Abstract
The [URE3] and [PSI+] prions are infectious amyloid forms of Ure2p and Sup35p. Several chaperones influence prion propagation: Hsp104p overproduction destabilizes [PSI+], whereas [URE3] is sensitive to excess of Ssa1p or Ydj1p. Here, we show that overproduction of the chaperone, Sse1p, can efficiently cure [URE3]. Sse1p and Fes1p are nucleotide exchange factors for Ssa1p. Interestingly, deletion of either SSE1 or FES1 completely blocked [URE3] propagation. In addition, deletion of SSE1 also interfered with [PSI+] propagation.
INTRODUCTION
The yeast nonchromosomal genes, [URE3], [PSI+], and [PIN+], are infectious proteins (prions) of Ure2p, Sup35p, and Rnq1p, respectively (Wickner, 1994; Derkatch et al., 2001). Each is a self-propagating amyloid form of the corresponding protein (Paushkin et al., 1996; Glover et al., 1997; King et al., 1997; Taylor et al., 1999; Sondheimer and Lindquist, 2000; Derkatch et al., 2001), and thus these systems bring the facile yeast molecular genetic tools to bear on the broader problem of amyloidoses.
Chaperones of the Hsp104, Hsp70, and Hsp40 groups are critical for propagation of [PSI+], [URE3], and [PIN+] (Chernoff et al., 1995; Newnam et al., 1999; Jung et al., 2000; Moriyama et al., 2000; Jones et al., 2004). Hsp104 can disaggregate denatured proteins aided by Hsp70 and Hsp40 (Glover and Lindquist, 1998), and Hsp104 probably breaks up large filaments to make smaller seeds (Eaglestone et al., 2000; Kryndushkin et al., 2003).
Mutation of the cytoplasmic Hsp70, Ssa1p, destabilizes the [PSI+] prion (Jung et al., 2000), and Ssa1p overproduction inhibits curing of [PSI+] by overproduced Hsp104 (Newnam et al., 1999). Overproduction of Ssa1p (but not the nearly identical Ssa2p) cures [URE3] (but not [PSI+]; Schwimmer and Masison, 2002), whereas mutation or deletion of SSA2 (but not SSA1) destabilizes [URE3] (Roberts et al., 2004). Overproduction of the Hsp40, Ydj1p, also cures [URE3] (Moriyama et al., 2000), whereas deletion of the G/F-rich domain of Sis1p cures [PIN+] (Sondheimer et al., 2001).
Hsp70s (including the cytoplasmic Ssa1 to Ssa4, Ssb1 and Ssb2) bind hydrophobic regions of proteins (“holdase” activity), preventing their aggregation and promoting their refolding (reviewed by Mayer and Bukau, 2005). Various cochaperones, such as Hsp40s (including Ydj1p, Sis1p), bind to both specific substrates and to Hsp70s bringing the substrates to be acted on by Hsp70s. Hsp70 action is mediated by ATP binding, hydrolysis and release: the Hsp·ATP complex is in an open conformation that rapidly binds and releases substrate, whereas Hsp·ADP is closed and binds tightly to substrate. Nucleotide exchange factors, such as Fes1p and Sse1p, promote the release of ADP by Hsp70, allowing ATP to bind, and thus affect the balance of binding, release, and refolding activities of Hsp70s. Sti1p, which interacts with both Hsp90s and Hsp70s, also stimulates the Ssa1p ATPase.
The key role of Hsp104 in prion propagation was discovered by screening for proteins whose overproduction cured [PSI+] (Chernoff et al., 1995). A similar screen using a hybrid of the Pichia methanolica Sup35 prion domain and the Saccharomyces cerevisiae Sup35 functional domain (Sup35PS) identified curing activity of several chaperones (Ydj1, Sis1, Sti1) as well as several factors that regulate chaperone activity at the transcriptional level (Kryndushkin et al., 2002). Here, we performed a similar screen on [URE3].
MATERIALS AND METHODS
Strains, Genetic Methods, and Plasmids
Strains used are listed in Table 1. Standard rich (YPD) or synthetic (SC) yeast media were used (Sherman, 1991). Adenine-poor medium (1/2 YPD) contains half the normal amount of yeast extract and was used for the red-white assay of ADE2 expression (Cox, 1965). Strains BY241 (used in the screening) and BY251 contain the ADE2 gene under control of the DAL5 promoter (Brachmann et al., 2005), allowing detection of the prion state of Ure2p and even different [URE3] prion variants by colony color. Three different [URE3] variants were characterized in those strains (Brachmann et al., 2005): v1, v2, and v3; BY241 v1, the same as [URE3-1] (Lacroute, 1971), was used in the screening. To measure [URE3] loss under chaperone overproduction, ∼30 yeast colonies from a transformation plate were inoculated in liquid YPD media and grown overnight to allow plasmid loss (which was ∼90% complete), and ∼104 cells were spread on a YPD plate. The ratio of red to white colonies was scored.
Table 1.
Yeast strains
| Strain name | Genotype | Reference |
|---|---|---|
| BY241 | MATa, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1,[URE3-1] | Brachmann et al. (2005) |
| BY241-HA | MATa, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1, URE2::URE2-HA | This study |
| BY241 ΔSSE1 | MATa, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1, sse1Δ::kanMX | This study |
| BY241 ΔFES1 | MATa, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1, fes1Δ::kanMX | This study |
| BY241 ΔSTI1 | MATa, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1, sti1Δ::kanMX | This study |
| BY251 | MATα, leu2, trp1, ura3, PDAL5:ADE2, PDAL5:CAN1, kar1,[URE3-1] | Brachmann et al. (2005) |
| 5V-H19 | MATa, ade2, SUQ5, leu2, ura3, [PSI+] | TerAvanesyan et al. (1994) |
| 5V-H19 ΔSSE1 | MATa, ade2, SUQ5, leu2, ura3, sse1Δ::kanMX | This study |
| 5V-H19 ΔFES1 | MATa, ade2, SUQ5, leu2, ura3, fes1Δ::kanMX | This study |
| 5V-H19-PS | MATa, ade2, SUQ5, leu2, ura3, SUP35::SUP35-PS,[PSI+PS]#1 | Kushnirov et al. (2000) |
| 74D-694 | MATa, ade1, leu2, trp1, ura3, his3, [PSI+] | Chernoff et al. (1995) |
| 74D-694 ΔSSE1 | MATa, ade1, leu2, trp1, ura3, his3, sse1Δ::kanMX | This study |
| 74D-694 ΔFES1 | MATa, ade1, leu2, trp1, ura3, his3, fes1Δ::kanMX | This study |
| 62δ-1DC | MATα, ade2, SUQ5, trp1, lys2, can1, kar1, [psi-] | D. Masison |
For [PSI+] experiments, strain 5V-H19 with strong or weak prion variants (Kryndushkin et al., 2003) or 74D-694 [PSI+] (Chernoff et al., 1995) was used. To detect the [PSI+] phenotype, strain 5V-H19 includes the ade2-1 UAA nonsense mutation and the SUQ5 tRNA suppressor, which suppresses ade2-1 only in combination with [PSI+]. [psi−] cells are Ade− and accumulate a red pigment related to impaired adenine biosynthesis. [PSI+] cells are Ade+ and form white (“strong” [PSI+] variants) or pink (“weak” variants) colonies. To enhance colony color development, only one third (6 mg/l) of normal adenine concentration was used in transformation plates. 5V-H19-PS, obtained from 5V-H19 by replacing SUP35 with the chimeric SUP35-PS allele encoding amino acids 1-186 of Sup35 of the yeast P. methanolica instead of amino acids 1-123 of Sup35 of S. cerevisiae, was used with the strong prion variant [PSI+ PS]#1. Four millimolar GuHCl was used to cure either [URE3] or [PSI+].
Plasmids and DNA Manipulations
Plasmids used are listed in Table 2. The S. cerevisiae genomic library (catalogue no. 37323) was purchased from American Type Culture Collection (Manassas, VA). Individual genes were tested for [URE3] curing by cloning into pRS425-based plasmids, using the Gateway Technology system (Invitrogen, Carlsbad, CA). The AttR1+CmR+ccdB+AttR2 block of pYES-Dest52 (Invitrogen) was amplified by PCR using primers p11 and p12 and transferred into pRS425 using XhoI and HindIII sites to produce a new destination vector (pRS425-Dest). Corresponding genes (SSE1, FES1, and YDJ1) were amplified from plasmids (genomic inserts in the case of SSE1 and YDJ1) or yeast genomic DNA (for FES1) using proofreading polymerase Pfx (Invitrogen) and primers p13 and p8 for SSE1, p14 and p9 for FES1, and p15 and p16 for YDJ1. Then these genes were inserted first by TOPO cloning into pENTR/D-TOPO vector (Invitrogen) containing AttL1,2 sites and then by recombination between AttL and AttR sites into pRS425-Dest vector using Clonase II Enzyme Mix (Invitrogen). Insertions were verified by sequencing. Sse1 K69M and G233D mutations were inserted via quick-change PCR, using p17 and p18 (for K69M) and p19 and p20 (for G233D) primers. Then mutated genes were inserted into pRS425-Dest similar to wild-type SSE1.
Table 2.
Plasmids
| Plasmid name | Markers | Reference |
|---|---|---|
| pRS425-SSE1 | LEU2, episomal | This study |
| pRS425-SSE1 K69M | LEU2, episomal | This study |
| pRS425-SSE1 G233D | LEU2, episomal | This study |
| pRS425-FES1 | LEU2, episomal | This study |
| pRS425-YDJ1 | LEU2, episomal | This study |
| Yeplac195-SIS1 | URA3, episomal | Kryndushkin et al. (2002) |
| Yeplac195-STI1 | URA3, episomal | Kryndushkin et al. (2002) |
| Yeplac195-SSB1 | URA3, episomal | Kryndushkin et al. (2002) |
| pFL44-HSP104 | URA3, episomal | Kryndushkin et al. (2002) |
| pH400-SSA1 | TRP1, episomal | This study |
| pH125-SSA1 | LEU2, episomal | This study |
URA3-based plasmids Yeplac195-SIS1, Yeplac195-STI1, Yeplac195-SSB1, and pFL44-HSP104 were kindly provided by M. Ter-Avanesyan (Kryndushkin et al., 2002). pH125 and pH400 were a kind gift of H. Edskes (Edskes and Wickner, 2000). pH125 is a pRS425-based episomal vector with inserted ADH1 cassette, containing ADH1 promoter and terminator, divided by multiple cloning sites. pH400 is the same as pH125 but contains TRP1 instead of LEU2. SSA1 was amplified from yeast genomic DNA using primers p21 and p22, containing BamHI and HindIII restriction sites for cloning. After overnight digestion BamHI-HindIII fragment was cloned into BamHI and HindIII linearized pH125 and pH400, resulted in pH125-SSA1 and pH400-SSA1 plasmids.
SSE1, FES1, and STI1 disruption cassettes were obtained by amplifying yeast genomic DNA of corresponding strains from the S. cerevisiae knockout collection (Winzeler et al., 1999) using primers: p1 and p2 for SSE1, p3 and p4 for FES1, and p5 and p6 for STI1 (see Table 3). Disrupted mutants were then obtained by transforming the resulting PCR fragment into yeast and selecting for G418-resistant colonies at a final concentration of 0.5 g/l. In each case disruption was confirmed with additional primers, one primer outside the cassette (p8 for SSE1, p9 for FES1, and p10 for STI1) and another inside the KanMX gene (p7 for all). The phenotypes (see Results) of each disruption were analyzed in three independent clones.
Table 3.
Primers
| Primer name | Primer sequence, 5′–nn–3′ |
|---|---|
| p1 | gcataatcatttgttacgtg |
| p2 | ATACGCGCTGGCATGTCCC |
| p3 | cagtgtactaacctatttcc |
| p4 | CTATTGAATGATGATCGAACG |
| p5 | caaactattgaactaaacgc |
| p6 | GTAAAGTTGTGCCAATTACG |
| p7 | tgattttgatgacgagcgtaat |
| p8 | GGTAGACTTTATGGATGGAG |
| p9 | CTGTTTACTTGTACTGATCC |
| p10 | ATGGAGCTTAGATATTCTCG |
| p11 | gttCTCGAGgctatcaaacaagtttgtac |
| p12 | CACaagcttATCGAACCACTTTGTACAAG |
| p13 | CACCaactagtataactcccatc |
| p14 | CACCaatgatgaaatgttatccc |
| p15 | CACCtgagttgtgtgtattcaac |
| p16 | ACAAAAAATGGTTTTCTGTGG |
| p17 | gccaacttgaTGagaattattggtttggattacc |
| p18 | GGTAATCCAAACCAATAATTCTCATCAAGTTGGC |
| p19 | ctgcgacaagcattttgAtggtagggacttcgatttgg |
| p20 | CCAAATCGAAGTCCCTACCAtCAAAATGCTTGTCGCAG |
| p21 | CAG GGATCC atgtcaaaagctgtcgg |
| p22 | GCA AAGCTT TTAATCAACTTCTTCAACGG |
For biochemical experiments the BY241-HA strain had URE2 with three C-terminal tandem hemagglutinin (HA) tags made by homologous recombination (Longtine et al., 1998), and the resulting Ure2-3HA was functional. [URE3] was selected on−ade medium and checked by curing with 4 mM GuHCl. This [URE3] was affected by overexpression of SSE1 or YDJ1 with efficiency similar to that of [URE3-1] in wild-type BY241.
Analysis of Yeast Cell Lysates and Electrophoresis.
Strain BY241-HA was grown in liquid media to an OD600 of 1.5. The cells were harvested, washed in buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol, and Complete protease inhibitor mixture; Roche Applied Science, Indianapolis, IN) and lysed by glass beads in the same buffer. Cell debris was removed by centrifugation at 10,000 × g for 10 min. Protein was measured with BCA reagent (Pierce, Rockford, IL). To determine the size distribution of Ure2p, the cell lysates (0.4 ml of 5 mg/ml) were fractionated by centrifugation through a 30% sucrose pad (0.5 ml) in the SW55 rotor (Beckman, Fullerton, CA) for 40 min at 43,000 rpm. Ure2-HA protein was detected by the anti-HA antibody (Roche Applied Science).
Agarose Gel Electrophoresis.
Agarose gel electrophoresis of prion aggregates of Ure2p was adopted from previously published procedures for Sup35p (Kryndushkin et al., 2003) or Rnq1p (Bagriantsev et al., 2006) prion polymers. Briefly, cell lysates of BY241-HA [URE3] were obtained as described above. Ure2p prion aggregates were sedimented through a 30% sucrose pad (0.5 ml) in the SW55 rotor (Beckman) for 40 min at 43,000 rpm. Resulted pellets were treated with 2% of SDS for 10 min at 37°C (prion polymers are resistant to such treatment in contrast to most cellular protein–protein interactions as shown in Kryndushkin et al., 2003) and were separated by horizontal 1.5% agarose gel electrophoresis in LB buffer with 0.1% SDS. As molecular weight markers, a preparation of chicken pectoralis extract containing abundant giant proteins titin (3 MDa) and nebulin (0.8 MDa) was used. After the electrophoresis, proteins were transferred from gels to Immobilon-P PVDF sheets (Millipore, Bedford, MA) by electric transfer unit (Invitrogen) for 1 h, followed by immunostaining. Because the polyclonal antibody obtained previously against recombinant Ure2p (Wickner, 1994) reacts poorly with aggregated Ure2p (Speransky et al., 2001), strain BY241-HA [URE3] was used. Ure2-HA protein was detected by the anti-HA antibody (Roche Applied Science).
RESULTS
A Screen for Prion-eliminating Factors
Strain BY241 carries ADE2 under control of the DAL5 promoter to monitor activity of Ure2p (Brachmann et al., 2005). Active Ure2p makes such a strain Ade− and red on adenine-limiting medium, but prion inactivation of Ure2p allows ADE2 transcription, resulting in pink or white colonies according to the level of transcription. We used the mitotically stable [URE3-1] prion variant (Lacroute, 1971) that results in a white colony phenotype. Strain BY241 [URE3-1] was transformed with a S. cerevisiae genomic library based on the multicopy plasmid YEp13. Red, pink, or red-sectored transformant colonies have lost [URE3], most spontaneously (∼1 per 1000 transformants). But in some cases the red colony color was due to the plasmid, either curing [URE3-1] or slowing prion conversion, causing increased levels of soluble Ure2p without eliminating the prion.
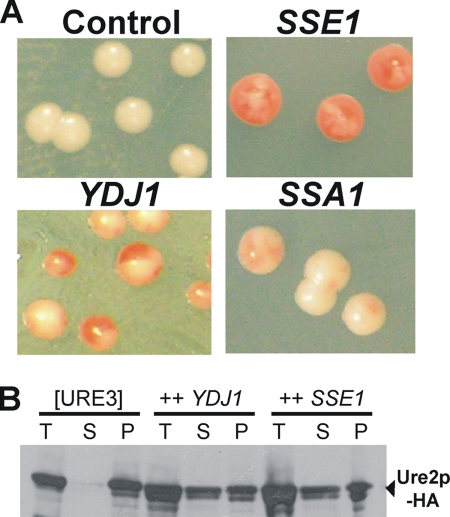
Among ∼150,000 transformants, ∼200 red “positives” were tested for spontaneous prion loss by crossing with strain BY251 carrying the same [URE3-1]. Plasmids isolated from transformants that were red or pink both as haploids and after mating with BY251 were transformed again into BY241 [URE3-1] to confirm the prion-curing effect. Confirmed plasmids were sequenced, and individual genes were tested again. Two proteins that cured [URE3-1] very efficiently were Ydj1p and Sse1p (Figure 1A, Table 4). Also, the lack of prion phenotype after the spontaneous loss of the plasmids, confirmed that [URE3-1] was indeed cured, ruling out mere suppression of the phenotype.
Figure 1.
Overproduced chaperones cure [URE3]. (A) Transformants of BY241 [URE3-1] with multicopy plasmids carrying the indicated genes. Control, pRS425 vector. (B) Centrifugation analysis of BY241-HA [URE3]. T, total lysate; S, supernatant fraction; P, pellet fraction. Fractions were analyzed by Western blot with anti-HA antibody.
Table 4.
Relative prion loss after chaperone overproduction
| Overproduced proteins | None | Ydj1p | Sse1p | Ssa1p |
|---|---|---|---|---|
| Relative [URE3] v1 loss (%) | 1 | 55 | 85 | 20 |
| Relative [PSI+PS]#1 loss (%) | 1 | 40 | 12 | 20 |
Data shown are the average of three experiments, with a variation of about ±10%.
Overproduction of either Ydj1p or Ssa1p is known to impair [URE3] propagation (Moriyama et al., 2000; Schwimmer and Masison, 2002), but the effect of Ssa1p is much smaller at least in our genetic background (Figure 1A, Table 1), so we did not identify it in the screening.
Sse1p is an Hsp110 family chaperone (Mukai et al., 1993), with 35% identity to Ssa1p and overall structure similarity, including an ATPase domain. The exact function of Sse1p in protein folding in vivo is not yet clear. Besides its “holdase” activity, Sse1p was recently found to be a nucleotide exchange factor (NEF) for yeast Hsp70 members Ssa1p and Ssb1p (Dragovic et al., 2006; Raviol et al., 2006), facilitating the release of a substrate from Hsp70.
We further tested overproduction of Ydj1p and Sse1p on [PSI+] and on [PSI+PS], based on a Pichia-Saccharomyces (Sup35-PS) hybrid. [PSI+PS] variant 1, in contrast to [PSI+], is cured by Ydj1p overexpression (Kryndushkin et al., 2002; Table 4). Overexpression of Sse1p slightly destabilized [PSI+PS]#1, but had no effect on [PSI+] (Table 4).
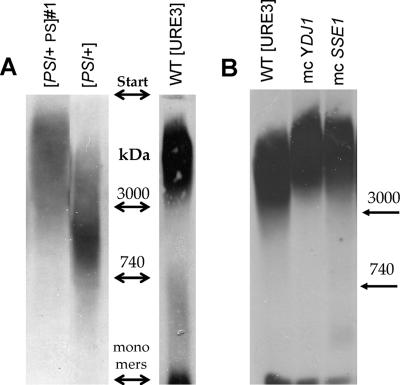
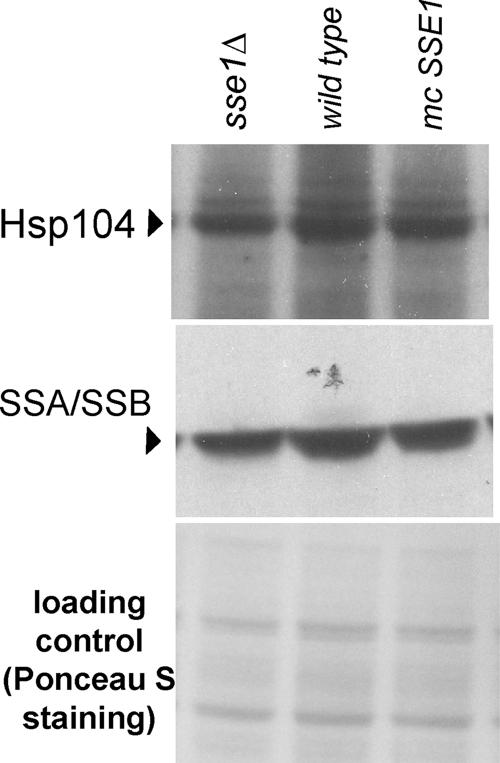
Ydj1p and Sse1p also showed high curing potency for two other [URE3] variants (BY241 v2 and v3; Brachmann et al., 2005) and in different host strains (BY251, BY241-HA [URE3] de novo induced). Supporting the genetic results, overproduction of either Ydj1 or Sse1 caused increased solubility of Ure2p (Figure 1B) and increased the length of Ure2p prion polymers (Figure 2B). Further, overexpression of Sse1p does not significantly change levels of Hsp104 or Ssa proteins (Figure 3).
Figure 2.
Agarose gel electrophoresis of prion polymers. Particulate fractions from sucrose pad centrifugation analyzed on 1.5% agarose gels and blotted to nitrocellulose. Immunostaining was with anti-Sup35 or anti-HA antibody. (A) Size comparison of [PSI+PS], [PSI+], and [URE3]. (B) Size differences of [URE3] prion polymers for wild-type cells and cells overproducing (mc) either Ydj1p or Sse1p.
Figure 3.
Levels of Hsp104p or Ssa proteins in BY241 [URE3-1] wild type, sse1Δ, and with overproduced (mc) Sse1p. Immunodetection was with anti-Hsp104 antibody (StressGen, San Diego, CA) or anti-Hsp70 (StressGen).
Sse1p ATPase Activity Is Needed To Cure [URE3]
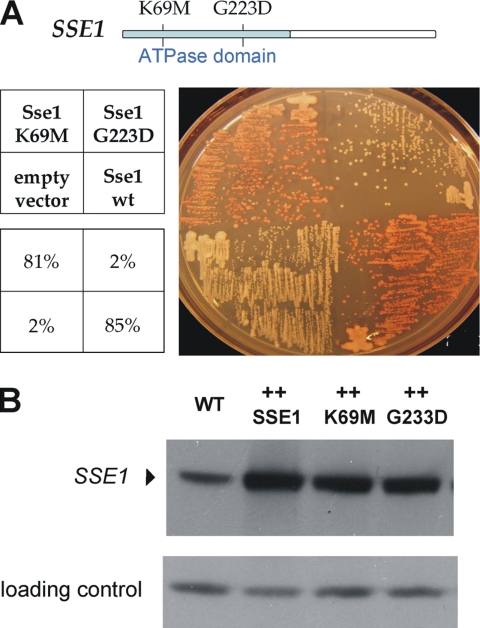
Because Sse1p is homologous to Ssa1p, it might act on [URE3] as a chaperone by direct interaction with soluble or filamentous Ure2p or some intermediate. Alternatively, Sse1p might act as a NEF for Ssa1p, affecting [URE3] indirectly. To distinguish between these two possibilities, we generated two point mutations within the ATPase domain of Sse1p. Sse1-G233D is known to reduce both ATP binding and hydrolysis and results in the loss of interaction and NEF function with Ssa1p (Shaner et al., 2004, 2005; Dragovic et al., 2006). In contrast, the Sse1p-K69M mutant can bind ATP (hydrolysis is defective) and retains both Ssa1p binding and NEF activity (Raviol et al., 2006). We found that overproduction of Sse1p-K69M efficiently cured [URE3-1], but Sse1p-G233D did not (Figure 4A). Because ATPase activity (not merely ATP binding) is generally essential for chaperone activity of Hsp70s, this suggests that Sse1p is not affecting [URE3] by a direct chaperone activity.
Figure 4.
Effect of Sse1 ATPase domain point mutations. (A) Strain BY241 [URE3-1] with overproducted wild-type Sse1p, Sse1-K69Q, Sse1-G233D, or vector streaked on ½ YPD (figures indicate relative prion loss). (B) Levels of normal and mutant Sse1p overproduction, measured with anti-Sse1 antibody and compared with vector alone (WT). The loading control is a nonspecific band recognized by anti-Sse1 antiserum, and the results were confirmed by Ponceau S staining.
Deletions of Either SSE1 or FES1 Disrupt [URE3] Propagation
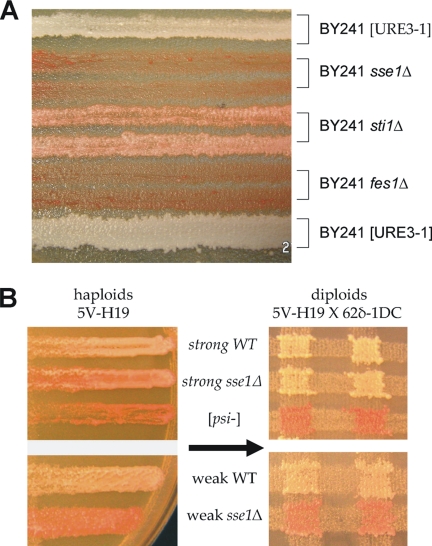
Because Sse1p apparently affects [URE3] stability via regulation of Ssa1p, we examined whether Fes1p, the other known NEF for Ssa1p (Kabani et al., 2002), affects [URE3]. Fes1p promotes ADP release upon binding to Ssa1p and may inhibit the ATPase activity of Ssa1p (Kabani et al., 2002). Several other Ssa1p cofactors (Ydj1p, Sis1p, Sti1p, Cpr7p, and others) stimulate ATPase activity of Ssa1p, pushing the reaction in the opposite direction (see Figure 5A). Unlike Ydj1p and Sse1p, overexpression of Sis1p, Sti1p, or Fes1p does not produce significant loss of [URE3-1] (data not shown). However, we found complete loss of prion propagation after deletion of SSE1 or FES1, and partial destabilization of prion propagation after deletion of STI1 (Figure 6A). The loss of [URE3] by sse1Δ was confirmed by mating BY241 sse1Δ with BY251 [URE3-1] and tetrad analysis: all sse1Δ spores had lost [URE3], but many SSE1 spores remained [URE3]. Similar data were obtained for BY241 fes1Δ× BY251.
Figure 5.
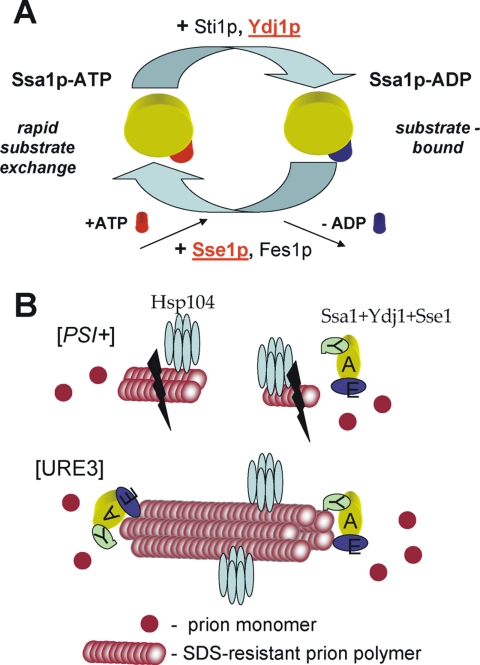
Schematic models. (A) Functional cycle of Ssa1p ATPase. Different cofactors either stimulate ATPase activity of Ssa1p or stimulate ADP exchange. (B) Possible difference between [PSI+] and [URE3] prion propagation: because of the relatively shorter polymers, [PSI+] is more sensitive to Hsp104 action; instead, [URE3] is more sensitive to the Ssa1/Ydj1/Sse1 complex.
Figure 6.
Deletion of either SSE1 or FES1 disrupt [URE3] propagation. (A) BY241 [URE3-1] deleted for the indicated genes streaked on ½ YPD. Red color indicates loss of [URE3]. (B) Strain 5V-H19 with [PSI+]strong or [PSI+]weak, sse1Δ derivatives and a guanidine-cured derivative and diploids with the wild-type [psi-] strain 62δ-1DC were streaked on ½ YPD. White color indicates the presence of [PSI+].
Deletion of SSE1 Can Also Affect [PSI+] Propagation
Using two independent strains (5V-H19 and 74D-694) and three different prion variants, deletion of SSE1 weakened the [PSI+] phenotype in all cases, but deletion of FES1 did not (although Jones et al., 2004 did observe both weakening and instability of [PSI+] in fes1Δ). Prion variant 335 (strong) in strain 5V-H19 sse1Δ showed white diploids after crossing with a [psi−] SSE1 partner, showing that the sse1Δ mutation only masked the [PSI+] phenotype, but variant 1111 (weak) in host strain 5V-H19 sse1Δ produced all red diploids, indicating complete prion loss (Figure 6B). Thus, the requirement for Sse1p is both prion and prion variant specific.
DISCUSSION
Sse1p Effects: Direct or Indirect?
Either the overexpression or depletion of Sse1p disrupts propagation of [URE3]. Our mutant results indicate that ATP binding, but not the ATPase of Sse1p are important for its prion-curing effects. Because the ATPase activity should be necessary for any Hsp70-like chaperone activity, this result suggests that prion curing is mediated by its association with and action on the Ssa proteins. Surprisingly, the ATPase-dead Sse1-K69Q mutant fully complements all phenotypes of an sse1Δ strain (Shaner et al., 2004), suggesting that many Sse1p functions are independent of its ATPase activity.
Optimal Ssa activity is required for stable propagation of [URE3], with overexpression of Ssa1p (Schwimmer and Masison, 2002) or deletion of SSA2 (Roberts et al., 2004) effectively curing [URE3]. We find that alteration of different cofactors can also efficiently disrupt [URE3] propagation. Sse1p also cooperates with Ssa1p in renaturing luciferase in vivo and in vitro (Dragovic et al., 2006), with either excess or deficient Sse1p impairing activity.
Although mutations of SSE1 suggest an indirect action through Ssa1p, differences in effects of NEFs (overproduction of Fes1p did not cure) might indicate an additional action for Sse1p. Masison's group showed that overexpression of Fes1p (Jones et al., 2004) and Sse1p (D. Masison, personal communication improved [PSI+] stability in an SSA1-21 background, whereas deletion of FES1 further destabilized it; overexpression and deletion of STI1 did the respective opposites. Their results imply that increasing the substrate-bound ADP form of Ssa1p impaired [PSI+] propagation, whereas the ATP form promotes [PSI+] (Jones et al., 2004). [URE3] appears to be more complex in that alteration of Ssa1 activity in either direction can damage prion propagation.
Interestingly, both NEFs for Ssa1p are required for [URE3]: independent deletions of either SSE1 or FES1 completely disrupted [URE3] propagation. Fes1p binds preferentially to Ssa1p·ADP, stimulating ADP release and inhibiting the ATPase activity of Ssa1p (Kabani et al., 2002). In contrast Ydj1p stimulates the Ssa1p ATPase. fes1Δ and ydj1-151ts suppress each other, suggesting antagonism (Kabani et al., 2002). Yjd1p overproduction cures [URE3] as does fes1Δ, consistent with this idea. Although fes1Δ restores growth at 37°C to ydj1-151 strains, sse1Δ makes the thermosensitive phenotype more severe (Goeckeler et al., 2002). These results argue against functional redundancy of Fes1p and Sse1p, though their functions may overlap. Sse1p forms specific complexes with Ssa1p and Ydj1p, as observed in vitro (Dragovic et al., 2006); Fes1p may form distinct complexes also involved in [URE3] propagation.
Chaperone-Typing of Prions
Either overproduction or depletion of Sse1p cured [URE3] mimicking the effects of Hsp104 on [PSI+] (Chernoff et al., 1995). Only [PSI+] was destabilized by Hsp104 overexpression, whereas [URE3] and hybrid [PSI+PS] showed sensitivity to overexpression of Hsp70 (Ssa1p) and its cofactors (Sse1, Ydj1). Such “chaperone-typing” indicates structural similarity between [URE3] and hybrid [PSI+PS] and distinction from [PSI+]. Although the structural differences between [URE3] and [PSI+] amyloids are unclear, SDS-resistant polymers on agarose gels are apparently smaller in many [PSI+] variants compared with the [URE3] and hybrid [PSI+PS] #1 (Figure 2A). Longer [URE3] polymers better adhere to each other to form large insoluble aggregates (Kryndushkin et al., 2003), some resistant to boiling in SDS. Perhaps this makes them more resistant to Hsp104 action than [PSI+] polymers.
The Ssa1p/Ydj1p/Sse1p complex must act on polymers by a different mechanism, possibly by binding to the ends of prion filaments. Depletion of Sse1p (or Fes1p) might shift Ssa's toward the ADP form, which binds substrates tightly. Such binding might block addition of new monomers to the fibril. The larger [URE3] aggregates have fewer ends compared with [PSI+] filaments and thus are more sensitive to Sse1p or Fes1p depletion. Overexpressed Sse1p might activate a disaggregating activity of Hsp70s on Ure2p polymers or stabilize Ure2p monomers, preventing their joining the amyloid filaments (Figure 5B).
In summary, Sse1p and Fes1p affect Ssa1p activity as NEFs, and, in addition, Sse1p cooperates with Ssa1p and Ydj1p in a stoichiometric complex in folding of certain proteins. Differences between Sse1p and Fes1p may be explained by their forming different complexes with Ssa1p, or by the fact that Sse1p is a more potent NEF.
ACKNOWLEDGMENTS
We thank D. Masison (National Institutes of Health, Bethesda, MD) for plasmids and comments on the manuscript, M. D. Ter-Avanesyan (Cardiology Research Center, Moscow, Russia) for plasmids and strains, P. Needham (National Institutes of Health, Bethesda, MD) for anti-Sse1 antibody, and members of our lab for critical reading of the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0128) on March 28, 2007.
REFERENCES
- Bagriantsev S. N., Kushnirov V. W., Liebman S. W. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B.-I., Inge-Vechtomov S. G., Liebman S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cox B. S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. Prions affect the appearance of other prions: the story of [PIN] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S. S., Ruddock L. W., Cox B. S., Tuite M. F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edskes H. K., Wickner R. B. A protein required for prion generation: [URE3] induction requires the Ras-regulated Mks1 protein. Proc. Natl. Acad. Sci. USA. 2000;97:6625–6629. doi: 10.1073/pnas.120168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J. R., Kowal A. S., Shirmer E. C., Patino M. M., Liu J.-J., Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Glover J. R., Lindquist S. Hsp104, Hsp70, and Hsp40, a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol. Cell. Biol. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G., Song Y., Chung S., Masison D. C. Propagation of yeast [PSI+] prion impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Jones G., Wegrzyn R. D., Masison D. C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Becherich J.-M., Brodsky J. L. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.-Y., Tittmann P., Gross H., Gebert R., Aebi M., Wuthrich K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D. S., Smirnov V. N., Ter-Avanesyan M. D., Kushnirov V. V. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V., Kryndushkin D. S., Boguta M., Smirnov V. N., Ter-Avanesyan M. D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A. R., Demarini D. J., Shah N. G., Wach A., Brachat A., Phillippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mayer M. P., Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes H. K., Wickner R. B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- Newnam G. P., Wegrzyn R. D., Lindquist S. L., Chernoff Y. O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. T., Moriyama H., Wickner R. B. [URE3] prion propagation is abolished by a mutation of the primary cytosolic Hsp70 of budding yeast. Yeast. 2004;21:107–117. doi: 10.1002/yea.1062. [DOI] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Trott A., Goeckeler J. L., Brodsky J. L., Morano K. A. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related Hsp70 family. J. Biol. Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200. [DOI] [PubMed] [Google Scholar]
- Shaner L., Wegele H., Buchner J., Morano K. A. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C., Fink G. R., editors. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego: Academic Press; 1991. pp. 3–21. [Google Scholar]
- Sondheimer N., Lindquist S. Rnq1, an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sondheimer N., Lopez N., Craig E. A., Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speransky V., Taylor K. L., Edskes H. K., Wickner R. B., Steven A. Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J. Cell Biol. 2001;153:1327–1335. doi: 10.1083/jcb.153.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K. L., Cheng N., Williams R. W., Steven A. C., Wickner R. B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- TerAvanesyan A., Dagkesamanskaya A. R., Kushnirov V. V., Smirnov V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [PSI+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]