward direction using compatible sticky ends at the BamHI and NheI sites, respectively. The same PCR fragment was ligated in the reverse orientation at the BglII and XbaI sites in pUASp-Nba-CS2-BgX. The CS-2 intronic sequence of ∼800 bp separates the two inverted copies (exons). Separating the inverted repeats helps propagate the plasmid in Escherichia coli and stabilize the transgene in the Drosophila genome. On GAL4-induced expression, the normal splicing machinery should remove the intron to generate double-stranded RNAi.
To overexpress the GFP-tagged Nopp140 isoforms, cDNAs encoding either full-length GFP-Nopp140-True or full-length GFP-Nopp140-RGG (Waggener and DiMario, 2002) were ligated into pUAST. These recombinant DNAs are designated pUAST-GFP-Nopp140-True and pUAST-GFP-Nopp140-RGG, respectively. All recombinant pUAST and pUASp plasmids were propagated in Sure E. coli cells (Stratagene, La Jolla, CA).
P-element–mediated Transformation, Nomenclature, and Inverse Genomic PCR
We used standard microinjection techniques (Spradling, 1986; Kiehart et al., 2000) to transform homozygous or hemizygous w1118 embryos with recombinant pUAST or pUASp constructs. We coinjected the helper plasmid, pUChsΔ2 that expresses transposase. pUAST and pUASp carry the mini-w+ gene; chromosomes harboring mini-w+ were determined using marked balancer stocks Sp1/CyO for chromosome 2, and ScmEt50/TM3 for chromosome 3. Both balancer stocks had a w− background.
Inverse genomic PCRs were performed as described (Huang et al., 2000; but see E. Jay Rehm's detailed protocol at http://www.fruitfly.org/about/methods/inverse.pcr.html) to identify the number and location of P-element insertions. Inverse PCR products were sequenced at the DNA Sequencing and Synthesis Facility, Iowa State University, Ames, IA.
RT-PCR
Total RNA was extracted from control and RNAi-expressing third instar larvae using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations and dissolved in RNAsecure Resuspension Solution (Ambion, Austin, TX). First-strand cDNA synthesis was performed using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's recommendations. Specific primer sets for the RT-PCRs were as follows: Nopp140-Common forward and reverse primers were 5′-CTAGCCAAGGTTTTCCAGCAGAAGAC-3′ and 5′-GCTGGCTTAGTCTCCTCATCGGAG-3′, respectively; actin 5C (Act5C) forward and reverse primers were 5′-CTCACCTATAGAAGACGAAGAAGTTGCTGCTCT-3′ and 5′-CTAACTGTTGAATCCTCGTAGGACTTCTCCAACG-3′, respectively; and ribosomal protein L32 (RpL32) forward and reverse primers were 5′-GTTGTGCACCAGGAACTTCTTGAATCCG-3′ and 5′-CTTCCAGCTTCAAGATGACCATCCGC-3′, respectively.
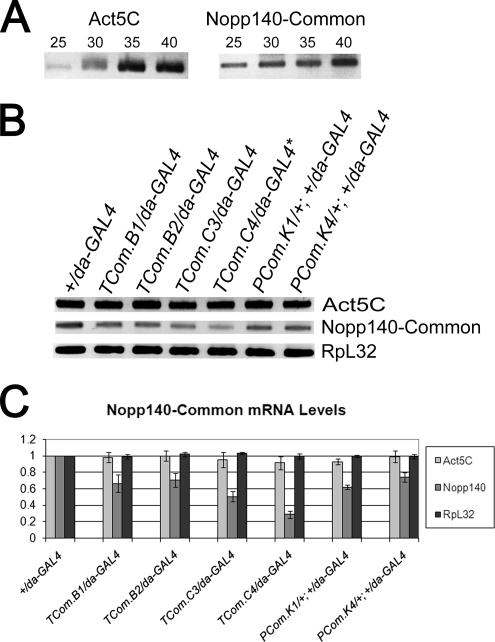
At least three independent experiments (i.e., three separate batches of larvae) were used for each described genotype. Thirty PCR cycles were used in the experiments presented in Figure 2. This number was determined empirically in separate experiments (Figure 2A) to ensure that the PCR was within the linear range of product accumulation (Figure 2A). PCR reactions for each batch of embryos were performed in triplicate. Band intensities for the RT-PCR products (Act5C, Nopp140, and RpL32) were measured using the Bio-Rad Quantity One software. These values were normalized to the respective RT-PCR products generated from control +/da-GAL4 larvae.
Figure 2.
RNAi-mediated knockdown of Nopp140 mRNAs in third instar larvae. (A) Thirty PCR cycles were used to amplify the first-strand cDNAs of actin 5C, Nopp140, and ribosomal protein L32 (not shown). Product accumulation was still within the linear range. (B) RT-PCR results showing the relative amounts of Nopp140 transcripts in nontransformed +/da-GAL4 larvae and the various larvae heterozygous for the indicated RNAi-encoding transgene and da-GAL4. (C) Band intensities for the RT-PCR products were measured using the Bio-Rad Quantity One software and normalized to +/da-GAL4. The most dramatic knockdown was observed in the TCom.C4/da-GAL4 larvae where Nopp140 mRNA levels were 30% of +/da-GAL4 levels. * TCom.C4 has two RNAi-expressing transgenes, one on the second chromosome and one on the third chromosome. Actin 5C and ribosomal protein L32 controls showed no more than a 10% variation in transcript levels for all larval progeny.
Antibody Production and Immunoblot Analysis
A chicken polyclonal antibody was prepared against the carboxy terminus of Nopp140-True. To express this peptide, the encoding cDNA segment was amplified using the forward primer, 5′-GACAAGGCCATGGACTTCACCTCCACACCT-3′ and the reverse primer, 5′-CACGGGTTTGTCGACCGCCAATTTGCA-3′. The forward primer encodes the carboxy terminus of Nopp140-True beginning at Asp586 (italicized GAC codon in the primer), whereas the reverse primer was designed to include ∼120 base pairs of the 3′ untranslated region beyond the normal stop codon used in the translation of Nopp140-True. The ends of the PCR product were trimmed with NcoI and SalI (underlined sites), and the product was ligated into the His·Tag-encoding pET30a (Novagen, Madison, WI) at the same restriction sites. We verified the open reading frame by DNA sequencing (Iowa State University, DNA Sequencing and Synthesis Facility) and then introduced the recombinant plasmid into competent BL21(DE3) E. coli cells that carried the pLysS plasmid. We used β-lactose (Sigma Chemical Co., St. Louis, MO; L-3750) at 0.1% (wt/vol) to induce fusion protein synthesis. The His-tagged peptide was enriched from E. coli sonicates using a His·Bind nickel chelation resin as described by the manufacturer (Novagen). The enriched peptide was resolved on SDS-12% polyacrylamide gels and electro-eluted. The chicken polyclonal IgY antibody was prepared against the peptide by Aves Labs (Tigard, OR).
A guinea pig antiserum was prepared against a unique peptide sequence within the carboxy terminus of Nopp140-RGG (-GRPDRSTWETNKFNGE-, residues 604–619). The peptide and polyclonal serum directed against it were prepared by Open Biosystems (Huntsville, AL).
For immunoblot analyses, adult ovaries and S2 culture cells were disrupted in SDS-sample buffer (Laemmli, 1970) using a Branson Digital Sonifier (Plainview, NY). A protease inhibitor cocktail (Sigma, P8340) was used at 1/100 dilution to help reduce proteolysis. Lysate proteins were resolved on SDS-polyacrylamide gradient (7.5–17%) gels and blotted onto nitrocellulose using the Bio-Rad semidry system (Richmond, CA). Blots were blocked with TTBS (0.9% NaCl, wt/vol, 100 mM Tris, pH 7.4, and 0.1% Tween 20) and incubated for 2 h with primary antibody. Primary antibodies included the chicken polyclonal anti-Nopp140-True diluted 1/500 in TTBS, the guinea pig anti-Nopp140-RGG serum diluted 1/100 in TTBS, or the mouse mAb G1C7 (100% hybridoma cell culture supernatant) that labels Nopp140 proteins (Cairns and McStay, 1995). The chicken antibody was detected using a peroxidase-conjugated rabbit affinity-purified anti-chicken IgG (ICN/Cappel, Irvine, CA) at 1/1000. The guinea pig antiserum and the mAb G1C7 and were detected using anti-guinea pig or anti-mouse Elite Vectastain ABC/peroxidase kits (Vector Laboratories, Burlingame, CA) according to the manufacturer's directions. Final peroxidase development used 3, 3′-diaminobenzidine (DAB) at 0.8 mg/ml, cobalt chloride at 0.4 mg/ml, and 0.1% H2O2 in 100 mM Tris, pH 7.5.
Fluorescence Microscopy
Immunofluorescence microscopy used the chicken and guinea pig antibodies described above to locate endogenous Nopp140-True or Nopp140-RGG, respectively, in control tissues and RNAi-expressing tissues. Larval and pupal tissues were dissected into Robb's minimal saline solution (Robb, 1969). Tissue squashes were prepared and fixed for immunofluorescence microscopy as described by Liu et al. (2006) except we used a final paraformaldehyde concentration of 0.5% in ∼86% ethanol for 10 min, followed by 0.5% paraformaldehyde in 1× PBS for an additional 10 min. After fixation slides were washed three times in 1× PBS, 5 min each. Squashed tissues were blocked for 30 min with 3% bovine serum albumin (BSA), 0.2% Tween 20, and 0.2% NP40 in 1× PBS, pH 7.4. The same blocking solution was used to dilute the chicken anti-Nopp140-True antibody to 1/200, and the guinea pig anti-Nopp140-RGG serum to 1/100. Primary antibodies were left on the slides for 1 h followed by three washes of 0.1% Triton X-100 in 1× PBS. An affinity-purified donkey anti-chicken IgG coupled to fluorescein isothiocyanate (ICN) or a goat anti-guinea pig IgG coupled to Alexa Fluor 546 (Molecular Probes, Eugene, OR; A-11074) were diluted to 1/250 with the same BSA blocking solution. These secondary antibodies were left on the respective slides for 30 min followed by three washes with 0.1% TX-100 in 1× PBS. The slides were stored in 1× PBS containing 0.04% sodium azide.
For GFP-Nopp140 localizations, larval and adult tissues were dissected directly into Brower's 2% paraformaldehyde solution (Brower, 1986; Blair, 2000) and allowed to fix for at least 10 min before mounting on microscope slides in a drop of the same fixative. Occasionally, tissues were stained with 4′-6-diamidino-2-phenylindole dihydrochloride (DAPI, Polysciences, Warrington, PA) (1.0 μg/ml) in the same solution before mounting. We used a Zeiss Axioskop equipped for phase contrast and fluorescence microscopy (Thornwood, NY), along with a SPOT SE camera and software for image acquisition (Diagnostic Instruments, Sterling Heights, MI).
RESULTS
RNAi Expression
Alternative splicing of the Nopp140 pre-mRNA yields two isoforms: Nopp140-True and Nopp140-RGG (Figure 1, A and B). The two proteins are identical in their first 583 amino acids, after which their carboxy termini differ significantly (Figure 1B). To reduce expression of both isoforms, we amplified a 450-base pair sequence from the Nopp140-RGG cDNA that encodes amino acids Thr52-Ser201 (amino terminal light gray bars in Figure 1B). Two copies of the 450-base pair sequence were ligated into pUAST (Brand and Perrimon, 1993) as inverted repeats with a 25-base pair spacer (Figure 1C). We also ligated two copies of the same segment in inverted orientation to either side of the intron-encoding DNA within pUASp-Nba-CS2-BgX (Zhu and Stein, 2004), a derivative of pUASp. Like pUAST, pUASp expresses transgenes from GAL4 UASs in most tissues, but it was designed to express more efficiently in the maternal germ line than does pUAST (Rorth, 1998; Duffy, 2002).
We present three separate transgenic insertions (founder embryos B, C, and K). Embryos B and C were injected with pUAST that contained the inverted 450-base pair common sequence (designated TCom.B and TCom.C), whereas embryo K was injected with the pUASp construct that contained the same inverted common 450-base pair sequence (designated PCom.K). An injected founder embryo (e.g., B) often produced several transgenic progeny. We therefore use an alpha-numeric suffix to refer to the injected embryo and its transgenic progeny line (e.g., TCom.B1 and B2) In most cases progeny from the same founder embryo shared the same insertion, but occasionally, sibling progeny contained a separate or additional insertion (see Table 1). TCom.C3, for instance, contained only one insertion at nucleotide 4806162 on chromosome arm 3R. Deep red eyes and genetic out-crossing showed that the sibling line, TCom.C4, contained two RNAi-encoding transgenes. One insertion mapped at the same site (nucleotide 4806162) on 3R, and the second insertion mapped on chromosome 2, but this site was impervious to repeated attempts to map it precisely by inverse PCR.
Table 1.
Genotypes and phenotypes of progeny trans-heterozygous for RNAi and da-GAL4 transgenes
| Genotypes of trans-heterozygous progeny X/X; 2/2; 3/3 (insertion chromosome and site of insertion) | Percent Nopp140 transcript levels | Phenotype of trans-heterozygous progeny |
|---|---|---|
| w1118/w−; +/+; +/da-GAL4 | 100 | Normal development and normal adult progeny |
| w1118/w−; +/+; TCom.B1(B2)/da-GAL4(Chromosome 3R at 107247) | ∼70 | 100% pupal eclosion; 95% of the adult progeny showed combinations of wing, leg, or tergite defects |
| w1118/w−; +/+; TCom.C3/da-GAL4 (Chromosome 3R at 4806161) | ∼50 | 100% early pupal lethality |
| w1118/w−; TCom.C4/+; TCom.C4/da-GAL4 (chromosome 2, site not determined) (chromosome 3R at 4806162) | ∼30 | Severe developmental delay; 100% third instar lethality |
| w1118/w−; PCom.K1(K4)/+; +/da-GAL4 (chromosome 2L at 13211775) | ∼65 | Severe developmental delay, 100% pupal lethality |
We present essentially four homozygous transgenic lines derived from the three founder embryos (B1/B2, C3, C4, K1/K4). For consistency, especially in the RT-PCR assays, we include sibling lines B2 (apparently identical to B1) and K4 (apparently identical to K1). There were no obvious developmental delays or morphological anomalies in the six homozygous lines. To induce RNAi expression, homozygous flies from these lines were crossed to flies homozygous for daughterless-GAL4 (da-GAL4) on chromosome 3. Due to the daughterless promoter (Caudy et al., 1988a, b), GAL4 expression in the heterozygous progeny is strong and nearly ubiquitous in all tissues and developmental stages.
RT-PCR determined the relative amounts of Nopp140 mRNA in third instar larvae that were heterozygous for the RNAi-encoding gene and da-GAL4 (Figure 2, Table 1). The period of lethality, however, differed for the various genotypic progeny. For example, lethality for TCom.C4/da-GAL4 progeny occurred late in the third larval instar, whereas lethality for TCom.C3/da-GAL4 progeny occurred in the pupal stage, and the vast majority of TCom.B2/da-GAL4 progeny eclosed as adults. To control for these various lethality periods (or lack of lethality), we isolated total RNA from early third instar larvae that still appeared healthy (mobile). We used the same first-strand cDNA synthesis to measure actin 5C (Act5C), Nopp140, and ribosomal protein L32 (RpL32) transcript levels (Figure 2B). In preliminary trials, we determined that 30 PCR cycles was within the linear range for the accumulation of PCR products (Figure 2A). The amounts of RT-PCR products from various RNAi-expressing larvae were compared with those from control +/da-GAL4 larvae at the same developmental stage. The abundance of Act5C and RpL32 transcripts did not change by more than 10% when compared with the respective transcript levels in control +/da-GAL4 larvae. We conclude that there was no significant lethality-induced degradation of mRNAs at the early third larval instar, most notably in TCom.C3/da-GAL4 and TCom.C4/da-GAL4 larvae (see below).
All six RNAi-expressing heterozygous larval genotypes showed a drop in Nopp140 transcript levels (Figure 2, B and C). Table 1 summarizes the phenotypes observed for these various progeny. TCom.B1(B2)/da-GAL4 progeny showed a 30% drop in Nopp140 mRNA levels, and they survived to adulthood, but the majority of these adults showed morphological defects in wings, legs, and cuticle (see below). TCom.C3/da-GAL4 larvae with a ∼50% drop in Nopp140 mRNA levels died as pupae, but they displayed no significant delay in advancing to the pupal stage. TCom.C4/da-GAL4 larvae with RNAi transgenes on the second and third chromosomes showed a ∼70% of loss of Nopp140 mRNA, and most died in the later third instar stage; the few individuals that advanced to the pupal stage did so with significant delays in development, but none of these pupae eclosed as adults. Finally, PCom.K1(K4)/+; +/da-GAL4 progeny with a 30–40% drop in Nopp140 transcript levels displayed significant delays in development, and most died as pupae. We conclude that survival is generally correlated with the percent loss of Nopp140 transcripts: a moderate loss of Nopp140 mRNA (∼30%) leads to adult deformities, whereas losses of ≥50% result in larval and pupal lethality.
Antibody Characterizations
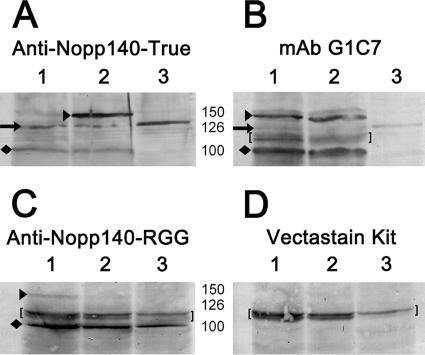
A chicken polyclonal serum was prepared against the carboxy terminus of Nopp140-True (Figure 3A). As a positive control, we tested the serum against GFP-Nopp140-True and GFP-Nopp140-RGG (tagged at their amino termini). These fusion proteins were expressed in separate transgenic lines described previously (McCain et al., 2006). We used ovary tissue from these flies because it is easy to hand isolate. We also used nontransfected S2 culture cells to help identify the endogenous Nopp140 isoforms. The chicken serum labeled the ∼150-kDa GFP-tagged Nopp140-True (black arrowhead in Figure 3A, lane 2), but not the GFP-Nopp140-RGG (Figure 3A, lane 1). The GFP tag is ∼27 kDa, thus the immunoreactive band at ∼126 kDa is likely the full-length endogenous Nopp140-True within the ovary (black arrow to the left of lane 1). A 126-kDa protein was the only band detected in S2 culture cell lysates (Figure 3A, lane 3). The faint bands just above 150 kDa observed primarily in lanes 1 and 3 could be hyperphosphorylated versions of endogenous Nopp140-True. Besides the 126-kDa band, we always observed a band at 100 kDa (diamond next to lane 1) in ovary lysates, but occasionally in S2 cell lysates. We believe this immunoreactive protein is a proteolytic product of exogenous GFP-Nopp140-True and endogenous Nopp140-True, both of which lack their amino termini.
Figure 3.
Immunoblots containing lysates of ovarian tissue and S2 culture cell were used to characterize isoform-specific antibodies. Lanes 1 and 2 in A–D contained lysates of adult ovary from transgenic flies that expressed either GFP-Nopp140-RGG (lane 1) or GFP-Nopp140-True (lane 2; see McCain et al., 2006). Lane 3 in all four panels contained lysates of nontransfected embryonic Drosophila S2 culture cells. (A) Chicken anti-Nopp140-True labeled the amino-terminal GFP-tagged Nopp140-True at ∼150 kDa (black arrowhead), and the endogenous Nopp140-True at 126 kDa (black arrow). The GFP tag is 27 kDa; thus we are confident the band at 126 kDa is the endogenous Nopp140-True isoform. The labeled band at 100 kDa (diamond) was likely a breakdown product of either the exogenous GFP-Nopp140-True or endogenous Nopp140-True, both lacking their amino termini. (B) Mouse mAb G1C7 labels Xenopus Nopp180 and nucleolin (Heine et al., 1993; Cairns and McStay, 1995). It labeled GFP-Nopp140-RGG and GFP-Nopp140-True at ∼150 kDa well, but endogenous Nopp140 at ∼126 kDa only weakly. MAb G1C7 labeled the 100 kDa band intensely. This 100-kDa band was typically prominent in ovary lysates. (C) The guinea pig anti-Nopp140-RGG serum labeled GFP-Nopp140-RGG, but not Nopp140-True. It also labeled the prominent 100-kDa band in ovary lysates and in this particular S2 cell lysate. (D) Vectastain kits were used to detect the primary antibodies in B and C. Kit reagents on their own labeled a close doublet of bands at ∼120 kDa (brackets in B–D).
We used mouse mAb G1C7 and aliquots of the same lysates used in Figure 3A to confirm the identity of bands detected by the anti-Nopp140-True serum (Figure 3B). This antibody labels a central basic domain of Xenopus Nopp180, an ortholog to Nopp140 (Cairns and McStay, 1995). MAb G1C7 also labels Xenopus nucleolin (Heine et al., 1993; Cairns and McStay, 1995). The amino terminal third of Xenopus nucleolin consists of alternating acidic and basic domains (Rankin et al., 1993; Meßmer and Dreyer, 1993) that are quite similar in sequence to those comprising the large central domain of Nopp140. According to Cairns and McStay (1995), mAb G1C7 recognizes an epitope within the first 88 amino acids of Xenopus nucleolin. This includes one acidic region with flanking basic regions. A sequence alignment between this region of Xenopus nucleolin and Drosophila Nopp140 suggests that -ATPAKATP- (residues 57–64 in Xenopus nucleolin and identical residues 402–409 in both isoforms of Drosophila Nopp140) is the common epitope recognized by mAb G1C7. As expected, mAb G1C7 labeled both GFP-Nopp140-RGG and GFP-Nopp140-True at 150 kDa (lanes 1 and 2, respectively, in Figure 3B, arrowhead next to lane 1). The endogenous Nopp140 band from the two ovary lysates and the S2 cell lysates was weakly detected at 126 kDa (arrow next to lane 1). Interestingly, mAb G1C7 labeled the 100-kDa band from the ovary lysates quite intensely (diamond next to lane 1). Because mAb G1C7 is weaker on Western blots than is the anti-Nopp140-True serum, we used a Vectastain ABC kit (Vector Laboratories) to detect G1C7 on the blots. Kit reagents, however, typically labeled a doublet of bands at ∼120 kDa (brackets in Figure 3, B and D).
We should note here that the 100- and 126-kDa proteins labeled by mAb G1C7 are not nucleolin. The closest candidate for a nucleolin homolog in Drosophila is Modulo, a nucleolar and heterochromatin protein characterized principally for its role in position effect variegation (Garzino et al., 1992; Perrin et al., 1998, 1999). Modulo has an apparent mass of 78 kDa on SDS gels (Krejci et al., 1989; Perrin et al., 1999), well below the 100–125-kDa region occupied by the endogenous Nopp140 proteins.
A guinea pig antiserum was prepared against a synthetic peptide segment within the carboxy tail of Nopp140-RGG (residues 604–619: -GRPDRSTWETNKFNGE-). We were restricted to these residues because of their predicted antigenicity and because Arg-Gly-Gly–rich domains are common to several other RNA-binding proteins. BLAST analysis indicated this peptide sequence is unique to Nopp140-RGG. GFP-Nopp140-RGG and GFP-Nopp140-True were again used to control for specificity in the Western blot analysis; the antiserum weakly labeled GFP-Nopp140-RGG but not GFP-Nopp140-True (Figure 3C, lanes 1 and 2, respectively). Of particular interest was the near absence of a 126-kDa endogenous band. Instead, the guinea pig serum strongly labeled the 100-kDa protein in the ovary lysates and in the particular S2 cell lysate used for this experiment. Although we cannot rule out specific processing events for the Nopp140 protein isoforms, especially Nopp140-RGG, we suggest that the 100-kDa immunoreactive bands are breakdown products of both Nopp140-True and Nopp140-RGG, despite the presence of proteolytic enzyme inhibitors during lysate preparation. Because anti-Nopp140-RGG labeled the 100-kDa protein almost exclusively, we suspect Nopp140-RGG is particularly unstable. Vertebrate nucleolin with a similar RGG domain is similarly unstable (Heine et al., 1993). By immunofluorescence microscopy, this guinea pig serum intensely labeled the ovarian nurse cell nucleoli (not shown), further supporting the 100-kDa band as the predominant Nopp140-RGG species present on the immunoblots. As with mAb G1C7, we used a Vectastain kit to detect the guinea pig antibody, thus the doublet at 120 kDa was again detected (brackets). From the positive controls displayed in the Figure 3, we conclude that the respective anti-sera are specific for Nopp140-True and Nopp140-RGG.
Loss of Nopp140 Protein: Immunofluorescence Microscopy
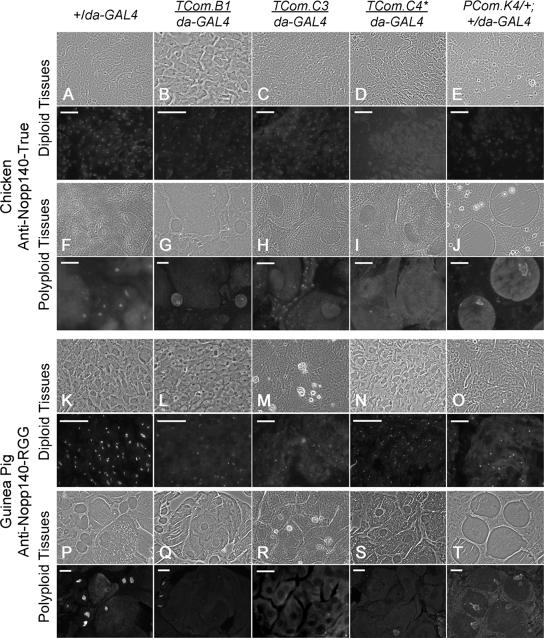
We next used the isoform-specific anti-sera and immunofluorescence microscopy to monitor the loss of endogenous Nopp140-True and Nopp140-RGG in larvae and pupae that expressed RNAi. Larval lethality likely results from RNAi expression in larval polyploid tissues, whereas adult deformities likely arise from RNAi expression in larval imaginal tissues. Immunofluorescence microscopy permitted a cell by cell examination of Nopp140 loss. Preliminary immunofluorescence observations showed that both endogenous Nopp140 isoforms appeared in all larval (e.g., Figure 4, A, F, K, and P) and adult tissues (not shown) examined to date.
Figure 4.
Immunofluorescence microscopy used the isoform-specific antibodies characterized in Figure 3 to detect endogenous Nopp140-True (A–J) and Nopp140-RGG (K–T) in control (+/da-GAL4) larvae or larvae and pupae heterozygous for an RNAi-expressing transgene and da-GAL4. In each set of figures, the immunofluorescence image is directly below the corresponding phase-contrast image. (A–E) Larval diploid imaginal (wing or leg) disk cells were probed with anti-Nopp140-True. Nucleolar staining was clearly evident in +/da-GAL4 control cells, weaker in TCom.B1/da-GAL4, TCom.C3/da-GAL4, and PCom.K4/+; +/da-GAL4 discs, but nearly absent in TCom.C4/daGAL4 disk cells. (F–J) Larval polyploid (midgut, Malpighian tubule, or salivary gland) cells stained with anti-Nopp140-True. Nucleolar staining was similar in the +/da-GAL4, TCom.B1/da-GAL4, and PCom.K4/+; +/da-GAL4 polyploid cells, but absent in the TCom.C3/da-GAL4 and TCom.C4/da-GAL4 polyploid cells. Notice that diploid imaginal island cells in the TCom.C3/da-GAL4 midgut retained nucleolar staining. (K–O) Larval imaginal diploid cells (K, L, N, and O) or pupal diploid cells (M) were probed with anti-Nopp140-RGG. Nucleoli in control +/da-GAL4 cells stained brightly, whereas nucleoli in the four RNAi-expressing lines were less intensely labeled. Most importantly, nucleoli in patches of cells failed to stain. (P–T) Larval polyploid cells (P, Q, S, and T) and pupal polyploid cells (R) were probed with anti-Nopp140-RGG. Nucleoli in control +/da-GAL4 cells labeled brightly, whereas nucleoli in polyploid cells stained weakly (PCom.K4/+; +/da-GAL4) or not at all. * TCom.C4 has two RNAi-expressing transgenes, one on the second chromosome and one on the third chromosome. Bars, 100 μm.
The chicken anti-Nopp140-True serum labeled nucleoli in all examined diploid and polyploid tissues of control +/da-GAL4 larvae (Figure 4, A and F). Conversely, TCom.B1/da-GAL4 larvae (eventual adult deformities) showed weaker nucleolar labeling in imaginal tissues (Figure 4B), whereas nucleoli in polyploid cells appeared to contain normal amounts of Nopp140-True (Figure 4G). TCom.C3/da-GAL4 larvae (eventual pupal lethality) displayed reduced labeling of Nopp140-True in their imaginal tissues (Figure 4C) and had nearly a complete loss of labeling in their polyploid nuclei, but not necessarily in their diploid nuclei (Figure 4H). TCom.C4/da-GAL4 larvae (larval lethality) showed the most significant declines of Nopp140-True in both their imaginal and polyploid tissues (Figure 4, D and I). PCom.K4/+; +/da-GAL4 larvae (delayed development with eventual pupal lethality) showed reduced labeling in their imaginal tissues (Figure 4E), but fairly normal labeling in their polyploid tissues (Figure 4J).
The guinea pig anti-Nopp140-RGG serum labeled nucleoli brightly in all diploid imaginal and polyploid tissues of control +/da-GAL4 larvae (Figure 4, K and P). Although the vast majority of imaginal cells in TCom.B1/da-GAL4 larvae contained labeled nucleoli, there were significant patches of these imaginal cells that simply failed to stain despite the presence of nucleoli (Figure 4L). A loss of Nopp140-RGG in clusters of imaginal cells may result in adult wing and leg malformations (see Figure 5). We never saw patches of unstained cells (i.e., nucleoli) in the control +/da-GAL4 tissues. In general, TCom.B1/da-GAL4 larvae displayed weaker labeling in their polyploid tissues (Figure 4Q). Based on fluorescence intensities, TCom.C3/da-GAL4 larvae (not shown) expressed nearly normal levels of Nopp140-RGG in imaginal and polyploid larval tissues. Conversely, early TCom.C3/da-GAL4 pupae displayed large patches of diploid and polyploid cells that lacked nucleolar staining (Figure 4, M and R). These animals died in the later pupal stages. TCom.C4/da-GAL4 larvae contained some Nopp140-RGG in their imaginal cells (Figure 4N), but nearly all their polyploid cells were deficient for the protein (Figure 4S); these animals died in the late third instar stage. Finally, PCom.K4/+; +/da-GAL4 larvae showed spotty labeling of Nopp140-RGG in the imaginal cells (Figure 4O); however, in general, their polyploid cells appeared to contain normal amounts (Figure 4T). These animals died in the early and late pupal stages. We conclude from these immunofluorescence studies that there is a good correlation between the severity of Nopp140-True and Nopp140-RGG loss in larval and pupal tissues and the degree and timing of larval/pupal lethality (C3, C4, K1/K4) versus adult deformities (B1/B2).
Figure 5.
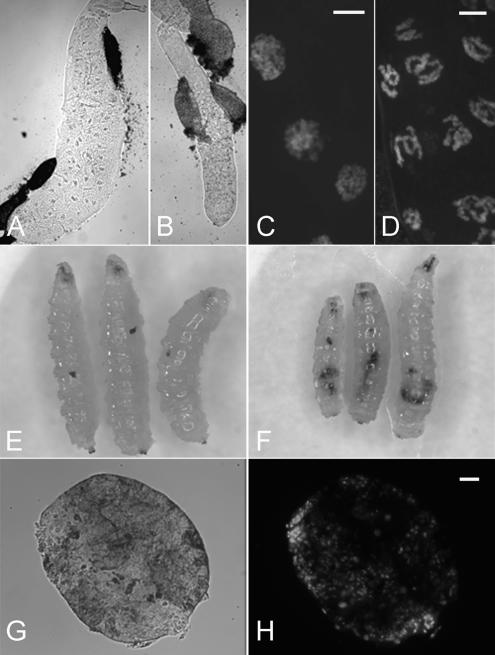
Severe phenotypes generated by RNAi-mediated depletion of Nopp140. (A) A salivary gland from a control +/da-GAL4 third instar larva. (B) Under identical magnification, the salivary gland from TCom.C4/da-GAL4 larva was about half the diameter of the normal gland. (C) DAPI-stained salivary glands from ±/da-GAL4 larvae showed chromatin distributed throughout much of the nucleus. (D) DAPI-stained salivary glands from TCom.C4/da-GAL4 larvae showed condensed chromatin and a greater density of nuclei per unit area, suggesting the cells were smaller than normal. (E and F) Melanotic tumors were observed in 5–10% of TCom.C4/da-GAL4 and PCom.K4/+; +/da-GAL4 larvae. (G) The melanotic tumors remained free of other tissues, and they were easily isolated by hand. (H) DAPI staining of the tumor showed a disorganized distribution of nuclei. Bars, 100 μm.
RNAi-induced Minute-like Phenotypes
The most interesting phenotypes (i.e., those relevant to the human TCS) were observed in TCom.B1(B2)/da-GAL4 progeny. These progeny contained ∼70% of normal Nopp140 mRNA levels (Figure 2C). There was no significant larval or pupal lethality associated with these progeny, and the vast majority eclosed as adults. Approximately 90–100% of these adults, however, displayed Minute-like phenotypes that included malformations in wings, legs, and tergites (Figure 5). Most TCom.B2/da-GAL4 flies displayed wing defects (Figure 5A) where the blade appeared crumpled, or portions of the wing margin were missing. Very similar wing defects were observed by silencing the Drosophila RpL14 gene using RNAi (Enerly et al., 2003). Although fluid-filled wing blisters can transiently occur in newly eclosed wild-type flies, their incidence and longevity were far greater in TComB2/da-GAL4 progeny. Often one of the two wings displayed a defect, whereas the other wing appeared quite normal.
Leg defects (Figure 5B) usually involved the meta-thoracic (hind) leg: many flies displayed knurled tarsi, and a few flies had severely bent femurs. Interestingly, Schultz (1929) noted that meta-thoracic legs were usually the most severely affected in Delta/Minute combinations. Similar leg defects were described for the temperature-sensitive Minute, M(3)80 (once called M(3)LS4Q-III; Sinclair et al., 1981, 1984) and for Minute M(1)7C (Andersson and Lambertsson, 1989). Disrupted tergite banding patterns, another typical Minute phenotype, were common in TCom.B2/da-GAL4 flies (Figure 5, C and D). The penetrance for these adult deformities was high (∼95% of the progeny displayed some defect) in three separate trials, but the actual defects were variable between flies; in fact most flies displayed combinations of defects. Although we expected to see the classic Minute phenotype of short and slender bristles, the bristles on most TCom.B2/da-GAL4 flies appeared normal. Occasionally we saw a fly with reduced bristle size, but in these few cases the entire thorax was underdeveloped. TCom.B1 had the same transgene insertion as TCom.B2 (Table 1), and as expected, TCom.B1/da-GAL4 adult progeny showed similar phenotypes, but interestingly, the penetrance was slightly less, with 71 and 85% in two separate trials.
TCom.C4/da-GAL4 progeny showed the most severe phenotypes. These larvae had salivary glands that were significantly smaller than those from control ±/da-GAL4 larvae (compare Figure 6, A and B). Cells within the TCom.C4/da-GAL4 salivary gland contained nucleoli, but they were not as readily discernible by phase-contrast microscopy. In addition, nuclear chromatin appeared unusually condensed (Figure 6D) compared with chromatin in control ±/da-GAL4 glands (Figure 6C). This condensed chromatin could result from the deteriorating general health of the larvae (e.g., apoptosis) or perhaps from the loss of yet undetermined extranucleolar functions of the Nopp140 isoforms. The close proximity of nuclei in the salivary glands from TCom.C4/da-GAL4 larvae suggested these cells were smaller, which would explain the reduced gland size. Associated with the larval lethality was the formation of melanotic tumors in ∼5–10% of TCom.C4/da-GAL4 and PCom.K4/+; +/da-GAL4 larvae (Figure 6, E and F, respectively). The tumors were freely floating within the larval body cavity, and we could easily isolate them (Figure 6, G and H). DAPI staining showed a rather disorganized arrangement of nuclei within the tumors (Figure 6H). This class of tumors is not believed to be neoplastic. Rather, these tumors arise from either a normal immune response to abnormal target tissues, or mutations in hematopoietic cells that then overgrow in the third larval instar (Sparrow, 1978; Watson et al., 1991). Although melanotic tumors are associated with many mutations in Drosophila (Sparrow, 1978), they have been associated with Minute mutations in the genes encoding ribosomal protein S6 (Watson et al., 1992) and Modulo (Garzino et al., 1992). Because the formation of melanotic tumors by the loss of S6 and Modulo is considered a Minute phenotype (Lambertsson, 1998), we conclude that the similar formation of tumors upon ≥50% depletion of Nopp140 transcripts is a Minute-like phenotype.
Figure 6.
RNAi-encoding TCom.B1/da-GAL4 progeny displayed a 30% drop in Nopp140 mRNA. This loss was associated with several defects that fall within the Minute class of mutations. (A) Crumpled wings, missing wing margins (arrow), or fluid-filled wing blisters (arrowhead) were prevalent defects. (B) Although less common, leg defects included bent femurs (arrow) and gnarled tarci. (C and D) Tergite deformities were common (arrows). Most flies displayed combinations of defects.
One transgenic line (PdsCom.K) expressed RNAi from UASp. PCom.K1/+; +/da-GAL4 and PCom.K4/+; +/da-GAL4 larvae (same transgene insertion) showed a 30–40% drop in Nopp140 mRNA levels (Figure 2, B and C). This reduction was slightly greater than the 30% observed for TCom.B1/da-GAL4 and TCom.B2/da-GAL4 progeny that eclosed as adults with various deformities. Instead of eclosing, however, 100% of the PCom.K1/+; +/da-GAL4 and PCom.K4/+; +/da-GAL4 progeny died as pupae after prolonged developmental delays in the larval stages. This slight difference in Nopp140 mRNA levels could theoretically explain pupal lethality in PCom.K4/+; +/da-GAL4 progeny versus leg, wing, and cuticle defects in TCom.B1/da-GAL4 adults, but a more likely explanation may be differences between promoter strength or tissue-type expression patterns between UASp versus UAST.
Overexpression of GFP-Nopp140 Isoforms Causes Lethality
From previous work (McCain et al., 2006), we knew that moderate transient expressions of both GFP-tagged isoforms from the Hsp70 promoter in embryos, larvae, or adults produced no adverse effects on development, health, or fertility. We have since established 19 separate transgenic lines that express GFP-Nopp140-True from UAST, which contains both the GAL4 UASs and the Hsp70 promoter (Figure 1C). As a basis for comparison, we heat-shocked third instar larvae homozygous for the second chromosome transgene, GFP-Nopp140-True.A9, and similar to the observations of McCain et al. (2006), we observed no adverse phenotypes. Nuclei and nucleoli in the salivary glands of these larvae appeared normal, with GFP-Nopp140-True properly localized to nucleoli (Figure 7, A–C).
Figure 7.
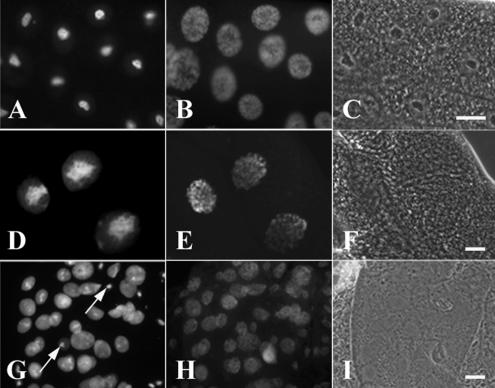
Overexpression of GFP-Nopp140-True (D–F) and GFP-Nopp140-RGG (G–I) in salivary glands leads to disrupted nucleolar (nuclear) structure and lethality. (A–C) Third instar larvae homozygous for the second chromosome insertion, pUAST-GFP-Nopp140-True.A9, were heat-shocked for 1 h and allowed to recover for 2 h. (A) Moderate expressions of GFP-Nopp140-True from the Hsp70 promoter failed to perturb nucleolar or nuclear morphology. (B) DAPI staining showed the polytenic chromatin distributed throughout the nuclei. (C) Phase-contrast microscopy showed the phase-dark nucleoli. (D–F) pUAST-GFP-Nopp140-True.A9/+; +/Sgs3-GAL4 third instar larvae overexpressed GFP-Nopp140-True in salivary glands. (D) Nucleoli appeared swollen and disorganized. (E) DAPI-stained chromatin. (F) Nucleoli were not readily discernible by phase-contrast microscopy. (G–I) pUAST-GFP-Nopp140-RGG.G4/+; +/da-GAL4 progeny overexpressed GFP-Nopp140-RGG that filled the nuclear volume. Most nucleoli appeared disrupted by fluorescence and phase-contrast microscopy. Arrows show a few nucleoli that remained normal in size. Most progeny died in the larval stages, but a few died as pupae. Bars, 100 μm
When heterozygous for da-GAL4, however, 15 of 19 UAST-GFP-Nopp140-True lines displayed larval lethality. For example, GFP-Nopp140-True.A9/+; +/da-GAL4 larvae died in the first larval instar. Examination of their salivary glands and midgut showed hypertrophied nucleoli (not shown), most likely due to the overexpression of Nopp140-True. Lethality may result from perturbed nucleolar function. To delay lethality, we used a salivary gland third chromosome GAL4 driver (Sgs3-GAL4) instead of the stronger, more ubiquitous da-GAL4 driver. As expected, the majority of GFP-Nopp140-True.A9/+; +/Sgs3-GAL4 larvae survived to the third larval instar, but their salivary gland nucleoli were again hypertrophied (Figure 7D), whereas their chromatin distribution appeared fairly normal (Figure 7E). We observed similar hypertrophied nucleoli previously when we overexpressed nucleolin in transfected mammalian CHO cells (Zhu et al., 1999). Meier (1996) further noted severe growth impairment in Saccharomyces cerevisiae when he overexpressed SRP40 (the yeast Nopp140 homologue). Finally, the third chromosome GFP-Nopp140-True.A1 line failed to show any lethality when crossed to da-GAL4. GFP was clearly expressed, but it accumulated in clusters within the cytoplasm of the Nopp140-True.A1/da-GAL4 larval salivary glands (not shown), suggesting that significant portions of the Nopp140 coding sequences were lost upon transgene integration. These progeny appeared normal, with no larval or pupal lethality. Overexpression of GFP, therefore, was not the cause of lethality observed in the other transgenic lines.
We also prepared 23 transgenic lines that overexpressed GFP-Nopp140-RGG, again from UAST. Sixteen of these lines displayed larval or pupal lethality when crossed to da-GAL4. Although overexpression of GFP-Nopp140-True caused nucleoli to swell, overexpression of GFP-Nopp140-RGG in Nopp140-RGG.G4/+; +da-GAL4 progeny disrupted nucleoli as observed in the salivary gland cells of second instar larvae (Figure 7, G–I). The arrows in Figure 7G show a few nuclei that remained morphologically normal. Most of the other nuclei in the field contained relatively large amounts of GFP-Nopp140-RGG distributed throughout the nuclear volume. Nucleoli were not readily discernible in these nuclei by either fluorescence or phase contrast microscopy. We conclude that overexpression of Nopp140-True and Nopp140-RGG disrupts nuclear integrity leading to lethality.
Mutual Rescue between RNAi and Overexpression
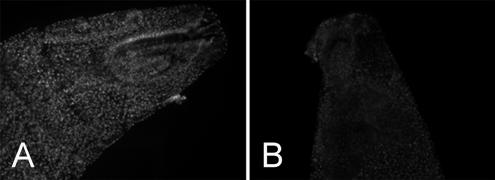
Expression of both RNAi and GFP-Nopp140-True in the same animal can partially rescue the lethality caused by expression of either transgene alone (Table 2; Figure 8). For example, overexpression of GFP-Nopp140-True in heterozygous Nopp140-True.A9/+; +/da-GAL4 larvae (Figure 8A) caused lethality in embryonic and first instar larval stages, whereas RNAi expression alone in TCom.C3/da-GAL4 progeny displayed 100% early pupal lethality. We generated double-homozygous stocks that carried the overexpressing Nopp140-True.A9 transgene on both second chromosomes and the RNAi-expressing TCom.C3 transgene on both third chromosomes. When crossed to the homozygous da-GAL4 driver, resulting heterozygous progeny survived to the pupal stage at least: in other words, there was no embryonic or first larval instar lethality due to the overexpression of GFP-Nopp140-True. We conclude from this observation that RNAi rescued the early larval lethality caused by the overexpression of GFP-Nopp140-True, but that GFP-Nopp140-True partially rescued the pupal lethality caused by RNAi. Approximately one third of these progeny eclosed as adults; some with small bodies (underdeveloped abdomens) and others with opaque wings. We conclude then that exogenous GFP-Nopp140-True partially rescued the early pupal lethality caused by RNAi expression from TCom.C3. Figure 8, A and B, shows the effects of RNAi on the exogenous expression of GFP-Nopp140-True: the GFP signal appeared significantly reduced in larvae that coexpressed RNAi.
Table 2.
Rescue between RNAi and exogenous Nopp140 expression
| Genotypes of heterozygous progeny X/X; 2/2; 3/3 | Phenotypes of heterozygous progeny |
|---|---|
| w1118/w−; +/+; TCom.C3/da-GAL4 | 100% died as early pupae |
| w1118/w−; Nopp140-True.A9/+; +/da-GAL4 | 100% died as embryos or first instar larvae |
| w1118/w−; Nopp140-True.A9/+; TCom.C3/da-GAL4 | No larval lethality. Approximately two thirds died as early and late pupae; remainder eclosed as adults with slight developmental delay, opaque wings, and small body size |
| w1118/w−; Nopp140-RGG.G4/+; +/da-GAL4 | 100% died as embryos or first and second instar larvae |
| w1118/w−; Nopp140-RGG.G4/+; TCom.C3/da-GAL4 | No larval lethality; 100% died as early and late pupae |
Figure 8.
First instar larvae expressing exogenous GFP-Nopp140 isoforms with or without the coexpression of RNAi. (A) GFP-Nopp140-True.A9/+; +/da-GAL4 larvae displayed bright nucleoli. These larvae died in the first instar. (B) GFP-Nopp140-True.A9/+; TCom.C3/da-GAL4 larvae showed diminished GFP labeling presumably from RNAi knockdown of exogenous mRNAs. These larvae survived to the pupal stage with a third of them eclosing as adults. Both images were captured using identical microscope and camera settings.
Similar to the overexpression of Nopp140-True, overexpression of GFP-Nopp140-RGG.G4 caused early larval lethality. Transheterozygous GFP-Nopp140R-GG.G4/+; TCom.3/da- GAL4 progeny survived to the pupal stage, indicating that RNAi rescued the early embryonic lethality caused by the overexpression Nopp140-RGG alone. Unlike exogenous Nopp140- True, however, exogenous Nopp140-RGG failed to rescue the RNAi-induced pupal lethality; 100% of these Nopp140-RGG.G4/+; TCom.C3/da-GAL4 pupae failed to eclose. We conclude that RNAi expressed from TCom.C3 can rescue the larval lethality caused by the overexpression of either Nopp140-True or Nopp140-RGG, and that exogenous expression of Nopp140-True can partially rescue RNAi-induced lethality.
DISCUSSION
There are over 50 known Minute mutations in Drosophila (Lambertsson, 1998; Grumbling et al., 2006). Most Minute mutations are considered haplo-insufficient, and their resulting phenotypes (see Introduction) are compatible with faulty protein synthesis. At least 13 Minute genes encode ribosomal proteins (e.g., Marygold et al., 2005), whereas others are thought to encode translation initiation factors (Kay and Jacobs-Lorena, 1987; Lambertsson, 1998; Sæbøe-Larssen et al., 1998). Uncharacterized Minute genes likely encode other ribosomal proteins or perhaps nucleolar ribosome assembly factors.
Mutations in a few Drosophila genes encoding ribosome assembly factors have been linked to Minute-like phenotypes. The first of these is minifly that encodes Nop60B (Phillips et al., 1998; Giordano et al., 1999), which is the Drosophila homologue of rat NAP57 (Nopp140-Associated Protein of 57 kDa) and of human dyskerin. Dyskerin is the nucleolar pseudouridine synthase, loss of which causes dyskeratosis congenita, a complex, multisystem (pleiotropic) disorder in humans that perturbs normal stem cell function (perhaps telomerase activity; reviewed by Meier, 2005). Complete loss of Nop60B in Drosophila is lethal, whereas a P-element insertion into minifly generated viable, but sterile adult males that display selective loss of germ line stem cells in their testes (Kauffman et al., 2003). The second Drosophila protein associated with ribosome assembly is Dribble, a predominantly nucleolar protein required for cell survival and normal pre-rRNA processing. Dribble contains a RNA-binding hnRNP K homology (KH) domain (Chan et al., 2001) and is homologous to the essential yeast KRR1p nucleolar protein required for 40S ribosome subunit assembly. Mutations in dribble lead to reductions in normal pre-rRNA processing intermediates and mature products and to the accumulation of an aberrant RNA processing intermediate. Larvae homozygous for various recessive dribble mutations arrest at the first larval instar stage (Chan et al., 2001). Finally, the Drosophila gene, pitchoune (meaning small), encodes a predominantly nucleolar DEAD-box RNA helicase. Mutations in pitchoune result in larvae that again arrest at the first larval instar stage, yet they survive for 7–10 d (Zaffran et al., 1998). Other known Drosophila genes (e.g., Nopp140, Fibrillarin, Nop56, and Nop5) and several conceptual genes encode potential ribosome assembly factors. Loss of these proteins could result in the loss of multiple nucleolar functions (Pederson, 1998; Olson, 2004) and thus to complex developmental errors.
Loss of Nopp140 Induces Minute-like Phenotypes
We used the inducible GAL4-UAS system in Drosophila to express interfering RNAs specifically designed to deplete Nopp140 transcripts. The most effective knockdown used a double-stranded RNA that targeted the common 5′ third of transcripts encoding either Nopp140-True or Nopp140-RGG. We made strong efforts to selectively target the isoform-specific 3′ coding sequences of the two Nopp140 mRNAs using the corresponding 3′-specific cDNA segments shown in gray in Figure 1B. Resulting phenotypes, however, were far weaker or nonexistent as compared with those obtained by targeting the 5′ end of the transcripts. For example, targeting the 3′ coding sequences of Nopp140-True transcripts in two separate lines (TTrue.S4/da-GAL4 and PTrue.T3/+; +/da-GAL4) displayed no lethality, and similar adult defects occurred with far less penetrance and severity than when we targeted the 5′ third of the transcripts: no more than 23% of the adult progeny showed mild tergite fusions, whereas only a few individuals (4–8%) showed slight wing defects. Targeting the 3′ coding sequences of the Nopp140-RGG transcripts produced no adverse effects; progeny from several transgenic lines when heterozygous with da-GAL4 developed normally. The isoform-specific antibodies labeled nucleoli in all tissues examined to date, thus both isoforms are coexpressed. Functional overlap between the two isoforms may explain the general failure in targeting the individual isoforms by RNAi. Finally, we were unable to measure the isoform specific mRNAs by RT-PCR: repeated efforts to amplify exon 3 by RT-PCR failed, and the RT-PCR product we obtained from the “RGG”-specific oligonucleotides simply proved to be incorrect by restriction enzyme analysis.
RNAi-induced Phenotypes
Minute-like phenotypes included larval and pupal lethality, delayed development, and adult leg, wing, and cuticle defects. Like many of the Minute mutations, loss of Nopp140 by RNAi seems to mimic haplo-insufficiency: a ≥50% loss of Nopp140 transcripts resulted in lethality, whereas a 30% loss of lead to adult deformities. Larval imaginal discs and pupal histoblast undergo rapid cell division, growth and differentiation, and thus may be particularly sensitive to reduced rates of protein synthesis. Insufficient protein synthesis, perhaps beneath a threshold (Crampton and Laski, 1994; Lambertsson, 1998), could perturb the normal development of adult wings and legs in the case of larval imaginal discs and cuticle in the case of pupal histoblasts.
Certain Minute mutations cause wings defects (Hart et al., 1993; Lambertsson, 1998) or enhance the dominant wing phenotypes of other mutations. For example, Alexander et al. (2006) showed that an EP transposition (EP 3614) into the first exon of the gene encoding ribosomal protein L13 (RpL13A) caused classic Minute phenotypes of shortened bristles and delayed development, but it also enhanced greatly the dominant negative wing phenotype of the Notch mutation, nd1. Likewise, the RpL19 Minute mutation, E(Dl)KP135, enhanced the notched wing phenotype of nd1 (Hart et al., 1993), but it also enhanced the wing phenotypes of Delta (Klein and Campos-Ortega, 1992) and the blistered wing phenotype of if3 (inflated3), a mutation of the PS2α integrin gene (Hart et al., 1993). Wing morphogenesis is dependent on Delta/Notch signaling and on integrin protein adhesion between the two wing epithelia. Loss of Notch, Delta, and integrin proteins by reduced ribosome production or function could explain the malformed wing phenotypes associated with these Minute mutations. Although we are just now pursuing this line of investigation, we predict that a loss of Nopp140 leads to reduced ribosome production or function in larval imaginal discs and pupal histoblasts and thus to the observed malformations in adults. A Nopp140 gene knockout (currently underway) should help determine how the loss of Nopp140 perturbs development, particularly in the genetic background of potential modifiers (e.g., mutations in the Minute, Delta, Notch, and integrin genes).
On the basis of an extensive study on segmental aneuploids, however, Lindsley et al. (1972) showed there are no haplo-insufficient genes in the region spanning 78A–79A of the third chromosome (Nopp140 maps to 78F4). Their results indicating that one copy of the Nopp140 gene provides sufficient product seems to be at odds with our observation that a 30% reduction in Nopp140 transcript levels yields adult deformities. Perhaps compensation in the haploid state provides sufficient transcripts by increasing transcription rates from the remaining gene and/or by enhancing the stability of the Nopp140 transcripts produced from the sole gene. RNAi would attack the transcript pool directly, thus bypassing any compensatory mechanisms at the gene level. A Nopp140 gene deletion should permit comparisons of potential haplo-gene phenotypes with those induced by RNAi.
A Potential Model the Human TCS
Leg, wing, and cuticle defects observed in adult TCom.B1/da-Gal4 progeny were reminiscent of the human craniofacial deformities associated with the human TCS. TCS results from haplo-insufficiency mutations in TCOF1 (Wise et al., 1997). Although ∼40% of the TCS cases are inherited (e.g., a family history exists), 60% of TCS cases result from spontaneous mutations that arise during gametogenesis or early development. Fifty-one mutations in the human TCOF1 gene have been documented: of these, 32 are deletions, 10 are insertions, and 7 are missense/nonsense mutations (Marszalek et al., 2002). Phenotypic expression is variable from patient to patient. Interestingly, four individuals had a defective TCOF1 gene, yet they showed no clinical manifestations of the disorder (Teber et al., 2004). Thus like many of the Minute mutations in Drosophila, TCS displays variable expressivity and incomplete penetrance. One hypothesis to explain this variability is unknown modifying factors (variable genetic backgrounds) affect phenotypic expression of TCOF1 mutations (Dixon and Dixon, 2004). What these genetic modifiers are in humans remain unknown.
The closest homologue to treacle in Drosophila is Nopp140; BLAST searches have failed to identify a TCOF1 gene in Drosophila. By pursuing the expression, functions, and interactions of Nopp140 in Drosophila, we hope to gain a better understanding of Minute-like phenotypes generated by its loss, especially in terms of penetrance and expressivity. Ultimately, we hope to test the effects of various modifiers on strains hypomorphic for Nopp140 function (gene mutation) and then extrapolate this understanding to model the penetrance and expressivity of the human TCS.
ACKNOWLEDGMENTS
We thank Dr. Craig Hart for excellent suggestions and critiques throughout the course of this work. We also thank LSU senior Colleen Craig and St. Joseph's Academy student Elisabeth Laughlin for their technical assistance. This work partially fulfills the requirements for Z.C.'s (Jennifer Cui) Ph.D. thesis in the Department of Biological Sciences, Louisiana State University, Baton Rouge. This work was supported by the National Science Foundation Grant MCB-0234245.
Abbreviations used:
- Nopp140
nucleolar phosphoprotein of 140 kilodaltons
- RNAi
RNA interference
- RGG
glycine and arginine–rich domain
- snoRNPs
small nucleolar ribonucleoprotein particles
- UASs
upstream activating sequences
- GFP
green fluorescent protein
- TCS
Treacher Collins syndrome.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0074) on March 28, 2007.
REFERENCES
- Andersson S., Lambertsson A. Characterization of a novel Minute-locus in Drosophila melanogaster: a putative ribosomal protein gene. Heredity. 1989;65:51–57. doi: 10.1038/hdy.1990.69. [DOI] [PubMed] [Google Scholar]
- Alexander S. J., Woodling N. S., Yedvobnick B. Insertional inactivation of the L13a ribosomal protein gene of Drosophila melanogaster identifies a new Minute locus. Gene. 2006;368:46–52. doi: 10.1016/j.gene.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Aris J. P., Blobel G. cDNA cloning and sequencing of human fibrillarin, a conserved nucleolar protein recognized by autoimmune antisera. Proc. Natl. Acad. Sci. USA. 1991;88:931–935. doi: 10.1073/pnas.88.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair S. Imaginal discs. In: Drosophila Protocols. In: Sullivan W., Ashburner M., Hawley R. S., editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. pp. 159–173. [Google Scholar]
- Brand A. H., Perrimon N. Targeted gene expression as a means of altering fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brower D. L. engrailed gene expression in Drosophila imaginal discs. EMBO J. 1986;5:2649–2656. doi: 10.1002/j.1460-2075.1986.tb04547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Cairns C., McStay B. Identification and cDNA cloning of a Xenopus nucleolar phosphoprotein, xNopp180, that is the homolog of the rat nucleolar protein Nopp140. J. Cell Sci. 1995;108:3339–3347. doi: 10.1242/jcs.108.10.3339. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vassin H., Brand M., Tuma R., Jan L. T., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the acheate-scute complex. Cell. 1988a;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Caudy M., Grell E. H., Dambly-Chaudiere C., Ghysen A., Jan L. Y., Jan Y. N. The maternal sex determination gene daughterless has zygotic activity necessary for the formation of peripheral neurons in Drosophila. Genes Dev. 1988b;2:843–852. doi: 10.1101/gad.2.7.843. [DOI] [PubMed] [Google Scholar]
- Chan H.Y.E., Brogna S., O'Kane C. J. Dribble, the Drosophila KRR1p homologue, is involved in rRNA processing. Mol. Biol. Cell. 2001;12:1409–1419. doi: 10.1091/mbc.12.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-K., Pai C.-Y., Huang J.-Y., Yeh N.-H. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol. Cell. Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.-M., Tsay Y.-G., Chang C.-J., Lee S.-C. Nopp140 is a mediator of the protein kinase A signaling pathway that activates the acute phase response α1-acid glycoprotein gene. J. Biol. Chem. 2002;277:39102–39111. doi: 10.1074/jbc.M205915200. [DOI] [PubMed] [Google Scholar]
- Crampton S. E., Laski F. A. string of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics. 1994;137:1039–1048. doi: 10.1093/genetics/137.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. J. Treacher Collins syndrome. Hum. Mol. Genet. 1996;5:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- Dixon J., Dixon M. J. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein treacle. Dev. Dyn. 2004;229:907–914. doi: 10.1002/dvdy.20004. [DOI] [PubMed] [Google Scholar]
- Dixon J., Jones N. C., Sandell L. L., Jayasinghe S. M., Crane J., Rey J.-P., Dixon M. J., Trainor P. A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Enerly E., Larsson J., Lambertsson A. Silencing the Drosophila ribosomal protein L14 gene using targeted RNA interference causes distinct somatic anomalies. Gene. 2003;320:41–48. doi: 10.1016/s0378-1119(03)00827-8. [DOI] [PubMed] [Google Scholar]
- Garzino V., Pereira A., Laurenti P., Graba Y., Levis R. W., Le Parco Y., Pradel J. Cell lineage-specific expression of modulo, a dose-dependent modifier of variegation in Drosophila. EMBO J. 1992;11:4471–4479. doi: 10.1002/j.1460-2075.1992.tb05548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano E., Peluso I., Senger S., Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales B., Henning D., So R. B., Dixon J., Dixon M. J., Valdez B. C. The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Hum. Mol. Genet. 2005;14:2035–2043. doi: 10.1093/hmg/ddi208. [DOI] [PubMed] [Google Scholar]
- Grumbling G., Strelets V. The FlyBase Consortium. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart K., Klein T., Wilcox M. A Minute encoding a ribosomal protein enhances wing morphogenesis mutants. Mech. Dev. 1993;43:101–110. doi: 10.1016/0925-4773(93)90028-v. [DOI] [PubMed] [Google Scholar]
- Hayano T., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., Takahashi N. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. J. Biol. Chem. 2003;278:34309–34319. doi: 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- Heine M. A., Rankin M. L., DiMario P. J. The gly/arg-rich (GAR) domain of Xenopus nucleolin facilitates in vitro nucleic acid binding and in vivo nucleolar localization. Mol. Biol. Cell. 1993;4:1189–1204. doi: 10.1091/mbc.4.11.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. M., Rehm E. J., Rubin G. M. Recovery of DNA sequences flanking P-element insertions: inverse PCR and plasmid rescue. In: Drosophila Protocols. In: Sullivan W., Ashburner M., Hawley R. S., editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. pp. 429–437. [DOI] [PubMed] [Google Scholar]
- Isaac C., Yang Y., Meier U. T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C., Marsh K. L., Paznekas W. A., Dixon J., Dixon M. J., Jabs E. W., Meier U. T. Characterization of the nucleolar gene product, Treacle, in Treacher Collins syndrome. Mol. Biol. Cell. 2000;11:3061–3071. doi: 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay M. A., Jacobs-Lorena M. Developmental genetics of ribosomal synthesis in Drosophila. Trends Genet. 1987;3:347–351. [Google Scholar]
- Kauffman T., Tran J., DiNardo S. Mutations in Nop60B, the Drosophila homolog of human Dyskeratosis congenita 1, affect the maintenance of the germ-line stem cell lineage during spermatogenesis. Dev. Biol. 2003;253:189–199. doi: 10.1016/s0012-1606(02)00013-1. [DOI] [PubMed] [Google Scholar]
- Kiehart D. P., Crawford J. M., Montague R. A. Quantitative microinjection of Drosophila embryos. In: Drosophila Protocols. In: Sullivan W., Ashburner M., Hawley R. S., editors. Cold Spring Harbor: 2000. pp. Cold Spring Harbor Laboratory Presspp. 345–359. [Google Scholar]
- Klein T., Campos-Ortega J. A. Second-site modifiers of the Delta wing phenotype in Drosophila melanogaster. Roux. Arch. Dev. Biol. 1992;202:49–60. doi: 10.1007/BF00364596. [DOI] [PubMed] [Google Scholar]
- Krejci E., Garzino V., Mary C., Bennani N., Pradel J. Modulo, a new maternally expressed Drosophila gene encodes a DNA-binding protein with distinct acidic and basic regions. Nucleic Acids Res. 1989;17:8101–8115. doi: 10.1093/nar/17.20.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The Minute genes in Drosophila and their molecular functions. Adv. Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by nucleotide sequence. Proc. Natl. Acad. Sci. USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L., et al. Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics. 1972;71:157–184. doi: 10.1093/genetics/71.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I., Girard C., Kuhn A. N., Watkins N. J., Schneider M., Bordonné R., Lührmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J. G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek B., Wojcicki P., Kobus K., Trzeciak W. Clinical features, treatment and genetic background of Treacher Collins syndrome. J. Appl. Genet. 2002;43:223–233. [PubMed] [Google Scholar]
- Marygold S. J., Coelho C.M.A., Leevers S. J. Genetic analysis of RpL38 and RpL5, two Minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics. 2005;169:683–695. doi: 10.1534/genetics.104.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCain J., Danzy L., Hamdi A., Dellafosse O., DiMario P. Tracking nucleolar dynamics with GFP-Nopp140 during Drosophila oogenesis and embryogenesis. Cell Tissue Res. 2006;323:105–115. doi: 10.1007/s00441-005-0044-9. [DOI] [PubMed] [Google Scholar]
- Meier U. T. Comparison of the rat nucleolar protein Nopp140 with its yeast homolog SRP40. J. Biol. Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Meier U. T. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U. T., Blobel G. A nuclear localization signal binding protein in the nucleolus. J. Cell Biol. 1990;111:2235–2245. doi: 10.1083/jcb.111.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U. T., Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Meßmer B., Dreyer C. Requirements for nuclear translocation and nucleolar accumulation of nucleolin of Xenopus laevis. Eur. J. Cell Biol. 1993;61:369–382. [PubMed] [Google Scholar]
- Miau L.-H., Chang C.-J., Tsai W.-H., Lee S.-C. Identification and characterization of a nucleolar phosphoprotein, Nopp140, as a transcription factor. Mol. Cell. Biol. 1997;17:230–239. doi: 10.1128/mcb.17.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O.J. Nontraditional roles of the nucleolus. In: Olson M.O.J., editor. The Nucleolus. New York: Kluwer Academic/Plenum Publishers; 2004. pp. 329–342. [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin L., Demakova O., Fanti L., Kallenbach S., Saingery S., Mal'ceva N. I., Pimpinelli S., Zhimulev I., Pradel J. Dynamics of the sub-nuclear distribution of Modulo and the regulation of position-effect variegation by nucleolus in Drosophila. J. Cell Sci. 1998;111:2753–2761. doi: 10.1242/jcs.111.18.2753. [DOI] [PubMed] [Google Scholar]
- Perrin L., Romby P., Laurenti P., Berenger H., Kallenbach S., Bourbon H.-M., Pradel J. The Drosophila modifier of variegation modulo gene product binds specific RNA sequences at the nucleolus and interacts with DNA and chromatin in a phosphorylation-dependent manner. J. Biol. Chem. 1999;274:6315–6323. doi: 10.1074/jbc.274.10.6315. [DOI] [PubMed] [Google Scholar]
- Phillips B., Billin A. N., Cadwell C., Buchholz R., Erickson C., Merriam J. R., Carbon J., Poole S. J. The Nop60B gene of Drosophila encodes an essential nucleolar protein that functions in yeast. Mol. Gen. Genet. 1998;260:20–29. doi: 10.1007/s004380050866. [DOI] [PubMed] [Google Scholar]
- Rankin M. L., Heine M. A., Xiao S., LeBlanc M. D., Nelson J. W., DiMario P. J. A complete nucleolin cDNA sequence from Xenopus laevis. Nucleic Acids Res. 1993;21:169. doi: 10.1093/nar/21.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb J. A. Maintenance of imaginal discs of Drosophila melanogaster in chemically defined media. J. Cell Biol. 1969;41:876–885. doi: 10.1083/jcb.41.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Gal4 in the Drosophila female germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Sæbøe-Larssen S., Lyamouri M., Merriam J., Oksvold M. P., Lambertsson A. Ribosomal protein insufficiency and the Minute syndrome in Drosophila: a dose-response relationship. Genetics. 1998;148:1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J. The Minute reaction in the development of Drosophila melanogaster. Genetics. 1929;14:366–419. doi: 10.1093/genetics/14.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A.R., Suzuki D. T., Grigliatti T. A. Genetic and developmental analysis of a temperature-sensitive Minute mutation of Drosophila melanogaster. Genetics. 1981;97:581–606. doi: 10.1093/genetics/97.3-4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D.A.R., Grigliatti T. A., Kaufman T. C. Effects of a temperature-sensitive Minute mutation on gene expression in Drosophila melanogaster. Genet. Res. Comb. 1984;43:257–275. [Google Scholar]
- Sparrow J. C. Melanotic “tumours”. In: Ashburner M., Wright T.R.F., editors. The Genetics and Biology of Drosophila. Vol. 2b. New York: Academic Press; 1978. pp. 277–313. [Google Scholar]
- Spradling A. C. P element-mediated transformation. In: Roberts D. B., editor. Drosophila: a Practical Approach. Oxford: IRL Press; 1986. pp. 175–197. [Google Scholar]
- Teber O. A., et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotype variation. Eur. J. Hum. Genet. 2004;12:879–890. doi: 10.1038/sj.ejhg.5201260. [DOI] [PubMed] [Google Scholar]
- The Treacher Collins Syndrome Collaboration Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nature. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Valdez B. C., Henning D., So R. B., Dixon J., Dixon M. J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggener J. M., DiMario P. J. Two splice variants of Nopp140 in Drosophila melanogaster. Mol. Biol. Cell. 2002;13:362–381. doi: 10.1091/mbc.01-04-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Query C. C., Meier U. T. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 2002;22:8457–8466. doi: 10.1128/MCB.22.24.8457-8466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson K. L., Johnson T. K., Denell R. E. Lethal(1)aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev. Genet. 1991;12:173–178. doi: 10.1002/dvg.1020120302. [DOI] [PubMed] [Google Scholar]
- Watson K. L., Konrad K. D., Woods D. F., Bryant P. J. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoitic system. Proc. Natl. Acad. Sci. USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise C. A., Chiang L. C., Paznekas W. A., Sharma M., Musy M. M., Ashley J. A., Lovett M., Jabs E. W. TCOF1 gene encodes a putative nucleolar phosphoprotein that exhibits mutations in Treacher Collins Syndrome throughout its coding region. Proc. Natl. Acad. Sci. USA. 1997;94:3110–3115. doi: 10.1073/pnas.94.7.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Isaac C., Wang C., Dragon F., Pogačić V., Meier U. T. Conserved composition of mammalian box H/ACA and box C/D/ small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell. 2000;11:567–577. doi: 10.1091/mbc.11.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Chartier A., Gallant P., Astier M., Arquier N., Doherty D., Gratecos D., Sémériva M. A Drosophila RNA helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development. 1998;125:3571–3584. doi: 10.1242/dev.125.18.3571. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Liu D., DiMario P. Nucleolin, defective for MPF phosphorylation, localizes normally during mitosis and nucleologenesis. Histochem. Cell Biol. 1999;111:477–487. doi: 10.1007/s004180050384. [DOI] [PubMed] [Google Scholar]
- Zhu X., Stein D. RNAi-mediated inhibition of gene function in the follicle cell layer of the Drosophila ovary. Genesis. 2004;40:101–108. doi: 10.1002/gene.20070. [DOI] [PubMed] [Google Scholar]