Abstract
Tripartite motif (TRIM)5α has recently been identified as a host restriction factor that has the ability to block infection by certain retroviruses in a species-dependent manner. One interesting feature of this protein is that it is localized in distinct cytoplasmic clusters designated as cytoplasmic bodies. The potential role of these cytoplasmic bodies in TRIM5α function remains to be defined. By using fluorescent fusion proteins and live cell microscopy, we studied the localization and dynamics of TRIM5α cytoplasmic bodies. This analysis reveals that cytoplasmic bodies are highly mobile, exhibiting both short saltatory movements and unidirectional long-distance movements along the microtubule network. The morphology of the cytoplasmic bodies is also dynamic. Finally, photobleaching and photoactivation analysis reveals that the TRIM5α protein present in the cytoplasmic bodies is very dynamic, rapidly exchanging between cytoplasmic bodies and a more diffuse cytoplasmic population. Therefore, TRIM5α cytoplasmic bodies are dynamic structures more consistent with a role in function or regulation rather than protein aggregates or inclusion bodies that represent dead-end static structures.
INTRODUCTION
The species-specific tropism of many retroviruses is determined by cellular restriction factors present in the cells of the host. One of the most well characterized restriction factors is the murine Fv1 gene, which prevents infection by particular strains of murine leukemia virus (MLV) (Best et al., 1996; for review, see Bieniasz, 2004; Goff, 2004). It is known that restriction by Fv1 involves the recognition of a capsid determinant in MLV (DesGroseillers et al., 1983; Kozak and Chakraborti, 1996). Analysis of Fv1 restriction indicates that restricted MLV strains can generate reverse transcription products but that they are unable to complete subsequent steps in infection (Pryciak and Varmus, 1992) and also that this restriction of infection is saturable, because it can be overcome with high levels of virus (Pincus et al., 1975; Duran-Troise et al., 1977).
A similar restriction to human immunodeficiency virus 1 (HIV-1) infection in primates has also been well characterized; and recently, the cellular factor present in the cells of Old World monkeys responsible for restricting infection by HIV-1 was cloned and identified as the cytoplasmic body component tripartite motif (TRIM)5α (Stremlau et al., 2004). Restriction of HIV-1 infection by TRIM5α resembles restriction of MLV infection by Fv1 in many ways. TRIM5α-mediated restriction is saturable with high multiplicity of infection (MOI), and the specificity of TRIM5α-mediated HIV-1 restriction is governed by the recognition of a capsid determinant in the restricted virus similar to the case of Fv1-mediated restriction of MLV infection (Towers et al., 2000; Cowan et al., 2002; Owens et al., 2003, 2004; Hatziioannou et al., 2004). However, TRIM5α-mediated restriction differs from Fv1-mediated restriction in that TRIM5α-mediated restriction is manifested earlier in the infectious pathway, before the accumulation of reverse transcription products (Shibata et al., 1995; Himathongkham and Luciw, 1996; Cowan et al., 2002; Munk et al., 2002).
Specifically, the TRIM5α proteins from rhesus monkey (rhTRIM5α) and African green monkey (agmTRIM5α) have been shown to restrict HIV-1 infection (Stremlau et al., 2004; Yap et al., 2004; Song et al., 2005c). Additionally, rhTRIM5α and agmTRIM5α as well as the human variant of TRIM5α (huTRIM5α) were subsequently identified as the factors responsible for restricting the infection by N-tropic MLV in cell lines derived from these species (Hatziioannou et al., 2004; Keckesova et al., 2004; Yap et al., 2004; Song et al., 2005c).
TRIM5α is a member of the family of proteins defined by the presence of a tripartite motif consisting of a RING domain, B-Box, and coiled-coil regions (Reymond et al., 2001). TRIM family members have been reported to be present in a variety of cellular structures, and they are known to form high-order–molecular-weight structures via their coiled coil regions (Reymond et al., 2001), although the function of this family of proteins remains poorly defined. TRIM5α also contains a SPRY domain that is critical in determining species-specific restriction of HIV-1 by Old World monkeys (Sawyer et al., 2005; Stremlau et al., 2005; Yap et al., 2005). Small amino acid substitutions between the SPRY domain of rhTRIM5α and huTRIM5α has been sufficient to derive huTRIM5α variants capable of restricting HIV-1 infection (Stremlau et al., 2005; Yap et al., 2005). The importance of this region is further underscored by evidence that these same residues that confer species-specific restriction within the SPRY domain have been positively selected during the course of primate evolution (Sawyer et al., 2005; Song et al., 2005b), suggesting that TRIM5α has been at the forefront of the battle between primates and retroviruses for millions of years.
Although the regions and residues within TRIM5α required for retroviral restriction have been identified, much less is known about the cell biology of TRIM5α and the mechanism by which it is able to inhibit infection by a retrovirus. To this end, we have constructed fusion proteins tethering variants of green fluorescent protein to the rhesus and human versions of TRIM5α. These fusion proteins functionally restrict retroviral infection in a manner identical to their nonfusion counterparts. We have used these proteins to examine the subcellular localization of rhTRIM5α and huTRIM5α. Using fluorescence, immunofluorescence, and electron microscopy, we confirm that both of these proteins form cytoplasmic bodies similar in appearance to the nuclear bodies formed by the TRIM family protein PML (Zhu et al., 1997). Live cell imaging reveals that these bodies are highly mobile and display two kinds of movements, short saltatory movement and longer, unidirectional movement. Furthermore, the movements of these bodies is microtubule dependent, as disrupting microtubules with nocodazole essentially abrogates long-distance movement of cytoplasmic bodies along with a substantial decrease in average velocity of particles. Additionally, TRIM5α is capable of assembling larger cytoplasmic bodies from several individual smaller bodies. We have examined these cytoplasmic bodies by using fluorescence recovery after photobleaching (FRAP) and photoactivation. We found that these bodies are not static, dead-end structures resulting from overexpression but rather dynamic structures that turn over rapidly due to exchange with free cytoplasmic TRIM5α as well as TRIM5α in similar neighboring bodies.
MATERIALS AND METHODS
Recombinant DNA Constructs
The huTRIM5α plasmid was a kind gift from Dr. Joseph Sodroski (Harvard School of Public Health). Yellow fluorescent protein (YFP)-huTRIM5α was constructed by inserting PCR-amplified YFP fragment between EcoRI and KpnI sites on the huTRIM5α plasmid by using the primers CCGGAATTCGTGAGCAAGGGCGAGGAGCTGTTC and CGGGGTACCCTTGTACAGCTCGTCCAT. The rhTRIM5α plasmid was a kind gift from Dr. Joseph Sodroski. The rhTRIM5α fragment was amplified by polymerase chain reaction (PCR) with the primers CGGGGTACCATGGCTTCTGGAATCCTG and CCCGCTCGAGTCAAGAGCTTGGTGAGCA. Then, the fragment was inserted between the KpnI and XhoI sites of the YFP-huTRIM5α clone. Photoactivatable (PA) green fluorescent protein (GFP) fusion proteins (PAGFP-rhTRIM5α and PAGFP-huTRIM5α) were constructed by replacing the YFP fragments on the YFP-rhTRIM5α and YFP-huTRIM5α with PCR-amplified PA-GFP fragment by using the YFP primers indicated above. To produce a cyan fluorescent protein (CFP)-α tubulin fusion protein, CFP was amplified from pECFP (Clontech, Mountain View, CA) by PCR by using the following primers: forward, 5′-CACCATGGTGAGCAAGGGCGAGGA-3′ and reverse, 5′-CTTGTACAGCTCGTCCATGC-3′. Tubulin was amplified from pEYFP-Tub (Clontech) using forward, 5′ GCATGGACGAGCTGTACAAG 3′, reverse, 5′ TGGATCCTTAGTATTCCTCT 3′. The primers were designed with overlapping regions and so the two segments could be fused in a joining PCR reaction by using the two original PCR products with the forward CFP primer and the reverse tubulin primer. The forward CFP primer was designed to commence with CACCATG allowing direction cloning into pENTR-D-TOPO (Invitrogen). Gateway cloning by LR recombination into pLgatewaySN (MLV vector) was then carried out. GFP-huTRIM5α cloning was carried out using a similar method. These constructs were introduced into cells using a vesicular stomatitis virus-G protein (VSV-G) pseudotyped Moloney MLV vector. The pmRFP-Dcp1a vector that expresses the mRNA-decapping enzyme involved in 5′-to-3′ mRNA degradation has been described previously (Kedersha et al., 2005; Gallois-Montbrun et al., 2007).
Cell Culture, Transfections, and Virus Production
HeLa, 293T, and human osteosarcoma cell (HOS) cells were cultured in complete DMEM containing 10% fetal bovine serum, penicillin (final concentration 100 U/ml), and streptomycin (final concentration 100 μg/ml). TE671 cells were cultured in DMEM containing 10% fetal calf serum, penicillin (final concentration 100 units/ml), and streptomycin (final concentration 100 μg/ml).
Transfections were performed with Effectene transfection reagent (QIAGEN, Valencia, CA) according to the manufacturer's protocol. HIV reporter virus was produced by poly(ethylenimine) transfection (Durocher et al., 2002) of 293T cells with 8 μg of pVSV-G and 12 μg of proviral construct R7Δ EnvGFP in which Nef gene was replaced with GFP. Simian immunodeficiency virus (SIV) reporter virus was made in similar manner. Virus was harvested as described previously 48 h posttransfection (Campbell et al., 2004). Vectors used for transduction were produced from 293T cells using calcium phosphate transfection of 7 μg of pVSV-G, 7 μg of vector (GFP T5 or CFP Tubulin), and 7 μg Moloney MLV Gag-Pol.
Stable Cell Lines
HeLa cells were transiently transfected with YFP-rhTRIM5α as mentioned above. Complete DMEM media with 200 μg/ml G-418 (Geneticin; Sigma-Aldrich, St. Louis, MO) was added to the cells 48 h after transfection for selection. FACS was performed to sort cells after 1-mo selection, and YFP-positive cells were subjected to single colony isolation. Colonies were subsequently screened for protein expression and viral restriction. These HeLa YFPrhTRIM5α cells were transiently transfected using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) as described previously (Gallois-Montbrun et al., 2007). Stable cell lines expressing both CFP-tubulin and GFP-huTRIM5α were generated by transduction of 8 × 104 TE671 cells in a 12-well plate by using Moloney MLV-based vectors with above-mentioned fragments at an MOI of ∼10. Typically, 99% of cells are transduced (as determined by flow cytometry).
Restriction Experiment
YFP-rhTRIM5α stable cells were plated on 96-well plates at 1 × 104 per well 1 d before infection. VSV-G pseudotyped GFP reporter HIV viruses (R7Δ EnvGFP) were used to infect cells by spinoculation (1200 × g for 2 h at 25°C) (O'Doherty et al., 2000). Virus was then removed, and fresh media were added to cells. 24 h later, cells were transferred to 24-well plates, and analyzed for GFP expression by FACS analysis 48 h postinfection.
Confocal Microscopy
HeLa cells expressing fluorescent protein fusions were fixed, the images were collected with a laser confocal scanning microscope (DM IRE2; Leica Microsystems, Deerfield, IL), and then they were processed with the LCS version 2.02 software (Leica Microsystems) and Adobe Photoshop version 6.0 (Adobe Systems, Mountain View, CA). Data are presented as Z-series compilations of 6–10 images in a stack (Gallois-Montbrun et al., 2007).
Electron Microscopy (EM)
HeLa cells were plated on 3.5-cm plate 1 d before the transient transfection with 1.176 μg of YFP-rhTRIM5α. Approximately 16 h posttransfection, cells were fixed with 2% glutaraldehyde (Sigma-Aldrich) in 0.1 M cacaodylate solution (Electron Microscopy Sciences, Hatfield, PA), washed with 0.1 M cacodylate buffer, postfixed with 1% glutaraldehyde in 0.1 M cacodylate solution, dehydrated with series of ethanol up to 100% absolute, and finally embedded with LX112 resin epoxy resin. Uranyl acetate and lead citrate were used to stain 200 mesh copper grids. EM pictures were collected on JEM 1220 TEM (JEOL, Tokyo, Japan) equipped with a Gatan digital camera.
Immunoblots
Cell lysates of transiently transfected HeLa cells or stable cells were subject to SDS-polyacrylamide gel electrophoresis, and transferred onto nitrocellulose membrane (Whatman Schleicher and Schuell, Keene, NH). YFP-huTRIM5α and YFP-rhTRIM5α were detected using a monoclonal anti-YFP antibody (1:5000; Clontech) and goat anti-mouse IgG secondary antibody (1:10,000; Pierce Chemical, Rockford, IL).
Tracking Particle Movement and Determining the Movement Rate
TE671 cells stably expressing GFP-huTRIM5α were taken as images in a 60-frame sequence captured at 1.325-s intervals by using live cell fluorescence microscopy with a 100× objective lens. To follow the track of particle movement manually, the 60 frames were combined into one image by using an image addition process on the basis that in the composite image, a given pixel is assigned the maximum intensity value observed throughout any of the 60 input images. Each track was manually verified as authentic by comparison with the original movies before further analysis. The interframe movement rates of the cytoplasmic bodies were further measured using the manual tracking function of the particle tracking software GMimPro (Mashanov and Molloy, National Institute for Medical Research, London, United Kingdom). This software can also automatically follow the movement of point sources of fluorescence (such as TRIM5α cytoplasmic bodies) within an image by using an automatic single particle tracking algorithm (Mashanov and Molloy, unpublished data). A particle will be tracked if above a given size and intensity. Tracks are made between nearest neighbors in consecutive frames. A minimum length of track (number of frames) can be specified as can the maximum movement allowed between frames (number of pixels). Because the time interval is fixed, the velocity can then be determined. This function of the software was used to automatically track the movement of TRIM5α cytoplasmic bodies.
Microinjection
HOS cells were plated on ΔT dishes (Bioptechs, Butler, PA). Cells were microinjected with a 9:1 ratio of eGFP-huTRIM5α (20 ng/μl):RNDextran (50 mg/ml) after the mixture was centrifuged at 16,000 × g for 10 min. One and a half hours after microinjection, cells were analyzed by deconvolution microscopy as described previously (McDonald et al., 2002). Images were acquired every 2 min for 1 h.
Fluorescence Recovery after Photobleaching
HeLa cells were plated on ΔT dishes and allowed to adhere for 6 h. Then, cells were transfected with 0.45 μg of YFP-huTRIM5α or YFP-rhTRIM5α by using Effectene transfection reagent (QIAGEN). 16 hours posttransfection, images were acquired on an inverted LSM510 Meta confocal microscope (Carl Zeiss, Thornwood, NY), with 40× water-immersion objective by using the 514-nm line of an argon laser. Fluorescence emission was collected using the 535- to 590-nm band-pass filter. Regions of interest were selected, and pretreatment images were collected before bleaching. Bleaching was performed using time series and bleach software (Carl Zeiss) with the laser at 100% intensity for 120 iterations. Postbleach images were obtained periodically every 30 s for 15 min. Analysis with a control region of interest (ROI) drawn a fair distance away from the bleached ROI indicated no significant bleaching while fluorescence recovery was monitored. Recovery is measured as the observed fluorescence at a given time point relative to the original observed prebleached fluorescence. The fluorescence recovery ration, R, is calculated as published previously (Molk et al., 2004): R = (F∞ − F0)/(Fpre − F0). F∞ is the average intensity of the bleached region after maximum recovery, F0 is the fluorescence intensity immediately after photobleaching (t = 0 s), and Fpre is the fluorescence intensity before photobleaching. Half-recovery time (t1/2) is the time necessary for the fluorescence to recover halfway between the fluorescence level after bleaching and the fluorescence at the plateau level.
Photoactivation
HeLa cells were transiently transfected with PAGFP-TRIM5α. A small discreet region of the cell is defined using Mosaic software (Photonic Instruments, Inc., St. Charles, IL), and 474-nm CFP light is directed to expose this region for 5 s, leading to the activation of the fluorescence of the GFP protein only in this defined region. Live cell imaging is taken every 1 min for 20 min to monitor the behavior of this fluorescent subpopulation of PAGFP-TRIM5α over time.
RESULTS
Fluorescently Tagged TRIM5α Proteins Restrict HIV-1 Infection
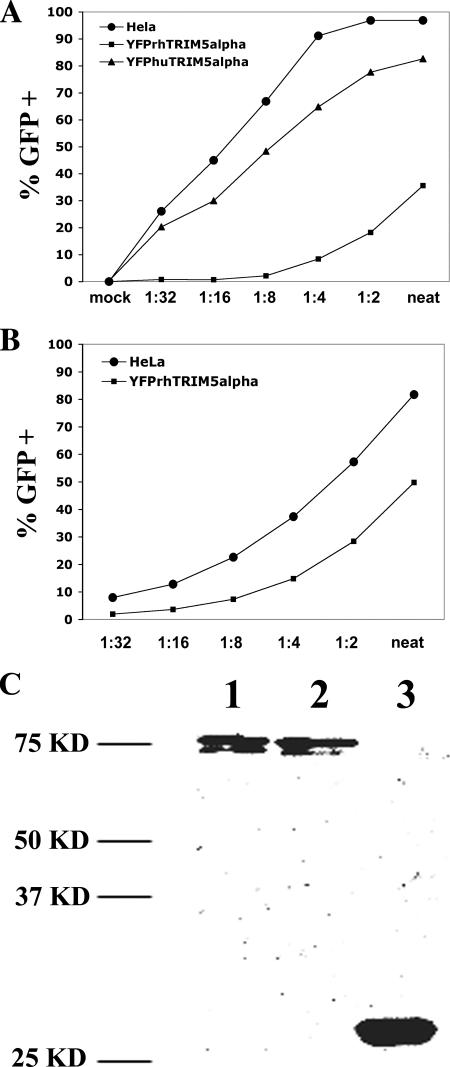
To initiate our studies, we generated fusion proteins tethering YFP to TRIM5α. Stable cell lines expressing YFP-rhTRIM5α or YFP-huTRIM5α were selected and characterized for their ability to restrict infection of an HIV-1 reporter virus that expresses GFP after infection of target cells. As seen in Figure 1A, a HeLa cell line stably expressing YFP-rhTRIM5α effectively restricted HIV-1 infection, whereas a much smaller degree of restriction was observed in a cell line stably expressing YFP-huTRIM5α. This restriction to infection by YFP-rhTRIM5α was specific to HIV-1 infection, because this cell line inhibited infection by SIV reporter virus to a much smaller degree (Figure 1B). We conclude that YFP-TRIM5α fusion proteins functionally restrict HIV-1 infection in a pattern similar to previous reports examining unfused TRIM5α variants (Stremlau et al., 2004; Stremlau et al., 2005). Western blot analysis indicated that YFP-rhTRIM5α and YFP-huTRIM5α were expressed as intact fusions in cells (Figure 1C).
Figure 1.
Functional validation of YFP-TRIM5α fusion proteins and the cellular localization of TRIM5α fusion proteins. Stable cell lines expressing YFP-TRIM5α fusion proteins were tested for their ability to restrict HIV-1 (A) or SIV (B) reporter viruses. YFP-rhTRIM5α (squares) effectively inhibited HIV-1 infection relative to HeLa control (circles), but it less effectively inhibited SIV infection. YFP-huTRIM5α (triangles) expression only minimally inhibited HIV-1 infection. (C) 293T cells were transfected with YFP-TRIM5α fusion proteins. YFP-huTRIM5α and YFP-rhTRIM5α fusion proteins were expressed as intact fusions when compared with unfused YFP.
Examination of TRIM5α Localization
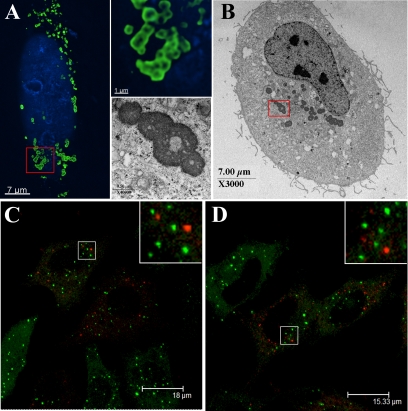
Having verified that fluorescently tagged versions of TRIM5α are expressed as intact, functional fusions, we next examined the localization of these proteins microscopically. As seen in Figure 2, A, C, and D, both human and rhesus versions of TRIM5α localized to cytoplasmic bodies as previously reported for hemagglutanin (HA)-tagged TRIM5α (Stremlau et al., 2004) and a number of other TRIM family members (Reymond et al., 2001). For the purpose of our study, any grouping of TRIM5α above diffuse cytoplasmic levels is referred to as cytoplasmic bodies. We find that as the level of TRIM5α expression increases, the cytoplasmic bodies become larger and more numerous (data not shown), in agreement with a previous report (Perez-Caballero et al., 2005a). Our observations also reveal that at low levels of expression, the HA-tagged human and rhesus TRIM5α have a diffuse cytoplasmic localization with fewer and smaller cytoplasmic bodies (data not shown). Low expression levels of the YFP-tagged versions also form small cytoplasmic bodies with some diffuse cytoplasmic localization. There seems to be a diffuse cytoplasmic population at both low and high expression levels, although it is more apparent when expression levels are low (data not shown). The TRIM5α cytoplasmic bodies we imaged were heterogeneous in size and shape, typically showing a spherical morphology. Cytoplasmic bodies with elongated wormy structures were also observed (data not show). Deconvolution microscopy revealed that the larger spherical cytoplasmic bodies were apparently hollow (Figure 2A, bottom inset), which was confirmed by analysis of YFP-huTRIM5α-transfected cells by using electron microscopy (Figure 2B, bottom inset). Similar patterns of localization to cytoplasmic bodies were also observed in stable HeLa cell lines (Figure 2, C and D). Importantly, these results demonstrate that these fluorescent fusion proteins have the same localization pattern as HA-tagged TRIM5α.
Figure 2.
(A) HeLa cells were transiently transfected with YFP-huTRIM5α, and the cells then fixed, stained for cellular DNA (blue), and TRIM5α localization (green) was examined. An enlargement highlighting the hollow body structures formed by YFP-hu-TRIM5α is shown in the top inset. (B) Electron microscopy confirms that YFP-huTRIM5α forms hollow structures, with a magnification of the structures observed shown in the bottom inset. The P-body markers mRFP-Dcp1a (C) and RFP-rck/p54 were expressed in cells stably expressing YFP-rhTRIM5α. Cells were fixed and analyzed by confocal microscopy. The merged images demonstrates the lack of TRIM5α (green) and Dcp1a (red) (C) or RFP-rck/p54 (red) (D) colocalization.
TRIM5α cytoplasmic bodies were not observed to associate with cellular markers of actin, early endosomes, the trans-Golgi network, or lysosomes by imaging (data not shown). TRIM5α cytoplasmic bodies did not coincide with processing bodies (P-bodies) (Figure 2, C and D), another class of cytoplasmic body that is involved in mRNA storage and metabolism (Anderson and Kedersha, 2006) when the fluorescent P-body markers mRFP-Dcp1A (Figure 2C) and RFP-rck/p54 (Figure 2D) (Gallois-Montbrun et al., 2007) were expressed in HeLa cells stably expressing YFP-rhTRIM5α. They also do not associate with vimentin as would be expected for aggresomes (data not shown) (Garcia-Mata et al., 2002).
TRIM5α Cytoplasmic Bodies Are Highly Motile
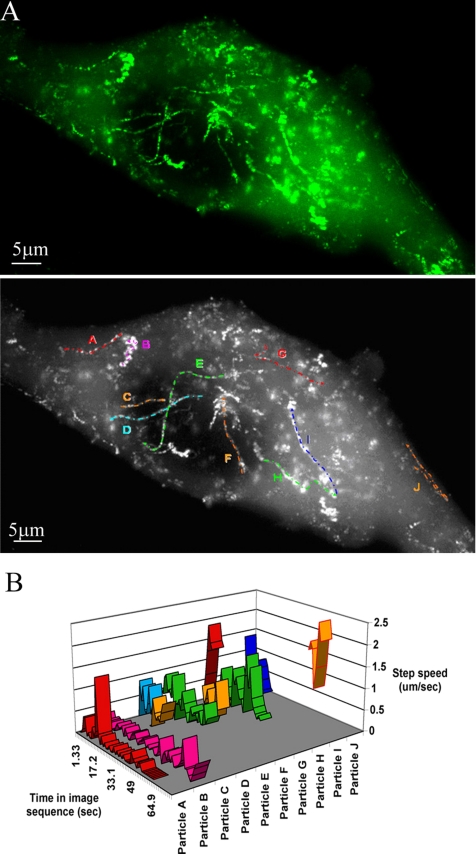
When live cell microscopy was used to examine TRIM5α behavior in living cells, it was clear that cytoplasmic bodies within these cells were highly motile. To record this movement, images were rapidly captured in sequence at the shortest possible time interval compatible with obtaining a sufficiently long exposure. Supplemental Movie 1A shows such a 60-frame sequence captured at 1.325-s intervals. The high degree of motility is clear. There seem to be two types of movement. Several of the bodies show multidirectional short saltatory movements, whereas others show rapid unidirectional long-distance movements. Using an image addition process, the 60 frames from Supplemental Movie 1A were combined into one image on the basis that in the composite image, a given pixel is assigned the maximum intensity value observed throughout any of the 60 input images (Figure 3A). This process clearly shows a number of tracks, which can be easily visualized in time-lapse format (Supplemental Movie 1A). Several such tracks and their direction are identified and labeled A–J (Figure 3A, bottom).
Figure 3.
TRIM5α particle movement and determination of the movement rates. (A) A live TE671 cell stably expressing GFP-huTRIM5α was imaged over 60 frames at 1.325-s intervals (0.75 frames/s). The 60 individual frames were summed using ImageJ on the basis that in the composite image, a given pixel is assigned the maximum gray scale value (0-256) observed in any of the 60 frames throughout the series. The tracks of a number of cytoplasmic bodies are shown in the top panel. The directionality and authenticity of these tracks were confirmed by comparison with Supplemental Movie 1. Several prominent tracks were identified for further analysis. These tracks are labeled A–J in bottom panel. (B) Velocities of particles A–J in A were measured throughout the time that they were observed during the 60-frame (80-s) sequence by using the manual tracking function of the particle tracking software GMimPro. The vertical y-axis shows the average speed of the particle between each consecutive frame (step speed; micrometers per second); the horizontal-z-axis shows the relative time during the image sequence in which the particular particle was observed.
The short-distance saltatory movements are exemplified by particle B, whereas the others show longer distance unidirectional movement. To better analyze and represent the movements of these particles, the interframe movement rates of the indicated bodies were measured using the manual tracking function of the particle tracking software GMimpro. This software allows the position of the particle to be recorded between each frame, and, because the time interval is fixed, the velocity can be easily determined. The data from such an analysis are shown in Figure 3B. The horizontal z-axis shows the time index in the image sequence during which the particle was observed. The vertical y-axis shows its step (interframe) speed. Using this approach, the fate of a particle throughout the entire sequence can be displayed.
Particle A shows periods of rapid movement followed by apparent stagnation. Over the time it was observed, the average speed of particle A was 0.1 μm/s; however, at least three bursts of rapid directional movement were observed of 0.4, 1.48, and 0.3 μm/s. These rapid movements can also be observed in Supplemental Movie 1. Particle B shows short, low-speed movements throughout most of the time it is observed; however, a number of more rapid movement bursts are observed toward the end of the movie up to 0.8 μm/s. Other particles, such as C, G, and J, show very rapid, although still variable, movements, throughout their observation time, with maximum speeds up to 2.3 μm/s observed. Particle J for example maintains a high average speed of 1.6 μm/s. Due to their very high motility, these particles could only be tracked for a short time before they left the focal plane. There was not an obvious preference for movement either to or from the cell periphery. Particles apparently moving in either direction were readily observed. Particle J, moreover, seemed to change direction during the time it was tracked, although not on exactly the same path.
It should also be noted that many more rapidly moving particles may exist, but they are not observed due to the limits of the detection technology (frame rate and camera sensitivity) used here. Such rapid long-distance directional movements of <1 μm/s suggest an active mechanism of movement, and they strongly implicate the microtubule network as the carrier.
Association of TRIM5α Cytoplasmic Bodies with the Microtubule Network
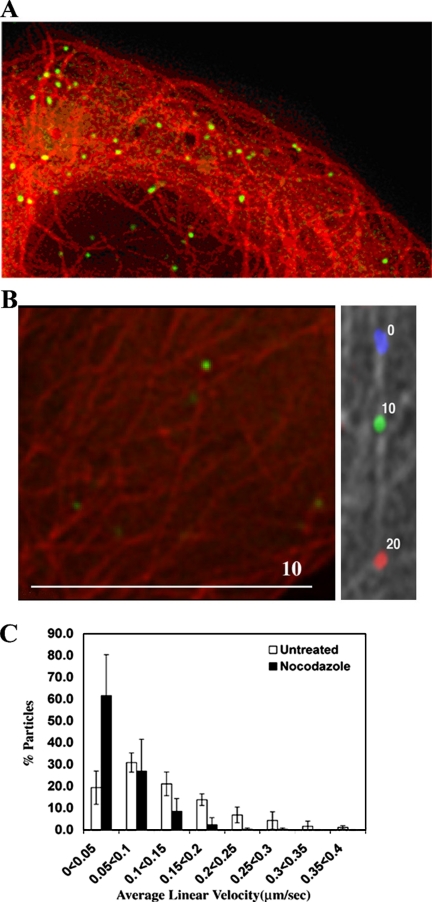
To determine whether TRIM5α cytoplasmic bodies can be observed in association with microtubules, CFP was fused to the N terminus of α-tubulin and subcloned into a retroviral vector. Stable cells were then derived by transducing TE671 cells with this construct as well as GFP-huTRIM5α. Live cells expressing both fusion proteins were observed using a fluorescence microscope. Cytoplasmic bodies can clearly be seen in association with the microtubule network (Figure 4A). If images are captured in sequence, movement on microtubules is apparent (Supplemental Movie 2), although this is hindered by the requirement to capture images for both CFP and GFP resulting in an increase in the frame rate to 10 s.
Figure 4.
Movement of individual cytoplasmic bodies upon microtubules and reduction in cytoplasmic body motility upon treatment with nocodazole. (A) A static image demonstrating the association of GFP-huTRIM5α (green) and CFP-labeled tubulin (red). (B) A live TE671 cell stably expressing CFP-labeled tubulin and GFP-huTRIM5α was imaged using fluorescence microscopy. Left, starting positions for a cytoplasmic body (green) and the microtubule network (red). Right, microtubule network in gray with the relative positions of the cytoplasmic body at 10-s intervals colored blue, green, and red. The relative time (seconds) is noted by the body. (C) The movement of GFP-TRIM5α stably expressed in TE671 cells was monitored by live cell microscopy. Cells were either left untreated or treated with 66 μM nocodazole for 2 h before imaging. Images were captured at a rate of 0.75 frames per second for a total of 60 frames. Particle tracks were analyzed using GMview, and they were verified manually to confirm authenticity. Five randomly chosen cells were analyzed in each case, resulting in 258 cytoplasmic body tracks for untreated cells and 306 tracks for nocodazole-treated cells. Velocities were grouped into 0.05 μm/s bins for each cell analyzed, and the percentage of particles in each bin was determined. The mean and SD values in each bin over five cells were then calculated.
Figure 4B shows movement of one cytoplasmic body on microtubules. To represent this movement clearly in print, the microtubule network and the first image in the sequence is shown in left panel, and the right panel shows the microtubule network in gray with the GFP signal from three successive frames shown in blue, green, and red, respectively.
Effect of Microtubule Disruption on Cytoplasmic Body Motility
To determine the importance of the microtubule network in the observed motility of TRIM5α cytoplasmic bodies, cells stably expressing GFP-huTRIM5α were treated with 66 μM nocodazole for 2 h before analysis by live cell fluorescence microscopy. Image sequences were again captured in sets of 60 at 1.325-s intervals. In these cells, long-distance movement of cytoplasmic bodies was essentially abolished (Supplemental Movie 1B). In an attempt to better quantify this effect, the automated particle tracking function of the GMimPro software was used (see Materials and Methods). This analysis has advantages and limitations. The principle advantages are that a huge amount of data can be analyzed relatively quickly and efficiently and that data sets can be compared without any unintentional bias on the part of the experimenter. The disadvantage is that the fastest moving particles are not tracked efficiently because rapid movement often means that nearest neighbors in consecutive frames are not derived from the same particle. This results in output skewed toward lower velocities. Nonetheless, the ability to examine objectively a large number of samples is ideal for comparing nocodazole-treated and untreated cells.
As shown in Figure 4C, image sequences of 60 frames were recorded for five cells in nocodazole-treated and untreated cells. This resulted in 258 tracks for untreated cells and 306 tracks for nocodazole-treated cells. Each track was manually verified. A significant reduction in average velocity of particles is apparent upon treatment with nocodazole. We conclude that the motility of TRIM5α cytoplasmic bodies requires an intact microtubule network.
Formation of Cytoplasmic Bodies of TRIM5α
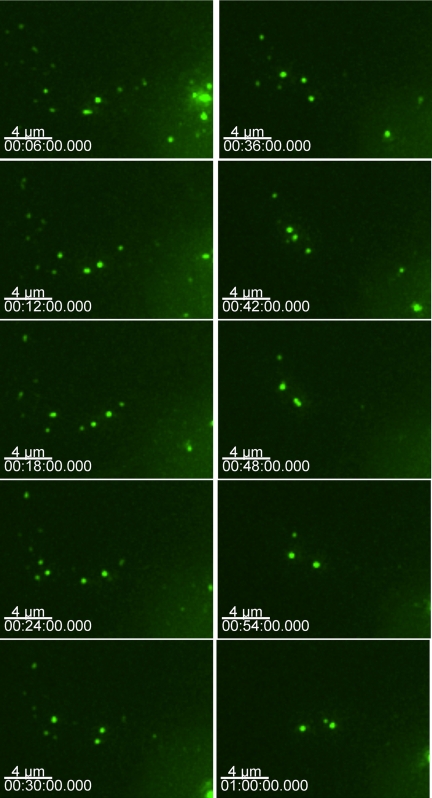
To gain insight into the expression, trafficking, and formation of TRIM5α cytoplasmic bodies, we observed cells expressing GFP-TRIM5α by using time-lapse deconvolution microscopy. To observe the first TRIM5α protein expressed in the cell, we microinjected GFP-TRIM5α expression plasmids directly into the nucleus of HOS cells and tracked the emerging GFP. This approach leads to the synchronous expression of protein and allows the first protein expressed to be followed. In the GFP-TRIM5α fusion proteins, expression was observed ∼90 min after DNA microinjection. In multiple experiments, small (∼1 μm) cytoplasmic bodies occurred immediately, along with a faint cytoplasmic GFP signal. The cytoplasmic bodies were highly mobile, and the smaller cytoplasmic bodies came together to form larger cytoplasmic bodies (Figure 5and Supplemental Movie 3). The individual frames of the time-lapse experiment are 2 min apart. In this sequence, smaller cytoplasmic bodies associate with each other to form larger bodies. However, in other time-lapse experiments, individual tubular cytoplasmic bodies can be observed collapsing into a single spherical body or separating to form two distinct, individual bodies (Supplemental Movie 4). These time-lapse experiments confirmed that TRIM5α cytoplasmic bodies are highly mobile and dynamic structures. In many ways, they behave like intracellular vesicular structures, although we do not have any data so far to demonstrate that cytoplasmic bodies associate with intracellular membranes.
Figure 5.
Smaller YFP TRIM5α cytoplasmic bodies combine to form larger bodies. HOS cells were microinjected with GFP-rhTRIM5α, and cells were imaged at 2-min intervals. At the onset of expression, small cytoplasmic bodies quickly became apparent, and these smaller bodies could combine to form larger bodies.
TRIM5α Continuously Exchanges between the Cytoplasmic Bodies and a Diffuse Form in the Cytoplasm
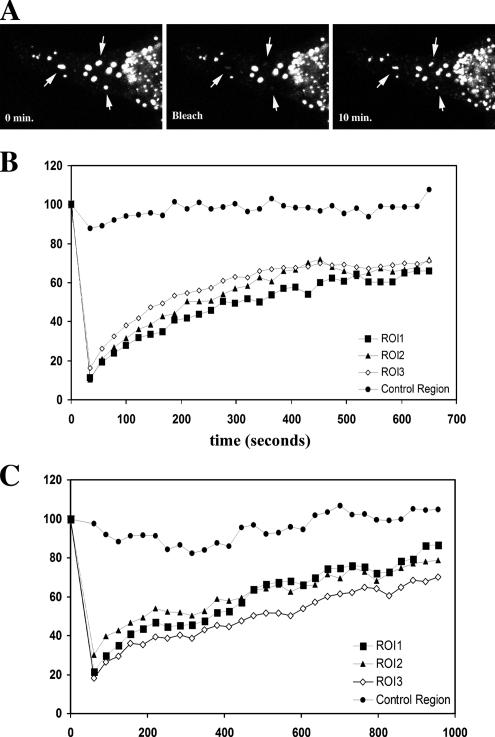
To gain insight into the dynamics of cytoplasmic bodies, we used FRAP as described previously (Steffens and Hope, 2004). Briefly, the confocal laser is projected onto a subregion of interest in the cell, in this case, a TRIM5α cytoplasmic body. The region is exposed with maximal laser intensity until the fluorescent signal in the targeted region is photobleached. After the signal is bleached, the cell is imaged over time, and the recovery of fluorescent signal is determined at each time point (Figure 6A). FRAP data are analyzed by plotting the percentage of recovery of the fluorescent signal as a percentage of the initial fluorescence observed before photobleaching. Our analysis with YFP-TRIM5α bodies reveals that the fluorescence of a photobleached cytoplasmic body significantly recovers within the 10-min time period of the experiment. For YFP-rhTRIM5α, R = 67.3 ± 3.0%; for YFP-huTRIM5α, R = 64.3 ± 2.1%. The time to recover 50% of the fluorescent signal (t1/2) is on the order of 4 ± 1 min in multiple experiments (Figure 6 and Supplemental Movie 5). Similar results were obtained when cells were kept at 37°C or at room temperature (∼23°C).
Figure 6.
FRAP analysis of TRIM5α cytoplasmic bodies. (A) Individual YFP-TRIM5α bodies were photobleached using a confocal laser, and fluorescence of the bleached body was monitored over time. Recovery of YFP-huTRIM5α (B) was similar to recovery observed in rhYFP-TRIM5α bodies (C).
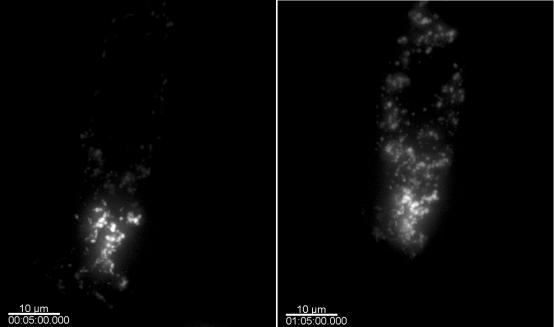
To examine the dynamics of TRIM5α bodies in another manner, we generated PAGFP-TRIM5α (Patterson and Lippincott-Schwartz, 2002). In these experiments, the fluorescence of the GFP protein is activated in a small discrete region of the cell, whereas the rest of the PAGFP-TRIM5α remains unactivated; therefore, it is minimally fluorescent. We examined TRIM5α dynamics by monitoring the behavior of this fluorescent subpopulation of PAGFP-TRIM5α in transiently transfected HeLa cells. A cell immediately after photoactivation and 60 min later is shown in Figure 7(Supplemental Movie 6). There is clearly a decrease in the fluorescent signal in the area of photoactivation and a concomitant increase in the GFP signal in the rest of the cytoplasmic bodies in the cell. These data confirm the results obtained using FRAP suggesting that TRIM5α cytoplasmic bodies are dynamic and proves that preexisting TRIM5α is able to shuttle between the cytoplasm and cytoplasmic bodies.
Figure 7.
Photoactivation analysis of TRIM5α cytoplasmic bodies. A HeLa cell transiently transfected with PAGFP-TRIM5α is activated in a discreet region, whereas the rest of the PAGFP-TRIM5α remains unactivated and therefore not fluorescent. TRIM5α dynamics is examined by monitoring the behavior of this fluorescent subpopulation of PAGFP-TRIM5α. A cell immediately after photoactivation (left) and 60 min later (right) is shown.
DISCUSSION
In this study, we examined the cellular behavior of the cellular restriction factor TRIM5α. We find that TRIM5α localizes primarily to cytoplasmic bodies when TRIM5α was fused to fluorescent protein for visualization. Importantly, fusion of TRIM5α to fluorescent proteins for visualization did not affect its ability to restrict infection (Figure 1).
A key issue in understanding the basic nature of cytoplasmic bodies is whether they are static or dynamic in protein composition. Using methods to analyze the dynamics of TRIM5α bodies in living cells, we have developed evidence that the latter is true. TRIM5α cytoplasmic bodies are highly mobile within a cell. We have demonstrated that this motility involves the microtubule cytoskeleton (Figure 4), demonstrating that TRIM5α cytoplasmic bodies exhibit active translocation within the cell. Other studies have identified a COS motif present in a subset of TRIM proteins that mediates microtubule binding (Short and Cox, 2006). Although TRIM5α does not possess this motif, it is nevertheless able to interact with the microtubule network to facilitate its intracellular transport. More work is required to determine the precise mechanism by which TRIM5 proteins achieve this intracellular motility.
Additionally, both FRAP and PAGFP studies presented in this article suggest that cytoplasmic bodies are dynamic and that they freely exchange protein between bodies and the cytoplasm. The rate of fluorescence recovery in TRIM5α cytoplasmic bodies observed using FRAP analysis indicates that these bodies turn over rapidly, demonstrating an ability of TRIM5α present in cytoplasmic bodies to be exchanged for TRIM5α present in the cytoplasm. Additionally, the extent of fluorescence recovery observed during FRAP analysis indicates that these cytoplasmic bodies turn over not just rapidly but almost completely in a relatively short time. The finding that TRIM5α present in cytoplasmic bodies can reenter the cytoplasmic pool is also supported by our studies using PAGFP-TRIM5α. We find that TRIM5α present in a given body rapidly diffuses into the cytoplasm and is capable of entering other bodies present in other areas of the cell. These findings suggest that TRIM5α cytoplasmic bodies do not represent static, dead end structures devoted to degrading overexpressed or misfolded proteins. Rather, it seems clear that TRIM5α cytoplasmic bodies represent dynamic structures that may provide insight into the mechanism of retroviral restriction by TRIM5α.
The ability of TRIM5α to form cytoplasmic bodies is not surprising, given that it has been reported that other TRIM family members form similar cytoplasmic bodies in cells, whereas others form multiprotein bodies localizing to the nucleus or to specific cytoplasmic components (Reymond et al., 2001). Moreover, the structure and dynamics of these bodies resembles in many ways the nuclear bodies formed by the most well-characterized TRIM family member, PML. The spherical, apparently hollow bodies we observed by EM in TRIM5α-expressing cells (Figure 2) resemble EM images of the nuclear bodies formed by PML (Zhu et al., 1997). In addition, the dynamic nature of these TRIM5α cytoplasmic bodies, as measured by FRAP, is similar to FRAP recovery performed on nuclear PML bodies (Rivera et al., 2003; Dong et al., 2004). In PML, it is thought that nuclear body formation is critical to PML function, because a well-characterized genetic translocation resulting in a fusion protein between PML and the retinoic acid receptor α results in the disruption of PML nuclear bodies and the induction of acute promelocytic leukemia (Song et al., 2005b). Although TRIM5α is not associated with the cytoplasmic P-bodies (Figure 2), both structures exhibit similar dynamics of protein exchanges between individual structures and associate with microtubules (Leung et al., 2006; Sweet et al., 2007). Such dynamic exchange of protein components may be a common feature of all such biological structures within cells.
It is possible that these bodies represent a functional unit involved in retroviral restriction. In this regard, it may be that preexisting bodies play a role in restriction, or rather that these bodies form around an incoming retrovirus from free TRIM5α in the cytoplasm. This would be consistent with reports suggesting that preexisting cytoplasmic bodies are not required for TRIM5α restriction (Perez-Caballero et al., 2005b; Song et al., 2005a) and with the observation that cytoplasmic body formation is correlated to expression level (data not shown; Perez-Caballero et al., 2005). Thus, TRIM5α may exist as a diffuse cytoplasmic protein and body formation may occur as the localized concentration of TRIM proteins increases around a cytoplasmic viral core. In this regard, we often observed fluorescently labeled virions localizing within TRIM5α cytoplasmic bodies (Cohen, 2005; Freed and Mouland, 2006) (our unpublished data). Alternatively, these bodies may represent storage compartments used to regulate the concentration of TRIM5α present in the cytoplasmic pool, similar to the mechanism used to regulate the activity of certain splicing factors in the nucleus, such as Tuftelin-interacting protein (TFIP)11 (Wen et al., 2005). TFIP11 is a splicing factor located in a subnuclear structure known as TFIP body. It has been shown that RNA polymerase II inhibitor treatment causes TFIP bodies to increase in size but decrease in number, and TFIP 11 resides at the surface of the sphere-like structure of the TFIP body with a hollow center. Therefore, TFIP bodies function as a storage/assembly/modification compartment for splicing components rather than the active sites, and TRIM5α bodies may represent a similar compartment.
Although more work is required to elucidate the precise nature of the role of cytoplasmic bodies in the cell biology of TRIM5α, these results clearly demonstrate that these bodies do not represent static, dead-end structures formed as a result of overexpression. Rather, they are mobile, highly dynamic structures that may help elucidate the poorly understood mechanism of TRIM5α restriction of retroviral infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health and the United Kingdom Medical Research Council. S.G.-M. is a Fellow of the European Molecular Biology Organization, and T.J.H. and M.H.M. are Elizabeth Glaser Scientists supported by the Elizabeth Glaser Pediatric AIDS Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1075) on March 28, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S., Le Tissier P., Towers G., Stoye J. P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Bieniasz P. D. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Campbell E. M., Nunez R., Hope T. J. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J. Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. Making a monkey out of HIV. More is becoming clear about a novel host factor that appears central to governing the species-specificity of retroviruses like HIV and could be a future antiviral target. IAVI Rep. 2005;9:9–12. [PubMed] [Google Scholar]
- Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3′ end of env. J. Virol. 1983;47:24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Stenoien D. L., Qiu J., Mancini M. A., Tweardy D. J. Reduced intranuclear mobility of APL fusion proteins accompanies their mislocalization and results in sequestration and decreased mobility of retinoid X receptor alpha. Mol. Cell Biol. 2004;24:4465–4475. doi: 10.1128/MCB.24.10.4465-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Troise G., Bassin R. H., Rein A., Gerwin B. I. Loss of Fv-1 restriction in Balb/3T3 cells following infection with a single N tropic murine leukemia virus particle. Cell. 1977;10:479–488. doi: 10.1016/0092-8674(77)90035-6. [DOI] [PubMed] [Google Scholar]
- Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O., Mouland A. J. The cell biology of HIV-1 and other retroviruses. Retrovirology. 2006;3:77. doi: 10.1186/1742-4690-3-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C. M., Byers H., Lynham S., Ward M., Malim M. H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Gao Y. S., Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- Goff S. P. Retrovirus restriction factors. Mol. Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himathongkham S., Luciw P. A. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- Keckesova Z., Ylinen L. M., Towers G. J. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., Chakraborti A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology. 1996;225:300–305. doi: 10.1006/viro.1996.0604. [DOI] [PubMed] [Google Scholar]
- Leung A. K., Calabrese J. M., Sharp P. A. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Vodicka M. A., Lucero G., Svitkina T. M., Borisy G. G., Emerman M., Hope T. J. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk J. N., Schuyler S. C., Liu J. Y., Evans J. G., Salmon E. D., Pellman D., Bloom K. The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell. 2004;15:1519–1532. doi: 10.1091/mbc.E03-09-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C., Brandt S. M., Lucero G., Landau N. R. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U., Swiggard W. J., Malim M. H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens C. M., Song B., Perron M. J., Yang P. C., Stremlau M., Sodroski J. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens C. M., Yang P. C., Gottlinger H., Sodroski J. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 2005a;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D., Hatziioannou T., Zhang F., Cowan S., Bieniasz P. D. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 2005b;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus T., Hartley J. W., Rowe W. P. A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1-sensitive and resistant cell cultures. Virology. 1975;65:333–342. doi: 10.1016/0042-6822(75)90039-2. [DOI] [PubMed] [Google Scholar]
- Pryciak P. M., Varmus H. E. Fv-1 restriction and its effects on murine leukemia virus integration in vivo and in vitro. J. Virol. 1992;66:5959–5966. doi: 10.1128/jvi.66.10.5959-5966.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera O. J., Song C. S., Centonze V. E., Lechleiter J. D., Chatterjee B., Roy A. K. Role of the promyelocytic leukemia body in the dynamic interaction between the androgen receptor and steroid receptor coactivator-1 in living cells. Mol. Endocrinol. 2003;17:128–140. doi: 10.1210/me.2002-0165. [DOI] [PubMed] [Google Scholar]
- Sawyer S. L., Wu L. I., Emerman M., Malik H. S. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R., Sakai H., Kawamura M., Tokunaga K., Adachi A. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 1995;76:2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- Short K. M., Cox T. C. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- Song B., Diaz-Griffero F., Park D. H., Rogers T., Stremlau M., Sodroski J. TRIM5alpha association with cytoplasmic bodies is not required for antiretroviral activity. Virology. 2005a;343:201–211. doi: 10.1016/j.virol.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Song B., Gold B., O'Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 2005b;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Javanbakht H., Perron M., Park D. H., Stremlau M., Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J. Virol. 2005c;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens C. M., Hope T. J. Mobility of the human immunodeficiency virus (HIV) receptor CD4 and coreceptor CCR5 in living cells: implications for HIV fusion and entry events. J. Virol. 2004;78:9573–9578. doi: 10.1128/JVI.78.17.9573-9578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M., Perron M., Welikala S., Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T. J., Boyer B., Hu W., Baker K. E., Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G., Bock M., Martin S., Takeuchi Y., Stoye J. P., Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Lei Y. P., Zhou Y. L., Okamoto C. T., Snead M. L., Paine M. L. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11) Cell. Mol. Life Sci. 2005;62:1038–1046. doi: 10.1007/s00018-005-4547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap M. W., Nisole S., Lynch C., Stoye J. P. TRIM5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap M. W., Nisole S., Stoye J. P. A single amino acid change in the SPRY domain of human TRIM5alpha leads to HIV-1 restriction. Curr. Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Zhu J., Koken M. H., Quignon F., Chelbi-Alix M. K., Degos L., Wang Z. Y., Chen Z., de The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.