Abstract
Previously, FRET confocal microscopy has shown that polymeric IgA-receptor (pIgA-R) is distributed in a clustered manner in apical endosomes. To test whether different membrane-bound components form clusters during membrane trafficking, live-cell quantitative FRET was used to characterize the organization of pIgA-R and transferrin receptor (TFR) in endocytic membranes of polarized MDCK cells upon internalization of donor- and acceptor-labeled ligands. We show that pIgA-R and TFR complexes form increasingly organized clusters during cotransport from basolateral to perinuclear endosomes. The organization of these receptor clusters in basolateral versus perinuclear/apical endosomes is significantly different; the former showing a mixed random/clustered distribution while the latter highly organized clusters. Our results indicate that although both perinuclear and apical endosomes comprise pIgA-R and TFR clusters, their E% levels are significantly different suggesting that these receptors are packed into clusters in a distinct manner. The quantitative FRET-based assay presented here suggests that different receptor complexes form clusters, with diverse levels of organization, while being cotransported via the polarized endocytic pathways.
INTRODUCTION
Understanding how proteins are correctly localized and why that process goes awry in diseases is a fundamental question in biomedical research. Many diseases, such as cystic fibrosis and hypercholesterolemia, are due to defects in protein sorting and transport. The mechanism of protein transport has been analyzed but the molecular and biophysical basis for clustering at all levels of membrane trafficking pathways remains to be defined. Here, quantitative Förster resonance energy transfer (FRET) confocal imaging has been used to address the dynamics of protein clustering during receptor-mediated endocytic membrane trafficking in live polarized epithelial cells.
Polarized endocytic pathways include several compartments with different biochemical and molecular properties; for example, basolateral and apical early endosomes (BEE and AEE, respectively), common endosomes (CE), and apical recycling endosomes (ARE; see Figure 1; Rojas and Apodaca, 2002; Mostov et al., 2003). For this study, filter-grown polarized epithelial Madin-Darby canine kidney (MDCK) cells, stably transfected with polymeric IgA-receptor (pIgA-R) and transferrin receptor (TFR; MDCK-PTR cells), are used because they express TFR and pIgA-R at moderate levels and have been extensively used to characterize the polarized trafficking of these receptors using their respective fluorophore-labeled ligands, i.e., holo-transferrin (Tfn) and dimeric IgA or pIgA-R ligand (Apodaca et al., 1994; Barroso and Sztul, 1994; Gibson et al., 1998; Brown et al., 2000; Wang et al., 2000; Rojas and Apodaca, 2002; Mostov et al., 2003; see Figure 1A). Both TFR and pIgA-R are involved in physiologically significant cellular processes, such as iron uptake for TFR (Lawrence et al., 1999; Cheng et al., 2004) and secretory immunity for pIgA-R (Rojas and Apodaca, 2002). TFR undergoes basolateral recycling via BEE, CE, and AEE (see Figure 1A, arrows 1, 2b, 2a, 3a, 3b, and 5b; Odorizzi et al., 1996; Gibson et al., 1998; Sheff et al., 1999; Leung et al., 2000). Entrance of basolaterally internalized TFR into the AEE suggests that these endosomes play a role in the basolateral recycling of TFR (Leung et al., 2000). Although, the majority of pIgA-R transcytoses to the apical PM via the CE and the ARE (see Figure 1A, arrows 3c and 6), a smaller, but significant minority, recycles back to the basolateral surface together with TFR (see Figure 1A, arrow 2a or 3a; Apodaca et al., 1994; Barroso and Sztul, 1994; Gibson et al., 1998; Brown et al., 2000; Leung et al., 2000; Wang et al., 2000). In contrast to TFR, which is predominantly located at the basolateral PM, pIgA-R is also found at the apical PM (Figure 1A, arrow 4). On apical internalization, pIgA-R is delivered to the AEE and CE (Figure 1A, arrows 4, 5a, and 5b) and recycled back to the apical PM via the ARE (Figure 1A, arrow 6; Barroso and Sztul, 1994; Leung et al., 2000). Basolateral-to-apical transcytosis and apical PM localization, and internalization of TFR are only achieved upon treatment with brefeldin A (BFA; see Figure 2A), a fungal metabolite that disrupts the polar sorting of endocytic receptors (Wan et al., 1992; Futter et al., 1998; Wang et al., 2001).
Figure 1.
Endocytic/transcytotic membrane trafficking pathways in polarized MDCK-PTR cells. (A) Arrows indicate natural endocytic/transcytotic membrane trafficking pathways available to fluorescently labeled pIgA-R ligand and Tfn in MDCK-PTR polarized cells. pIgA-R ligand and Tfn bind pIgA-R and TFR, respectively, at the basolateral PM and those receptor-ligand complexes are delivered to the BEE (arrows 1), and then to the CE (arrows 2b). Whereas TFR complexes are recycled back to the basolateral PM together with a minor fraction of pIgA-R complexes (arrow 2a), the majority of pIgA-R complexes are transcytosed to the apical PM via the CE and ARE (arrows 3b and 6). On internalization from the apical PM, pIgA-R complexes are delivered to the AEE (arrow 4) and CE (arrow 5b) and recycled back to the cell surface (arrows 5a and 6). Cell images (Figure 2B) are collected using confocal microscopy at different cell heights: the basolateral (blue rectangle), perinuclear (green rectangle), and apical (red rectangle) regions. Cy3- or Alexa555-labeled Tfn-TFR complexes, red arrows; Alexa488-labeled pIgA-R-ligand complexes, green arrows. AEE, apical early endosome; ARE, apical recycling endosome; BEE, basolateral early endosome; CE, common endosome; PM, plasma membrane; TJ, tight junction. (B) Cross sections of the polarized endocytic morphology show that the majority of the BEE localizes at the periphery of the cell in the basolateral region, whereas the CE localizes predominantly in the perinuclear region and the ARE above the nucleus and below the apical PM. This model also shows the potential contamination of perinuclear and apical endosomes with peripherally localized BEE and AEE, respectively.
Figure 2.
Characterization of polarized endocytic compartments. Alexa555-Tfn (red staining) was cointernalized for 30 min at 37°C into the basolateral surface of polarized MDCK cells in the presence [Treatment (+BFA), f–k] or absence [Control (-BFA), a–e] of BFA. Then, immunofluorescence was performed using anti-Rab11 (a–c and f–h), a ARE marker, or anti-EEA1 (d–e and i–k), an early endosomal marker, and secondary antibodies labeled with Alexa488 (green staining). Then, confocal imaging was performed and images were collected at different cell regions. Apical, 10–9 μm above the insert filter (a, d, f, and i); perinuclear, 6–7 μm above the insert filter (b, d, g, and j); and basolateral, 2–4 μm above the insert filter (c, e, h, and k). Bar, 5 μm. Yellow staining indicates overlap between the TFR and the Rab11 or EEA1 distribution patterns.
An essential prerequisite for efficient membrane protein trafficking is the segregation of protein molecules within continuous membrane sheets to form microdomains with specific protein compositions. The sorting of membrane components from fluid-phase molecules has been suggested to depend on the endosomal geometry and iterative fractionation (Geuze et al., 1987; Dunn et al., 1989; Mayor et al., 1993). On the other hand, membrane-bound receptors may undergo a signal-mediated enrichment upon exiting the PM or endosomes to be delivered to their specific destinations (Maxfield and McGraw, 2004). Thus, an important step in endocytic receptor trafficking could be the concentration of receptor-ligand complexes in transport intermediates.
The organization of receptors in endocytic membranes has been analyzed using electron microscopy (Geuze et al., 1984; Stoorvogel et al., 1989). Demonstration that gold-labeled Tfn or gold-labeled pIgA-R ligand concentrate in discrete packaging sites, such as in clathrin-coated buds, in polarized MDCK cells has been hard to achieve because of technical difficulties in loading the endosomal system close to saturation (Geuze et al., 1984; Futter et al., 1998; Gibson et al., 1998). In polarized MDCK cells, some studies have suggested that receptors are organized in a nonuniform manner in endocytic membranes (Futter et al., 1998), whereas other studies have indicated that receptors are distributed in a random manner throughout endocytic tubules and vesicles (Gibson et al., 1998).
In this study, we used quantitative FRET confocal microscopy in live cells to investigate the nature of receptor organization in membranes of polarized endosomes Membrane-bound receptors can either be assembled in clusters, distributed randomly or possibly exhibit a continuum between these two states. The relationship between FRET and the geometric distribution of labeled membrane components is highly nonlinear. Thus, mathematical modeling has played a key role in identifying relevant parameters and developing criteria to reveal the presence of clustered receptors (Kenworthy and Edidin, 1998; Varma and Mayor, 1998; Kenworthy et al., 2000; Tcherkasskaya et al., 2002). The quantitative FRET assay presented here uses the efficiency of the energy transfer (E%) and its relationship to donor and acceptor (A) fluorescence intensities to establish whether the close proximity between membrane proteins is due to random associations (“molecular crowding”) or specific nonrandom clustering (Zimet et al., 1995; Kenworthy and Edidin, 1998; Kenworthy et al., 2000; Pentcheva and Edidin, 2001; Zacharias et al., 2002; Wallrabe et al., 2003a,b, 2006; Bhatia et al., 2005; Wallrabe and Barroso, 2005). Our results show that clusters, comprising these two receptor-ligand complexes during cotransport, are formed with increasingly tighter organizations along the polarized endocytic pathway of live MDCK-PTR cells.
MATERIALS AND METHODS
Culture of MDCK-PTR Cells on Filter Inserts
MDCK-PTR cells stably transfected with rabbit pIgA-R and human TFR (Brown et al., 2000; Wang et al., 2000) were grown to confluence in 100-mm cell culture dishes, trypsinized, centrifuged, and resuspended in DMEM/10% fetal bovine serum/Pen-Strep (Barroso and Sztul, 1994). Approximately 300,000 cells were placed on top of an inverted Transwell Clear insert (Corning Costar, Cambridge, MA), which allowed for their direct visualization through a coverslip using an inverted microscope (Brown et al., 2000; Wallrabe et al., 2003a). After 3–4 d in culture, the fully polarized monolayer was immediately used according to different internalization protocols (Barroso and Sztul, 1994; Wallrabe et al., 2003a). Cellular polarity was established by the reduced apical uptake and basolateral staining of apically internalized fluorophore-labeled Tfn (<10%) and ricin (<5%), using confocal microscopy (Barroso and Sztul, 1994).
Fluorophore-labeled Ligands
Alexa488 (donor) was conjugated to pIgA-R pseudoligands ([Fab]2 fragments raised against rabbit pIgA-R extracellular domain) according to the manufacturer's (Invitrogen, Carlsbad, CA) protocol (Barroso and Sztul, 1994; Wallrabe et al., 2003a). Conjugation of Cy3 (acceptor) was performed, according to the manufacturer's (GE Healthcare, Waukesha, WI) instructions, to iron-free human Tfn (apo-Tfn; Sigma, St. Louis, MO), followed by iron loading with ferric ammonium citrate as described previously (Barroso and Sztul, 1994). Alexa488 (donor) or Alexa555 (acceptor) bound to human Tfn were purchased from Invitrogen and preloaded with iron. Previously, we have shown that MDCK cells expressing only pIgA-R (MDCK-PWE cells) bound iron-loaded human Tfn at very low levels because of the inability of dog TFR to recognize human Tfn (Barroso and Sztul, 1994). Furthermore, iron-loaded dog Tfn was internalized at low levels into MDCK cells in a TFR-dependent and pIgA-R-independent manner (Barroso and Sztul, 1994). Significant apical re-endocytosis upon release of ligands into the apical media does not happen under our internalization conditions (Barroso and Sztul, 1994; Wallrabe et al., 2003a).
Internalization of Fluorophore-labeled Ligands
Inserts with a fully polarized MDCK-PTR cell monolayer were washed with PBS2+, equilibrated with MEM/HEPES/BSA at 37°C, and pretreated 15 min with or without 10 μM BFA. Then, these cells were internalized for 30 min at 37°C with different amounts of Alexa488-pIgA-R ligands and/or Cy3-Tfn/Alexa555-Tfn (40–100 μg/ml) from the basolateral and/or apical PM in the presence or absence of 10 μM BFA. High concentrations of pIgA-R ligand and Tfn helped minimize the presence of empty receptors. Cells were washed and fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS), as described previously (Barroso and Sztul, 1994; Wallrabe et al., 2003a,b, 2006). For live cells, the protocol was exactly the same, except that each insert followed the regimen individually and was imaged immediately at room temperature. As a positive control for random distribution of substrate-bound proteins, Alexa488-Tfn and Alexa555-Tfn were incubated for 30 min at room temperature onto polylysine-coated glass coverslips, fixed with 4% paraformaldehyde, washed in PBS, and imaged within 48 h using confocal microscopy. As a positive control for membrane protein clustering, Alexa488-Tfn and Alexa555-Tfn were internalized for 30 min at 37°C into polarized MDCK-PTR cells, as described previously (Wallrabe et al., 2006).
For FRET assay purposes (filter-bound cells or polylysine-bound coverslips), three different samples were used: double-labeled specimens, containing D- and A-labeled ligands (pIgA-R and/or Tfn) and two single-label specimens containing either D- or A-labeled ligands; the single-label reference samples were used to establish spectral-bleedthrough (SBT) levels.
Immunofluorescence of Polarized Cells
Polarized MDCK-PTR cells were internalized from the basolateral PM with Alexa555-Tfn for 30 min at 37°C and treated with or without BFA, as described above. Then, cells were washed, fixed with 4% paraformaldehyde, and processed for immunofluorescence, as described previously (Andrade et al., 2004). Monoclonal antibodies against EEA1 (BD Biosciences, San Jose, CA) and rabbit polyclonal against Rab11 (Invitrogen/Zymed, South San Francisco, CA) were used at 1:100 and 1:40 dilutions, respectively. Cells were visualized using a Zeiss 510META LSM microscope (Thornwood, NY, as described below), and Z-series with a 0.5-mm interval were collected. Image analysis of confocal images was performed using Adobe Photoshop 5.5 (San Jose, CA). To allow intensity comparisons, we used similar conditions to collect and manipulate images within each antibody experiment.
Laser Scanning FRET Microscopy
For data collection, the specimen was positioned in a small chamber created by a coverslip between two metal rings, filled with a small amount of PBS (fixed cells) or MEM/HEPES/BSA media (live cells) and placed on the microscope stage. Nikon PCM 2000 (Melville, NY) or Bio-Rad Radiance 2100 laser scanning confocal microscopes (Richmond, CA), respectively, mounted on a Nikon TE200 or TE300 epifluorescence microscopes and equipped with Argon (488 nm) and Green HeNe (543 nm) lasers and emission filters, 515/30 nm and LP590 nm, were used with a 60× water immersion lens, 1.2 NA to collect images for FRET processing. For image acquisition and processing, SimplePCI software (Compix, Cranberry Township, PA) was used to drive the Nikon hardware, and LaserSharp2000 was used for the Bio-Rad microscope.
Data Collection
Two-color Z-series with a 0.5-μm vertical step were collected to check cell height (10–15 μm) and to select focal planes at different cell heights. Optimal PMT and accumulation settings and laser power levels were established in this pre-FRET-acquisition phase. With the zoom setting at ≈2×, images of the double-labeled specimen were taken with the Green HeNe laser, 543-nm excitation, i.e., acceptor excitation, and the acceptor emission channel (LP590 nm) followed by imaging with the argon laser 488-nm excitation, i.e., donor excitation, and the donor (515/30 nm) or the acceptor (LP590 nm) emission channels. The single-labeled acceptor specimen followed the same protocol. The image of the single-labeled donor specimen at acceptor excitation wavelength was collected to verify the instrument cross-talk in both channels. Images of all three types of specimen were taken under identical imaging conditions; PMT gain and black-level were set at identical values to collect data simultaneously in both channels into 1024 × 1024- or 512 × 512-pixel eight-bit images. Bleaching was undetectable during the short exposure to collect the final image. The Bio-Rad Radiance 2100 confocal system was used for live cell image acquisition. A custom macro was used to toggle between donor and acceptor excitation lasers and thus minimize the delay in switching lasers. The Nikon PCM2000 microscopes were used to collect images from fixed cells.
Postacquisition Data Generation
First, images were background-subtracted and processed by the PFRET software, which removed donor and acceptor SBT pixel-by-pixel on the basis of matched fluorescence levels between the double-label specimen and single-label reference specimens, using seven images: two single-label donor reference images (donor excitation/donor channel and acceptor channel); two single-label acceptor reference images (donor and acceptor excitation, both in the acceptor channel); three double-label images (donor excitation/donor and acceptor channel, acceptor excitation/acceptor emission; Elangovan et al., 2003; Wallrabe et al., 2003a, b). The three double-labeled images were named as follows: quenched donor (qD), i.e., the donor excitation/donor emission; acceptor (A), i.e., acceptor excitation/acceptor emission; and uncorrected FRET (uFRET), i.e., donor excitation/acceptor emission. The pixel-by-pixel correction used to generate the processed FRET (PFRET) image was actually based on the average value of narrow fluorescence ranges, for more efficient running of the correction algorithm (Elangovan et al., 2003). In our case, we chose the average of 12 fluorescence units per range, i.e., 0–12, 13–24, etc., continuing to the highest fluorescent units in the image. Using the average of even narrower ranges did not improve the sensitivity.
Postacquisition Data Analysis
Our measurements fall into the category of “apparent” E%, which is dependent on Förster-type energy transfer E (Wouters et al., 1998; Lakowicz et al., 1999), but is also influenced by the concentrations of those donor or acceptor molecules that are not involved in energy transfer; for brevity we will use E%, instead of “apparent” E%. E% is an expression of the energy transfer as a percentage of the unquenched donor, d = qD + γ·PFRET, as described in the following equation: E% + 100·(γ·PFRET)/D, i.e., E% = 100·1 − (qD/D) (Elangovan et al., 2003; Wallrabe et al., 2003a, 2006; Bonamy et al., 2005; Wallrabe and Barroso, 2005). The γ factor, which is a function of the quantum yield of the fluorophores and the spectral sensitivity of the detection setup, plays a crucial role in recording precise E% and distances between fluorophores. Because the excitation efficiencies (ε), quantum yields of the fluorophore molecules and the detection efficiencies (Q) remain constant throughout the experiments, and the γ factor does not affect the answers that FRET-based clustering analysis seeks. Therefore, for simplicity we used γ = 1, as described previously (Elangovan et al., 2003; Wallrabe et al., 2003a,b, 2006). Nevertheless, different microscopes using different imaging collection instruments and settings will by definition have distinct γ factors. Therefore, the relative E% values differ for data collected using distinct microscope systems. Such a case is demonstrated when comparing the perinuclear data set collected using different microscopic settings (cf. Figures 4C vs. 6B). Thus, different data sets can only be compared when collected using identical microscopy settings, for example, Figures 4, C and D, and 5D or Figures 6, A and B, 7, C and D, and 8, A and B.
Figure 4.
FRET-based assay distinguishes between clustered and random membrane protein distributions. (A) A random organization model, E% is dependent on acceptor levels but not on D/A ratios. (B) Alexa488- and Alexa555-Tfn are bound to polylysine-covered coverslips, imaged by confocal microscopy and processed for FRET analysis. The Acceptor, D/A and E% values were extracted for a wide variety of ROIs and plotted against Acceptor levels at D/A ≈ 1 (squares) or D/A ≈ 2 (triangles). E% shows a clear dependency on Acceptor levels and independency from D/A. (C) In a clustered organization model, E% is independent from acceptor levels but rises with decreasing D/A ratios. (D) Alexa488- and Alexa555-Tfn are bound to TFR at the basolateral PM and internalized for 30 min at 37°C, imaged by confocal microscopy and processed for FRET analysis. The Acceptor, D/A and E% values were extracted for a wide variety of ROIs and plotted against Acceptor levels as described for B. In B and D, trendlines are shown as visual helpers (D/A ≈ 1, dotted line; D/A ≈ 2, dashed line). E% is largely independent from Acceptor levels and decreases with increasing D/A ratios. (E) Acceptor values at D/A ≈ 1 were split into three groups (low, medium, and high), and the respective average E% values were plotted for these Acceptor groups. Although the E% values from polylysine-bound Tfn samples positively correlated to A levels, E% from TFR-bound Tfn remains similar at all Acceptor levels, as expected for random and clustered distributions, respectively. Error bars, 95% confidence interval.
Figure 6.
Perinuclear TFR and pIgA-R complexes show a clustered organization upon internalization from opposite PMs in the presence of BFA. (A) The organization of pIgA-R and TFR complexes cointernalized from opposite PMs in the presence of BFA was assayed at the perinuclear region (green rectangle) using FRET confocal microscopy. (B) Live-cell confocal FRET imaging was performed at the perinuclear region of polarized MDCK-PTR cells upon internalization of Cy3-Tfn and Alexa488-pIgA-R ligand from the basolateral and apical surfaces, respectively, in the presence of BFA. Pseudocolor images depict the pixel-by-pixel distribution of D/A (a) Acceptor (b), and E% (c) levels at the perinuclear region. Examples of selected ROIs are shown as white squares. Bar, 5 μm. (C) E%, D/A and Acceptor levels were calculated for a wide range of perinuclear ROIs. E% values were plotted as a function of Acceptor levels for D/A ≈ 1 (□) and for D/A ≈ 2 (▵). Trendlines are shown as visual helpers (D/A ≈ 1, dotted line; D/A ≈ 2, dashed line). E% is largely independent of Acceptor levels and increases with decreasing D/A ratios, suggesting that TFR and pIgA-R complexes show a clustered organization in perinuclear endosomes, independently of BFA and internalization protocols. (D) Perinuclear E% using cointernalization from basolateral PM in the absence of BFA (◇; Figure 4C) or from opposite surfaces in the presence of BFA (○) were plotted as a function of Acceptor levels for D/A ≈ 1. Trendlines are shown as visual helpers (−BFA Bas. PM inter., dotted line; +BFA Opp. PM inter., dashed line). These two data sets are not significantly different (p > 0.001) using an ANCOVA statistical analysis (Table 2).
Figure 5.
pIgA-R and TFR complexes form clusters in perinuclear, but not in basolateral endosomes. (A) The organization and distribution of pIgA-R and TFR complexes in endosomes that are localized at the basolateral (blue rectangle) and perinuclear (green rectangle) regions was assayed using FRET confocal microscopy after cointernalization of Cy3-Tfn and Alexa488-pIgA-R ligand from the basolateral PM. (B) Live-cell confocal FRET imaging was performed on polarized MDCK-PTR cells basolaterally cointernalized with Alexa488-pIgA-R ligand and Cy3-Tfn. Pseudocolor images, processed using the PFRET correction algorithm, depict the pixel-by-pixel distribution of Acceptor (b and e), D/A (a and d), and E% (c and f) levels at the basolateral (d–f) and perinuclear (a–c) regions. Examples of selected ROIs that were selected according to the definitions of basolateral and perinuclear endocytic regions are shown as white squares. Bar, 5 μm. (C and D) E%, D/A and Acceptor levels were calculated for a wide range of ROIs (white squares). E% values were plotted as a function of Acceptor levels for D/A ≈ 1 (□) and for D/A ≈ 2 (▵). In C and D, trendlines are shown as visual helpers (D/A ≈ 1, dotted line; D/A ≈ 2, dashed line). (C) E% is largely independent of Acceptor levels and increases with decreasing D/A ratios, suggesting a clustered organization of TFR and pIgA-R complexes in perinuclear endosomes. (D) E% shows a variable dependency on Acceptor levels depending on D/A, indicating a mixed/random organization of TFR and pIgA-R complexes in basolateral endosomes.
Figure 7.
BFA affects receptor organization in basolateral endosomes but not in perinuclear endosomes. (A) E% values at D/A ≈ 1 were plotted as a function of Acceptor levels for basolateral ROIs collected from images generated upon internalization from basolateral PM in the presence (diamonds) or absence (circles) of BFA. Trendlines are shown as visual helpers (−BFA, dotted line; +BFA, dashed line). As expected, E% is dependent of Acceptor levels in basolateral endosomes. These two data sets are significantly different (p > 0.001) using an ANCOVA statistical analysis (Table 2) with +BFA E% levels slightly higher than −BFA E% levels. (B) E% values at D/A ≈ 1 were plotted as a function of Acceptor levels for perinuclear ROIs collected from images generated upon internalization from basolateral PM in the presence (diamonds) or absence (circles) of BFA. Trendlines are shown as visual helpers (−BFA, dotted line; +BFA, dashed line). As expected, E% is independent of Acceptor levels in perinuclear endosomes. These two data sets are not significantly different (p > 0.001) using an ANCOVA statistical analysis (Table 2).
Figure 8.
Perinuclear and apical TFR and pIgA-R complexes show a clustered organization. (A) The organization of pIgA-R and TFR complexes cointernalized from basolateral PMs in the presence of BFA was assayed at the apical (red rectangle) and perinuclear (green rectangle) regions using FRET confocal microscopy. (B) Live-cell confocal FRET imaging was performed at the perinuclear and apical regions of polarized MDCK-PTR cells upon internalization of Cy3-Tfn and Alexa488-pIgA-R ligand from the basolateral PM, in the presence of BFA. Pseudocolor images depict the pixel-by-pixel distribution of Acceptor (b and e), D/A (a and d), and E% (c and f) levels at the apical (a–c) and perinuclear (d–f) regions. Examples of selected ROIs are shown as white squares. Bar, 5 μm. (C and D) E%, D/A and Acceptor levels were calculated for a wide range of ROIs. E% values were plotted as a function of Acceptor levels for D/A ≈ 1 (□) and for D/A ≈ 2 (▵). Trendlines are shown as visual helpers (D/A ≈ 1, dotted line; D/A ≈ 2, dashed line). E% is largely independent of Acceptor levels and increases with decreasing D/A ratios, suggesting that TFR and pIgA-R complexes present a clustered organization in perinuclear and apical endosomes.
A custom-written analysis program was able to select pixels of the eight-bit gray-scale fluorescence intensities of uFRET, A and qD images ranging between 0 at the lower bound and at the higher bound one below [255 minus background value] to exclude any saturated pixels. Under our imaging conditions, there were very few saturated pixels (Wallrabe et al., 2003a, 2006). Then, appropriate regions of interest (ROIs) were visually selected from the uFRET image. These ROIs were subsequently applied to the other images to extract the fluorescence values. The values which include PFRET (actual energy transfer levels as per the PFRET SBT correction algorithm), qD and A levels were transferred to an Excel spreadsheets (Microsoft, Redmond, WA) for calculation of additional parameters such as E%, D, and D/A ratios. These values were averaged over ROIs containing 50–500 pixels. For further FRET clustering analysis, E% values that correspond to A or D values of 10–80 Gy-scale units per pixel were selected to avoid the noise of very high or very low A or D fluorescence intensities on E% and to exclude outlier values (<5%), i.e., individual values that lie outside the overall observed range (Bhatia et al., 2005; Wallrabe et al., 2006).
Statistical Analysis
To provide insights as to whether E% is affected by increasing levels of A at specific D/A ranges, the data were arranged into several D/A and A ranges. For D/A ranges, we used the following ranges: D/A ≈ 1 ∈ [1/] and D/A ≈ 2 ∈ [], which corresponds to categories with a twofold increase, centered around 1 and 2, respectively. Thus, D/A ≈ 1 ranged from D/A values of 0.7–1.4, whereas D/A ≈ 2 ranged from values of 1.4–2.8. In bar charts, the gray-scale intensity cohorts for A were defined by splitting in three the range defined by the lowest and highest value of A (Low = [20;40], Medium = [40;60], High = [60;80]). This approach can only be used to compare E% values for different data sets that show overlap between the high, medium and high ranges of A (Figures 3E and 8B).
Figure 3.
A morphology-based system to discriminate between polarized endosomes. (A) Basolateral-to-apical endocytic/transcytotic pathway of basolaterally cointernalized TFR and pIgA-R complexes in the presence (+) or absence (−) of BFA, which missorts TFR to the apical PM, as shown by the red arrows 3b, 5a, and 6. (B) Alexa488-pIgA-R ligand (green staining) and Cy3-Tfn (red staining) were cointernalized for 30 min at 37°C into the basolateral surface of polarized MDCK cells in the presence (+BFA, b, d, and f) or absence (−BFA, a, c and e) of BFA. Then, confocal imaging was performed and images were collected at different cell regions. Apical, 10 μm above the insert filter (a and b); perinuclear, 7.5 μm above the insert filter (c and d); and basolateral, 3 μm above the insert filter (e and f). Bar, 5 μm. Yellow staining indicates overlap between the pIgA-R ligand and TFR distribution patterns. (C) BFA effect on basolateral, perinuclear, and apical endosomes containing TFR-Tfn and pIgA-R-ligand complexes. The relative amount of fluorescently labeled pIgA-R ligand and Tfn is evaluated by the average fluorescence intensity of a wide variety of ROIs that were collected and discriminated into basolateral, perinuclear, and apical endocytic regions according to the morphology-based system based on their location versus the filter insert, PM, and nucleus (Figure 1B). ROIs include endocytic punctate structures and contain 50–500 pixels. In the presence of BFA, a significant portion of TFR complexes redistributes to the apical region (+BFA, TFR bars), due to the elimination of their basolateral polarized sorting, which is clearly detected in the absence of BFA (−BFA, TFR bars). Error bars, 95% confidence interval.
To statistically analyze whether or not E% was dependent of the level of A at specific D/A ranges, we used the correlation coefficients (r value) and the slope (s values) as indicators (Table 1). The closer the r values are to 0, the less E% is dependent on A levels; the closer r values are to 1, the more E% depends on A levels. Another important parameter to determine whether E% depends on A levels is the slope of the linear regression, because a slope close to zero may have a high correlation, but suggest that E% does not depend on A. To analyze whether the E% cohorts at different A levels (10–19, 20–29, 30–39, 40–49, etc.) were significantly different or not, we used the ANOVA with a single-factor analysis from the Excel data analysis software package to establish p values between groups; significance of the statistic differences between the groups was indicated by p < 0.001.
Table 1.
Summary of the FRET data to assay the organization of TFR/pIgA-R receptor-ligand clustering in basolateral, perinuclear, and apical endosomes
| Ligands |
||||||||
|---|---|---|---|---|---|---|---|---|
| Polylysine-bound Tfn |
TFR-bound Tfn |
pIgA-R ligand and Tfn bound to pIgA-R and TFR at the PM |
||||||
| NA |
Basolaterala |
Basolateral |
Perinuclear |
Apical |
||||
| −BFA | −BFA | −BFAa | +BFAa | −BFAa | +BFAa | +BFAa | +BFAb | |
| E% vs. A | r = 0.72c | r = 0.10c | r = 0.77c | r = −0.22c | r = −0.32c | |||
| at D/A ≈ 1 | s = 0.50c | s = 0.01c | s = 1.05c | s = −0.13c | s = −0.07c | |||
| r = 0.48d | r = 0.40d | r = 0.06d | r = 0.01d | r = 0.13d | ||||
| s = 0.39d | s = 0.40d | s = 0.04d | s = 0.01d | s = 0.09d | ||||
| p = 10−161 (+)c | p = 5.5 × 10−1 (−)c | p = 5.6 × 10−4 (+)c | p = 2.6 × 10−1 (−)c | p = 1.9 × 10−1 (−)c | ||||
| p = 1.5 × 10−5 (+)d | p = 1.0 × 10−3 (+)d | p = 6.6 × 10−1 (−)d | p = 7.8 × 10−1 (−)d | p = 5.7 × 10−1 (−)d | ||||
| E% vs. D/A | Independent | Dependent | Mixed behavior | Mixed behavior | Dependent | Dependent | Dependent | Dependent |
| Organization | Random | Clustered | Random/clustered | Random/clustered | Clustered | Clustered | Clustered | Clustered |
Endocytic regions: basolateral, perinuclear, and apical; treatments, −BFA and +BFA. NA, not available. r, correlation r-value; s, slope of the linear regression; p, critical value obtained from an ANOVA single-factor; (+), significant with p < 0.001; (−), not significant.
a Internalization from basolateral PM.
b Internalization from opposite PMs.
c Data from image sets collected with the Bio-Rad confocal microscope (live-cell samples).
d Data collected with the Nikon confocal microscope (fixed samples).
To compare different data sets (for example, treated and untreated with BFA or different endosome groups), we did an ANCOVA using [R] to assess whether the treatment (alone) has an effect or not on the distribution. In a first model, we verified that the treatment did not significantly modify the slope of E% = f(A) (cf. p value for A × Variable in Table 2). Then, in a reduced model, where the data are fitted with a common slope, we assessed whether the treatment had an effect affected, by testing if the intercept at the origin was modified (cf. p value for Variable in Table 2). It is important to stress that directly comparable data sets were collected and processed under identical microscopy settings and FRET conditions (Figures 4, C and D, and 5D or Figures 6, A and B, 7, C and D, and 8, A and B; Table 2); data sets that are not collected and processed under identical microscopy settings and FRET conditions were not directly compared (Figures 4C vs. 6B). The combination of correlation coefficients, slope values, and ANOVA and ANCOVA analyses allows us to make some powerful deductions about the nature of the distribution of receptor-ligand complexes in endocytic membranes (Wallrabe et al., 2003a,b, 2006).
Table 2.
Effect of BFA treatment, endocytic region differences, and internalization protocols on TFR/pIgA-R receptor-ligand clustering
| Basolateral±BFAad |
Perinuclear±BFAad |
Perinuclear −BFAac vs. +BFAcb |
Perinuclear vs. Apical +BFAad | |||||
|---|---|---|---|---|---|---|---|---|
| D/A ≈ 1 | D/A ≈ 2 | D/A ≈ 1 | D/A ≈ 2 | D/A ≈ 1 | D/A ≈ 2 | D/A ≈ 1 | D/A ≈ 2 | |
| A × variable | p = 9.3 × 10−1 (−) | p = 4.9 × 10−1 (−) | p = 7.1 × 10−1 (−) | p = 7.8 × 10−1 (−) | p = 4.0 × 10−1 (−) | p = 4.2 × 10−2 (−) | p = 4.0 × 10−1 (−) | p = 3.3 × 10−1 (−) |
| Variable | p = 2.3 × 10−6 (+) | p = 1.3 × 10−9 (+) | p = 3.4 × 10−1 (−) | p = 3.5 × 10−1 (−) | p = 7.0 × 10−3 (−) | p = 3.2 × 10−3 (−) | p = 3.8 × 10−6 (+) | p = 1.7 × 10−5 (+) |
| Conclusions: effect of BFA on clustering | Increases clustering | Increases clustering | No effect | No effect | No effect and internalization on clustering | No effect and internalization on clustering | Decreased clustering in the apical region | Decreased clustering in the apical region |
Endocytic regions: basolateral, perinuclear, and apical; treatments, −BFA and +BFA. A × variable, interaction between A level and variable (i.e., treatment, internalization region, or endocytic region) monitored (test if slopes are identical); Variable, effect of variable in a reduced model where A × variable interaction is removed (test if regression with common slope have the same intercept); p, critical value obtained from an ANCOVA; (+), significant with p < 0.001; (−), not significant.
a Internalization from basolateral PM.
b Internalization from opposite PMs.
c Data from image sets collected with the Bio-Rad confocal microscope (live-cell sample).
d Data collected with the Nikon confocal microscope (fixed samples).
RESULTS
Previously, we have used quantitative confocal FRET to demonstrate that pIgA-R-ligand complexes are organized in a clustered manner in the apical endosomes of polarized MDCK cells (Barroso and Sztul, 1994; Elangovan et al., 2003; Wallrabe et al., 2003a,b, 2006; Wallrabe and Barroso, 2005). These results strongly suggest that the clustering of membrane-bound receptor-ligand complexes is an important part of their protein sorting and transport in polarized cells. To test this hypothesis, quantitative confocal FRET has been used to determine whether TFR and pIgA-R complexes form differently organized clusters in various polarized endosomes during their trafficking in live MDCK-PTR cells.
Morphology-based Assay To Discriminate between Different Polarized Endosomes
A strategy based on cell morphology has been developed to identify distinct endocytic compartments in polarized epithelial cells using FRET confocal microscopy (Barroso and Sztul, 1994; Brown et al., 2000; Wang et al., 2000, 2001; Maxfield and McGraw, 2004; Rodriguez-Boulan et al., 2005). A two-step approach was used: Various endocytic compartments were identified in the polarized MDCK-PTR cell system in the presence or absence of BFA by determining their “geographical” localization in comparison to two well-known endocytic markers, Rab11, a well-characterized ARE marker (Casanova et al., 1999; Leung et al., 2000; Wang et al., 2001), and EEA1, an effector of Rab5 function that has been associated with early endosomes (Mu et al., 1995; Simonsen et al., 1998b; Leung et al., 2000), in particular the AEE (Leung et al., 2000). A selection criterion based on the relationship between basolateral, perinuclear, and apical endocytic structures located at different cell heights and subcellular regions and different polarized compartments, such as AEE/BEE, CE and ARE, was applied to a ROI-based quantitative FRET approach to assign FRET values to specific endocytic compartments. The main objective is to assay FRET events located at different endocytic regions while avoiding the technical challenges that may arise from introducing an additional fluorescent endocytic marker into a FRET-based assay in view of potential spectral overlap and/or availability of laser lines.
To identify various polarized endocytic compartments by colocalization with well-known markers, MDCK-PTR cells were basolaterally internalized with Alexa555-Tfn for 30 min at 37°C in the presence or absence of BFA, which missorts Tfn to the apical PM (Wang et al., 2001). Because Tfn remains attached to TFR during its intracellular trafficking, detectable fluorescence staining indicates the presence of Tfn-TFR receptor-ligand complexes; referred to, for simplicity, as TFR complexes. Then, cells were fixed and immunofluorescence was performed using anti-Rab11 or anti-EEA1 and Alexa488-secondary antibodies. Cells were imaged using a Zeiss 510META LSM confocal microscope and z-series of double-label images were collected at 0.5-μm intervals. Finally, the distribution of Rab11 and EEA1 was overlaid with that of Tfn-TFR complexes, and three images were selected at different cell heights: 2–4 μm (basolateral region), 6–7 μm (perinuclear region), and 10–9 μm (apical region) above the insert filter (Figure 2).
As shown previously (Wang et al., 2001), TFR complexes undergo a dramatic shift in distribution upon treatment with BFA. In untreated cells, TFR complexes accumulate in punctate structures predominantly at the basolateral and perinuclear regions, with a reduced amount found in the apical region (Figure 2, a–e). In BFA-treated cells, TFR complexes redistribute mainly to the apical and perinuclear regions, with few punctate structures found in the basolateral region (Figure 2, i–k). Although the distribution of TFR complexes remain the same in the presence or absence of BFA in the basolateral (periphery) and perinuclear (dispersed throughout) regions, in the apical region it shifts from a dispersed peripheral pattern in the absence of BFA to a more centralized distribution in the presence of BFA. In untreated cells, Rab11's centralized apical distribution shows a reduced colocalization with the weak Tfn staining (Figure 2c; Wang et al., 2001). In contrast, in the apical region of BFA-treated cells, the TFR distribution pattern shows a significant overlap with the Rab11 staining (Figure 2h). These results suggest that BFA induces the movement of TFR complexes into the ARE as it is being delivered to the apical PM via the transcytotic pathway (Wang et al., 2001).
Our results also confirm previous data showing that basolaterally internalized TFR complexes are delivered to the AEE (Leung et al., 2000), because colocalization is detected between them and that of EEA1, predominantly in the cell periphery of the perinuclear and apical regions (Figure 2, d and e). These results suggest that AEE may play a role in the basolateral recycling of TFR complexes (Leung et al., 2000). Interestingly, in BFA-treated cells, only a reduced level of colocalization is detected between the peripherally disperse EEA1 punctate structures and the TFR complexes concentrated in the central area of the apical region (Figure 2k). These results suggest that in the absence of BFA, the TFR complexes participating in the basolateral recycling pathway have access to the BEE, CE, and AEE at the basolateral, perinuclear, and apical regions, respectively. In contrast, in the presence of BFA, the TFR complexes are shifted to the basolateral-to-apical transcytotic pathway in which they are mainly delivered to the BEE, CE, and ARE at the basolateral, perinuclear, and apical regions, respectively.
To develop a morphology- and ROI-based quantitative imaging approach to localize FRET signal to different endocytic compartments in polarized MDCK-PTR cells, Alexa488-pIgA-R ligands and/or Cy3-Tfn were internalized jointly from the basolateral surface for 30 min at 37°C in the absence or presence of BFA (Figure 3A) and imaged at different cell heights using confocal microscopy (Figures 1 and 2). As mentioned above, detectable fluorescence staining indicates the presence of pIgA-R and/or TFR receptor-ligand complexes, referred to, for simplicity, as pIgA-R and TFR complexes. Our preparation of [Fab]2 derived from affinity-purified polyclonal antibodies against secretory component shows significantly higher transcytotic ability than monovalent Fabs (50–60 vs. 25–30%; Breitfeld et al., 1989; Barroso and Sztul, 1994) but lower than dIgA (50–60 vs. 70–80%; Apodaca et al., 1994; Brown et al., 2000) and does not accumulate intracellularly (Breitfeld et al., 1989; Barroso and Sztul, 1994). This difference in transcytotic/recycling behavior is probably due to the fact that the binding of dIgA induces the dimerization of pIgA-R (Singer and Mostov, 1998; Rojas and Apodaca, 2002; Van IJzendoorn et al., 2002; Mostov et al., 2003), resulting in the stimulation of the receptor transcytosis, at least threefold (Song et al., 1994; Luton et al., 1999; Luton and Mostov, 1999; Giffroy et al., 2001). [Fab]2 may partially cross-link pIgA-R, thus mimicking the ligand-induced receptor dimerization and allowing the partial induction of pIgA-R transcytosis. However, there is no abnormal cross-linking between [Fab]2 and the pIgA-R because that process would generate large complexes that would mask the FRET signal and be directed to the lysosomes. This targeting to the lysosomes would result in high levels of degradation (Weissman et al., 1986), which does not occur under our experimental conditions (Barroso and Sztul, 1994; Wallrabe et al., 2003a). Here, we show that there is significant colocalization between basolaterally internalized TFR and pIgA-R ligand complexes in BFA treated or untreated cells (Figure 3B, a–d) as well as between TFR complexes and Rab11 in BFA-treated cells (Figure 2h). These results suggest that TFR and pIgA-R complexes are part of the recycling/transcytotic pathway and not of the late endosome/lysosomal pathway.
A wide variety of ROIs were collected using a selection criterion based on their subcellular location in relationship to: 1) insert surface, i.e., the basal surface of the cells; 2) the nucleus; and 3) the cell center or periphery (Figures 1–3). In Figure 1B, a schematic representation of the different endocytic morphologies of cross sections at the basolateral, perinuclear, and apical locations is shown to help understand the ROI selection; basolateral ROIs are collected at the periphery of the cell across the lower part of the nucleus, whereas perinuclear ROIs are collected at the upper part of the nucleus away from the PM and the apical ROIs at the center of the cell above the nucleus. Finally, the ROI fluorescence intensities were subjected to a quantitative analysis to determine the response of these three endosome groups to BFA (Figure 3C). On the basis of the TFR/Rab11 and TFR/EEA1 colocalization distribution pattern (Figure 2), we propose the following: The basolateral ROIs represent the BEE and the perinuclear ROIs the CE. The apical region contains both the AEE and the ARE. In the nontreated cells, apical TFR complexes colocalize with EEA1 and a weak colocalization with Rab11. In contrast, in the BFA-treated cells, the TFR complexes exhibit a weak corelation with EEA1 distribution and high levels of colocalization with Rab11, clearly identifying the ARE.
In Figure 3B, four images are shown from a Z-series of double-label images collected with a 0.5-μm vertical step using confocal microscopy. Basolaterally cointernalized pIgA-R and TFR complexes are found in peripherally located basolateral endosomes (Figure 3, A and B, e and f). In the absence of BFA, both pIgA-R and TFR complexes traffic as far as the perinuclear region (Figure 3, A and Bc) and pIgA-R is predominantly detected at the apical location (Figure 3, A and Ba). In the presence of BFA, both pIgA-R and TFR complexes are detected in endocytic structures located mainly at the perinuclear and apical region (Figure 3, A and B, b and d).
In Figure 3C, three groups of endosomes (basolateral, perinuclear, and apical) are distinguishable based on their location, relative amount of TFR and pIgA-R and response to BFA, confirming the qualitative results of Figures 2 and 3B. This quantitative fluorescence analysis is consistent with previous reports indicating that BFA disrupts polar sorting (Hunziker et al., 1991; Wan et al., 1992; Prydz et al., 1992; Barroso and Sztul, 1994; Futter et al., 1998; Wang et al., 2001) and supports our morphology-based approach.
FRET To Discriminate Clustered and Random Protein Distributions
Mathematical models have been used to discriminate a clustered from a random membrane protein organization based on the relationship between E% and A levels at specific ranges of D/A ratios, which are experimentally determined using quantitative FRET (Zimet et al., 1995; Kenworthy and Edidin, 1998; Kenworthy et al., 2000; Zacharias et al., 2002; Wallrabe et al., 2003a,b, 2006; Wallrabe and Barroso, 2005). In a random situation, the likelihood of an acceptor colocalizing with a given donor is positively correlated with A levels and leads to an increase in E% (Figure 4A). Conversely, in a clustered scenario where molecules by definition are in proximity either in dimer or higher-order oligomeric complexes, E% is largely independent of A levels, and it does not decrease to zero when A trends to zero (Kenworthy and Edidin, 1998; Kenworthy et al., 2000; Pentcheva and Edidin, 2001; Spiliotis et al., 2002; Figure 4C). Furthermore, E% versus D/A is used to provide further insights into the tightness of the cluster organization (Wallrabe et al., 2003a,b, 2006). An overall negative dependency between E% and D/A at high and low A levels suggests a tight cluster organization (Figure 4C), whereas a general independency, highlights a random organization (Figure 4A). In a mixed random/clustered organization, where an assortment of clusters and randomly distributed proteins are found, E% may be positively correlated to A levels, but at a given A level it may be negatively dependent on D/A (Pentcheva and Edidin, 2001; Bhatia et al., 2005).
As a positive control for randomly organized proteins, different ratios of donor-Tfn and acceptor-Tfn bound to polylysine-coated coverslips were subjected to FRET confocal imaging (Figure 4B). A strong positive relationship between E% and A levels at both D/A ≈ 1 and D/A ≈ 2 is detected for Tfn-bound to polylysine-covered coverslips (Figures 4, B and E), with E% values trending to zero with decreasing A levels and correlation coefficients of r = 0.72 (slope s = 0.5) at D/A ≈ 1 and r = 0.85 at D/A ≈ 2 (Table 1). ANOVA test on E% at different A levels at D/A ≈1 yields a p value indicating significant evidence that the means are not equal (p < 0.001; Table 1). Furthermore, the E% versus A relationship behaves independently of D/A, i.e., at both low and high A levels E% values are not dependent on D/A (Figure 4B). These results indicate that donor-Tfn and acceptor-Tfn show a random organization upon binding to polylysine-coated coverslips.
For a clustered organization, donor-Tfn and acceptor-Tfn were bound to the TFR homodimer (Lawrence et al., 1999; Cheng et al., 2004) at the PM and internalized into MDCK-PTR cells (Wallrabe et al., 2006). Identical confocal imaging settings were used on both the polylysine- and the cell-based FRET assays (Figure 4, B and D). No significant positive dependence of E% on A levels at D/A ≈ 1 and D/A ≈ 2 is detected (Figure 4, D and E). In agreement, the E% values do not trend to zero with decreasing A levels and correlation coefficients show r = 0.08 (slope s = 0.04) at D/A ≈ 1 and r = −0.04 at D/A ≈ 2 (Table 1). ANOVA test on E% at different A groups at D/A ≈ 1 yields a nonsignificant p value (p > 0.001; Table 1). Furthermore, E% shows a negative dependence on D/A, because at both high and low A levels, E% rises with decreasing D/A (Figure 3D). These results confirm the clustered distribution of TFR complexes in endocytic membranes.
pIgA-R and TFR Complexes Show Distinct Organizations in Basolateral versus Perinuclear Endosomes.
To test whether pIgA-R and TFR complexes cluster together along the endocytic pathway, we have used FRET to assay the organization of pIgA-R and TFR complexes at the basolateral and perinuclear endosomes. Live-cell FRET confocal microscopy is performed on images collected at 2–4 μm above the filter (basolateral; blue rectangle) upon cointernalization of donor-pIgA-R ligand and acceptor-Tfn for 30 min at 37°C into the basolateral surface of polarized epithelial MDCK-PTR cells (Figure 5A). Pseudocolor images depict D/A, A, and E%, respectively, in a pixel-by-pixel manner, showing the morphology patterns of these parameters. The typical basolateral morphology with peripherally localized punctate endocytic structures and a centrally located nucleus (N) is clearly detected across all images (Figure 5B, d–f). E% images show significant energy transfer between TFR and pIgA-R complexes in the basolateral region (Figure 5Bf).
To perform a FRET quantitative analysis, a large number of ROIs (>50 pixels) from several images are selected, each representing endocytic punctate structures as described in Figure 1B. For clarity, only four ROIs (white rectangles) are shown as an example in Figure 5B, d–f. To avoid potential contamination by perinuclear endosomes, basolateral ROIs are mainly collected at the periphery of the cell (Figure 1B). Then, the average values for E%, D, and A are calculated for these ROIs. The data are split into D/A ≈1 and D/A ≈ 2 ranges, and E% is then plotted against A levels. In the basolateral region, E% shows a clear dependence on A levels at the D/A ≈ 1, but not at the D/A ≈ 2 range (Figure 5D). Correlation analysis substantiates these conclusions with values of r = 0.77 (slope s = 1.05) and r = −0.11, respectively for D/A ≈ 1 and D/A ≈ 2 (Table 1). Further analysis shows that the rising average A level is clearly matched by equally rising average E% values; ANOVA analysis demonstrates the positive dependency of E% on increasing A levels at D/A ≈ 1 with a significant p value (p < 0.001; Table 1). Moreover, E% appears to behave independently from D/A at low A values, whereas E% clearly increases with decreasing D/A ratios, at high A levels (Figure 5D). Thus, applying the model concepts expressed in Figure 4 (Pentcheva and Edidin, 2001; Wallrabe et al., 2003a, 2006; Bhatia et al., 2005), these results suggest that TFR and pIgA-R complexes show a mixed random/clustered behavior in basolateral endosomes.
To determine whether pIgA-R and TFR show a random or clustered distribution in perinuclear endosomes, quantitative confocal FRET is performed on images collected from the same live cells at 6.0–8.0 μm above the filter under identical imaging settings (Figure 5A, green rectangle). The typical perinuclear shape of irregular and punctate endocytic structures surrounding the upper region of the nucleus (N) is detected across all images (Figure 5B, a–c). Significant energy transfer levels are also detected between TFR and pIgA-R complexes in the perinuclear region (Figure 5Bc). Again, a broad set of ROIs is selected from these structures (Figure 5B, a–c, white squares). To avoid potential contamination with basolateral endosomes, perinuclear ROIs are not collected at the cell periphery (Figure 1B). At both D/A ranges, E% shows a clear independence from A levels (Figure 5C). Correlation analysis substantiates these conclusions with values of r = −0.22 (slope s = −0.13) and r = −0.31, respectively, for D/A ≈ 1 and D/A ≈ 2 (Table 1). ANOVA single-factor comparing E% values at different A levels within the D/A ≈ 1 range yields a nonsignificant p value (p > 0.001; Table 1). Furthermore, E% displays a negative dependence on D/A at both high and low A levels (Figure 5C). These results clearly suggest that pIgA-R and TFR complexes are organized in a clustered distribution in perinuclear endosomes.
Perinuclear Distribution Investigated in the Presence of BFA by Internalizing pIgA-R and TFR Complexes from Opposite PMs
A caveat of the studies described above is the potential contamination of the perinuclear region with basolateral endosomes, which may occur upon the cointernalization of pIgA-R and TFR complexes from the basolateral surface. To address this potential problem, we have investigated the perinuclear organization of these receptor-ligand complexes, cointernalized from opposite PMs in the presence of BFA (Figure 6A, green rectangle), because the majority of the pIgA-R and TFR complexes colocalizes at the perinuclear and apical regions (Figure 6B).
Analyzing the perinuclear receptor organization based on two different internalization schemes (basolateral cointernalization vs. from opposite PMs), both show typical perinuclear endocytic pattern (Figure 5B, a–c, and 6B, a–c) and E% behavior being independent of A levels at all D/A ranges (Figure 5C and 6C). At the perinuclear region upon internalization from opposite PMs, the correlation coefficients, r = −0.32 (slope s = −0.07) and r = −0.16, confirm the E%'s independence from A levels at D/A ≈ 1 and D/A ≈ 2, respectively (Figure 6C; Table 1). Furthermore, ANOVA analysis comparing E% values at different A levels at D/A ≈ 1 with a nonsignificant p value (p > 0.001) suggests that there is no difference between groups (Table 1). Furthermore, E% shows a clear negative relationship with D/A; E% increases with decreasing D/A ratios at both high and low A levels (Figure 6C). These results indicate a highly organized clustered distribution for pIgA-R and TFR in perinuclear endosomes upon internalization from opposite PMs.
To analyze the impact or otherwise of BFA on the organization of pIgA-R and TFR complexes in perinuclear endosomes, we compared above FRET data obtained upon internalization from opposite PMs in the presence of BFA (Figure 6C) with that obtained upon internalization from the basolateral surface in the absence of BFA (Figure 5C). As shown in Figure 6D, perinuclear E% at D/A ≈ 1 behaves in a largely independent manner from increasing A levels in the presence and absence of BFA using different internalization protocols. Furthermore, an ANCOVA analysis comparing the perinuclear E% data obtained using different internalization protocols in the presence and absence of BFA yields a nonsignificant p value, p > 0.001, at all the A ranges (Figure 6D; Table 2).
pIgA-R and TFR Clusters Are Differentially Affected by BFA in Polarized Endosomes
To assay whether BFA affects the organization of pIgA-R and TFR complexes in different endocytic regions, we have compared FRET data collected at the basolateral and perinuclear regions upon cointernalization of pIgA-R and TFR complexes into the basolateral surface for 30 min at 37°C in the absence or presence of BFA. Although basolateral E% at D/A ≈ 1 rises with increasing A levels in the presence and absence of BFA with correlation coefficients and slope values of r = 0.40, s = 0.40 and r = 0.48, s = 0.39, respectively (Figure 7A), perinuclear E% at D/A ≈ 1 behaves in a largely independent manner from increasing A levels in the presence and absence of BFA with correlation coefficients and slope values of r = 0.01, s = 0.01 and r = 0.06, s = 0.04, respectively (Figure 7B). Interestingly, an ANCOVA analysis comparing the E% data in the presence and presence of BFA shows a significant p value for basolateral endosomes (p < 0.001) and a nonsignificant p value (p > 0.001) for perinuclear endosomes (Table 2).
In summary, BFA clearly does not affect the nature of receptor clustering in the perinuclear endosomes, whether internalized from opposite PMs (with BFA) or cointernalized from the basolateral PM (with or without BFA). Thus, these results suggest that pIgA-R and TFR complexes form highly organized clusters in perinuclear endosomes, independently of internalization protocols and/or BFA treatment. However, our data also suggest that BFA may affect indirectly the nature of the organization of the pIgA-R and TFR complexes in the basolateral endosomes.
pIgA-R and TFR Complexes Show Different Clustered Organizations in Apical and Perinuclear Endosomes
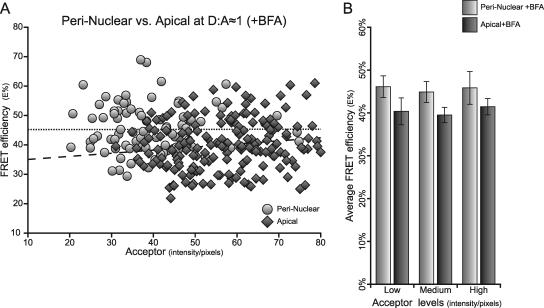
Here, we compare the perinuclear and apical organization of pIgA-R and TFR complexes internalized from basolateral PMs in the presence of BFA. Images are collected at 6–8 μm above the basal substrate for perinuclear localization and at 9–11 μm for apical localization (Figure 8A; green and red rectangles, respectively). Comparing FRET events at the perinuclear and apical endosomes is only possible in the presence of BFA, whereas TFR complexes are redistributed to the apical region showing strong colocalization with ARE's Rab11 as well as with transcytosing pIgA-R; in contrast, in untreated cells, the apical TFR complexes colocalize mainly with the AEE's EEA1 and not with Rab11. As expected, in the apical region of untreated cells, FRET between TFR and pIgA-R complexes is undetectable, i.e., <5% (data not shown) because they are found in different compartments, AEE and ARE, respectively. With BFA, perinuclear endosomes show a typical perinuclear distribution surrounding the upper nuclear region (Figure 8B, d–f), whereas the apical endocytic punctate structures are localized in the center of the cell, above the nucleus (a–c). Again, a large number of ROIs are selected to compare perinuclear and apical endosomes, considering the selection criteria described in Figure 1B (Figure 8B, white squares). The dependency or otherwise of E% on A levels for a range of apical and perinuclear ROIs is plotted on Figure 8, C and D. Both perinuclear and apical regions demonstrate a low r at D/A ≈ 1 (r = 0.01 and 0.13, respectively) with also low slope values, s = 0.01–0.09, and at D/A ≈ 2 (r = 0.03 and 0.43, respectively; Table 1), indicating that E% is not dependent on A levels for both perinuclear and apical endosomes. Single-factor ANOVA comparing E% values at D/A ≈ 1, yields nonsignificant p values (p > 0.001) for both perinuclear and apical receptor clusters corroborating the r and slope analysis (Table 1). Furthermore, perinuclear and apical E% show a clear negative dependency on D/A at both high and low A levels (Figure 8, C and D), all parameters indicating a highly clustered organization.
In Figure 9A, we compare the E% data for perinuclear and apical in the presence of BFA. As established above, E% at D/A ≈ 1 behaves in a largely independent manner from increasing A levels both for the apical and the perinuclear endosomes (Figure 9A). However, a significant difference is detected between the apical and perinuclear E% at D/A ≈ 1 using an ANCOVA analysis (p < 0.001; Table 2). To confirm this indication, the data sets are split into three groups with low, medium, and high A levels, and the average E% is plotted versus respective A groups (Figure 9B). Similarly, a significant difference (p < 0.001) is detected between apical and perinuclear E% at D/A ≈ 1 at all A groups. Thus, although the pIgA-R and TFR complexes show a clustered organization in both, the perinuclear and apical endosomes, their E% levels are significantly different. Because E% is an expression of distance, the proximities within a cluster may be different in perinuclear versus apical endosomes.
Figure 9.
Perinuclear and apical E% show significant differences. (A) E% values at D/A ≈ 1 were plotted as a function of Acceptor levels for perinuclear (circles) versus apical (diamonds) ROIs collected from images generated upon internalization from basolateral PM in the presence of BFA. Trendlines are shown as visual helpers (perinuclear, dotted line; apical, dashed line). As expected, E% is independent of Acceptor levels in perinuclear and apical endosomes. These two data sets are significantly different (p > 0.001) using an ANCOVA statistical analysis (Table 2) with perinuclear E% levels slightly higher than apical E% levels. (B) Perinuclear + BFA (light grey bars) and apical + BFA (dark grey bars) E% averages were plotted versus different groups of Acceptor (low, medium, and high) upon cointernalization from basolateral PM in the presence of BFA are shown to be significantly different (p < 0.001) by an ANOVA single-factor statistical analysis. The presence of significantly higher E% levels in the perinuclear endosomes at the same D/A ≈ 1 suggests a different overall organization of the receptor-ligand complexes in the perinuclear versus the apical endosomes in the presence of BFA. Error bars, 95% confidence interval.
DISCUSSION
Previously, we have used FRET to demonstrate that pIgA-R-ligand complexes are distributed in a clustered manner in apical endosomes of polarized MDCK-PTR cells (Wallrabe et al., 2003a,b). These results suggested that the formation of clusters is an essential part of the sorting and trafficking of receptor-ligand complexes via polarized endocytic pathways. Several testable predictions result from this hypothesis: one is that different membrane-bound receptors may form clusters when traveling together to the same destination. Another is that receptor clusters may show distinct organizations in various polarized endocytic compartments, possibly influenced by their location, final destination, or other regulatory mechanisms. To test these predictions, we have used live-cell confocal FRET to characterize the organization of TFR and pIgA-R complexes in polarized endosomes of MDCK cells.
Intracellular Localization of the FRET Signal
To localize the intensity-based FRET signal within polarized cells, we have developed a method to differentiate the basolateral, perinuclear, and apical endosomal locations, representing the BEE, CE, and ARE compartments, respectively (Apodaca et al., 1994; Barroso and Sztul, 1994; Brown et al., 2000; Leung et al., 2000; Wang et al., 2000; Hoekstra et al., 2004; Rodriguez-Boulan et al., 2005). These endosomes, which to some extent overlap with one another, were identified using a geographical criteria based on the subcellular localization of the staining pattern of fluorophore-labeled pIgA-R and TFR complexes, Rab11, an ARE marker (Casanova et al., 1999; Leung et al., 2000; Wang et al., 2001) and EEA1, an early endosome marker (Mu et al., 1995; Simonsen et al., 1998a), in particular the AEE (Leung et al., 2000). This methodology allows the assignment of FRET signal to specific endocytic compartments, overcoming confocal FRET's inability to identify the reference space in which the energy transfer is taking place.
Of the three endocytic regions, the basolateral endosome is in some ways the easiest to identify predominantly as the BEE. Imaging at a focal plane across the lower part of the nucleus at a consistent distance from the basal substrate and analyzing only peripherally located fluorescence largely avoids “contamination” by the CE. Isolating the perinuclear endosomes by imaging them across the upper part of the nucleus at the center of the cell and away from the periphery to avoid contamination with the BEE/AEE and ARE, is somewhat more difficult, but doable with a relatively high degree of confidence. Reduced perinuclear and strong apical colocalization of Rab11 with TFR complexes suggests low overlap between perinuclear and apical endosomes, as expected for CE and ARE. Although some colocalization between EEA1 and TFR complexes is detected at the perinuclear region, it occurs mainly in the cell periphery, suggesting that contamination by the BEE/AEE can be reduced by collecting ROIs away from the cell periphery. Interestingly, in the presence of BFA, only a weak colocalization is detected between EEA1 and TFR complexes at the perinuclear region, providing a clear distinction between CE and early endosomes. The apical region is narrowed down by imaging close to the apical PM, clearly above the nucleus and at the center of the cell. Morphological identification of the apical endosomes as the ARE, is achieved by the staining with pIgA-R ligand, but not Tfn, in the absence of BFA, and by the strong colocalization of TFR complexes with Rab11 or pIgA-R in the presence of BFA. The apical endosomes should be comprised mainly of ARE because imaging at the center of the cell and close to the apical PM will avoid contamination with AEE and CE, respectively. Furthermore, in the presence of BFA, TFR complexes show a reduced colocalization with EEA1 and a strong overlap with Rab11 at the apical region suggesting that they are shifted to a basolateral-to-apical transcytotic pathway where they are delivered to the BEE, CE and ARE at the basolateral, perinuclear and apical regions, respectively. These results suggest that while BEE, AEE, and CE are all subdomains of a wider network of endosomal compartments (Odorizzi et al., 1996; Leung et al., 2000), the ARE is part of a separate endosomal compartment, which acts as a direct route to the apical PM. Furthermore, BFA causes a significant alteration in the localization of TFR complexes throughout the endocytic pathway of polarized MDCK cells but not in the distribution of Rab11 and EEA1, suggesting as previously indicated by Wang et al. (2001), that BFA disrupts the polar sorting of Tfn-TFR complexes but not the overall sequential localization of polarized endocytic compartments as well as their ability to remain distinct from each other. Therefore, the definition of basolateral, perinuclear and apical endosomes as BEE, CE, and ARE, respectively, is insensitive to BFA and independent of the transport pathways and cargo that may cross them.
Quantitative FRET to Discriminate Random and Clustered Receptor Distributions
Because of its low resolution (≈200 nm), qualitative fluorescence microscopy used widely in cell biology cannot unequivocally confirm colocalization just because the staining pattern of two fluorophores appears to be overlapping. In contrast, FRET confocal microscopy with all the described correction steps can provide this answer as FRET only occurs when proximities are between 1 and 10 nm; it also allows additional analyses and modeling steps that give further insights into the nature of the colocalization.
A statistical approach based on correlation, slope and ANOVA/ANCOVA statistical analysis of the E% versus A relationship at D/A ≈ 1 was developed to distinguish between a random and a clustered receptor organization. The E% versus A at D/A ≈ 1 relationship was selected because of its high sensitivity to changes in receptor organization (Wallrabe et al., 2006). E% versus D/A is used to provide further insights into the tightness of the receptor cluster organization (Wallrabe et al., 2003a,b, 2006).
To verify the veracity of the FRET-based clustering assay, a negative control, where donor and acceptor-Tfn molecules were applied to polylysine-coated coverslips, was assayed using FRET confocal microscopy. In the absence of any receptor-mediated organization, the data shows a dependence of E% on A levels with a high (close to one) positive correlation and slope values and a significant ANOVA p value (Table 1). A strong dependency of E% on A levels together with an overall independence of E% from D/A indicates a clear random receptor distribution upon binding to polylysine-covered slides. A positive control where proteins by definition are clustered was used by taking advantage of the dimeric nature of TFR (Lawrence et al., 1999; Cheng et al., 2004). The data show a clear independent behavior of E% from A levels with a low (close to zero) correlation and slope values and a nonsignificant ANOVA p value (Table 1). The two parameter data, with E% showing a clear independence from A levels and a negative dependence on D/A ratio, fits a clustered organization (Wallrabe et al., 2006). In summary, these results provide a higher and lower bound, respectively, for the organization level of receptor-ligand complexes in endocytic membranes (Wallrabe et al., 2006).
pIgA-R and TFR Complexes Form Increasingly Tighter Clusters along Polarized Endocytic Trafficking
Here, quantitative FRET confocal microscopy indicates a basic distinction between basolateral and perinuclear endosomes (Sheff et al., 1999; Brown et al., 2000). In both basolateral and per-nuclear endosomes, E% shows a positive dependency on A levels. However, an unequivocal overall independence of E% from D/A ratio at all A levels is detected in the perinuclear but not in the basolateral endosomes (Table 1). We propose that early in endocytic trafficking pIgA-R and TFR complexes are more loosely organized, showing a mixed random/cluster organization, where clusters comingle with randomly organized receptors. On reaching the perinuclear and apical endosomes together, pIgA-R and TFR complexes show a well-organized clustered distribution with the majority of complexes in clusters.
A gradient of increasing receptor clustering is detected from the basolateral to perinuclear endosomes (Figure 10), suggesting that receptor clustering correlates strongly with endocytic transport. Thus, multiple cycles of tubulovesicular formation and fusion, which are required for iterative endocytic membrane sorting (Mayor et al., 1993), may lead to tighter receptor clusters during recycling and transcytosis, providing a justification for why clustering would become more organized as the two cotransported receptors move deeper into the cell.
Figure 10.
Model for distinct organizations of receptor clusters throughout the endocytic pathways of polarized cells.
Another important point to address is the generality of the receptor clustering phenomenon during membrane trafficking. These experiments were performed in MDCK-PTR cells, which express moderate amounts of human TFR and rabbit pIgA-R. MDCK-PTR and their respective parental cells show similar TFR and pIgA-R transport kinetics and regulation (Apodaca et al., 1994; Brown et al., 2000; Babbey et al., 2006). Therefore, we expect FRET events between TFR and pIgA-R complexes to be minimally affected by the overexpression of these receptors. In agreement, we have recently demonstrated the ability of endogenous low-density-lipoprotein receptor (LDL-R) to cluster in the sorting endosomes of nonpolarized MDCK-PTR cells (unpublished results). Thus, we posit that the clustering of receptor-ligand complexes is an integral part of their membrane trafficking pathways.
Effect of BFA on the Organization of Receptors during Polarized Trafficking
BFA has been shown to induce significant alterations in the polarized endocytic trafficking of TFR and pIgA-R, without affecting endosomal identity (Hunziker et al., 1991; Wan et al., 1992; Prydz et al., 1992; Barroso and Sztul, 1994; Futter et al., 1998; Wang et al., 2001). In particular, BFA increases the basolateral-to-apical transcytosis of TFR and reduces its basolateral recycling (Wan et al., 1992, 2001; Futter et al., 1998; Figure 3A). Other research suggests that although the basolateral-to-apical transcytosis of pIgA-R is impaired by BFA, detectable amounts of pIgA-R are still able to reach the apical PM in the presence of BFA (Hunziker et al., 1991; Barroso and Sztul, 1994; Wang et al., 2001). BFA's molecular mechanism is related to its ability to inhibit the membrane association of coat proteins and to block the formation of transport vesicles.
Here, we have tested whether BFA-induced disruption of polarized endocytic sorting leads to a block in receptor clustering. Our results suggest that this is not the case and that thus, BFA-dependent coats are not required for receptor clustering. However, these results do not rule out a possible involvement of BFA-insensitive coats, such Hrs (Sachse et al., 2002; Yan et al., 2005). Other molecules may play a role in the receptor clustering process, such as, sorting nexins, (Carlton et al., 2004, 2005), phosphoinositide-binding proteins, such as epsin (Ford et al., 2002; Legendre-Guillemin et al., 2004), or alternate endocytic adaptors or clathrin-associated sorting proteins (Traub, 2005).
BFA's induced disruption of polarized sorting affects the organization of receptor-ligand complexes in the basolateral endosomes, resulting in an increase of E% levels while maintaining the mixed random/clustered distribution. This data suggests that polarized sorting of pIgA-R and TFR is initiated at the BEE by spatially segregating transcytosing from recycling receptors. This would predict that in BFA-treated cells, an increase in BEE E% levels, while maintaining a mixed random/cluster organization, may be due to an increase availability of pIgA-R and TFR to randomly interact with each other and/or to form a slightly higher number of receptor clusters. It is important to note that spatial segregation within a 10–100-nm range would be enough to be detected using a FRET assay; this range would not be easily distinguished using standard fluorescence confocal microscopy. On the other hand, a similar tightly clustered behavior in perinuclear endosomes in the presence or absence of BFA suggests that sorting occurs mainly before the arrival of receptor-ligand complexes to the CE.
In summary, polarized membrane trafficking can be discriminated into two steps. One is the BFA-independent formation of receptor clusters, which is initiated at the BEE. Another step is the BFA-dependent spatial segregation of trancytosing from recycling receptors; this sorting would determine the composition of receptor clusters.
pIgA-R and TFR Complexes Form Distinctly Organized Clusters in Perinuclear and Apical Endosomes
As mentioned above, the FRET data collected at the apical region should originate mainly from receptor clusters localized at the ARE, as indicated by the colocalization between TFR complexes and Rab11, whereas the FRET signal collected at the perinuclear region should reflect mainly the CE and not the ARE and BEE/CEE considering the reduced colocalization between TFR complexes and Rab11 and EEA1. Our results indicate that although both types of endosomes show the existence of receptor clusters, there are significant differences in E% values between them, suggesting different pIgA-R and TFR cluster organizations. Therefore, the difference detected between FRET data collected at apical and perinuclear endosomes in the presence of BFA should reflect ARE and CE different receptor cluster organizations and subsequent functional differences between these two endosomes.
E% is very often an expression of distance and higher levels are associated with shorter distances. In the context of a cluster, this could mean tighter packing. Modeling, using the known geometries of both pIgA-R and TFR would be one way to address this question in the future. However, the three-dimensional configurations of the TFR and pIgA-R complexes may allow for a number of packing modalities increasing the complexity of such a modeling analysis. Nevertheless, the lower E% levels detected in the ARE suggests a reduced clustering organization of pIgA-R and TFR complexes. Several hypotheses may explain such behavior; for example, the higher tubulation of the CE even in the absence of BFA may be related to their tighter receptor cluster organization. Differences in clustering platforms or clustering facilitators might also impact the cluster organization.
Biological Implications
The FRET-based quantitative assay presented here suggests that different receptor-ligand complexes form clusters, with distinct levels of organization, while being cotransported toward a common destination along the polarized endocytic pathways of MDCK cells (Figure 10). Several interesting questions about the biological basis of this distinct organization in endocytic receptor clusters can be raised. An appealing possibility is that increased cluster organization is a consequence of the progression of membrane trafficking along the endocytic pathway; may be due to multiple iterative steps of tubulovesicular formation and fusion. Another hypothesis is that distinct receptor cluster organizations are caused by differences in the molecules responsible for the establishment of the platform where clustering takes place and/or for facilitating the actual clustering process. Yet another interesting hypothesis is that membrane microdomains showing increased clustering of receptor-ligand complexes may overlap with regions of high curvature and/or tubulation. This scenario is possible because the oligomerization of transmembrane proteins with a conical shape has been proposed to induce membrane deformation and curvature (McMahon and Gallop, 2005). Finally, a model describing apical targeting via lipid rafts proposes that a clustering event is necessary to convert apical proteins associated with small lipid rafts into functional apical sorting platforms (Hannan et al., 1993; Paladino et al., 2004; Rodriguez-Boulan et al., 2005). The exact mechanism generating this clustering remains to be determined. Nonetheless, the findings presented here underline the importance of endocytic receptor clustering and its key role in membrane trafficking and sorting in polarized cells. The assay described here should provide another useful approach to extend the exploration in this area.
ACKNOWLEDGMENTS
We thank Dr. K. Dunn (University of Indiana) for the MDCK-PTR cells and Dr. E. Sztul (University of Alabama) and Dr. J. Mazurkiewicz (Albany Medical College) for their insightful comments.
Abbreviations used:
- A
acceptor intensity levels
- D
unquenched donor intensity levels
- D/A
unquenched donor/acceptor ratios
- FRET
Förster resonance energy transfer
- pIgA-R
polymeric IgA-receptor
- TFR
transferrin-receptor
- Tfn
holo-transferrin
- pIgA-R complexes
pIgA-R-pIgA-R ligand complexes
- TFR complexes
TFR-Tfn complexes.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0700) on April 4, 2007.
REFERENCES
- Andrade J., Zhao H., Titus B., Timm P. S., Barroso M. The EF-Hand Ca2+-binding protein p22 plays a role in microtubule and endoplasmic reticulum organization and dynamics with distinct Ca2+-binding requirements. Mol. Biol. Cell. 2004;15:481–496. doi: 10.1091/mbc.E03-07-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G., Katz L. A., Mostov K. E. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J. Cell Biol. 1994;125:67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbey C. M., Ahktar N., Wang E., Chen C. C., Grant B. D., Dunn K. W. Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 2006;17:3156–3175. doi: 10.1091/mbc.E05-08-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso M., Sztul E. S. Basolateral to apical transcytosis in polarized cells is indirect and involves BFA and trimeric G protein sensitive passage through the apical endosome. J. Cell Biol. 1994;124:83–100. doi: 10.1083/jcb.124.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S., Edidin M., Almo S. C., Nathenson S. G. Different cell surface oligomeric states of B7–1 and B7- 2, implications for signaling. Proc. Natl. Acad. Sci. USA. 2005;102:15569–15574. doi: 10.1073/pnas.0507257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonamy G. M., Guiochon-Mantel A., Allison L. A. Cancer promoted by the oncoprotein v-ErbA may be due to subcellular mislocalization of nuclear receptors. Mol. Endocrinol. 2005;19:1213–1230. doi: 10.1210/me.2004-0204. [DOI] [PubMed] [Google Scholar]
- Breitfeld P. P., Harris J. M., Mostov K. E. Postendocytotic sorting of the ligand for the polymeric immunoglobulin receptor in Madin-Darby canine kidney cells. J. Cell Biol. 1989;109:475–486. doi: 10.1083/jcb.109.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. S., Wang E., Aroeti B., Chapin S. J., Mostov K., Dunn K. W. Definition of distinct compartments in polarized Madin-Darby canine kidney (MDCK) cells for membrane-volume sorting, polarized sorting and apical recycling. Traffic. 2000;1:124–140. doi: 10.1034/j.1600-0854.2000.010205.x. [DOI] [PubMed] [Google Scholar]
- Carlton J., Bujny M., Peter B. J., Oorschot V. M., Rutherford A., Mellor H., Klumperman J., McMahon H. T., Cullen P. J. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr. Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Carlton J. G., Bujny M. V., Peter B. J., Oorschot V. M., Rutherford A., Arkell R. S., Klumperman J., McMahon H. T., Cullen P. J. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J. Cell Sci. 2005;118:4527–4539. doi: 10.1242/jcs.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. E., Wang X., Kumar R., Bhartur S. G., Navarre J., Woodrum J. E., Altschuler Y., Ray G. S., Goldenring J. R. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zak O., Aisen P., Harrison S. C., Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Dunn K. W., McGraw T. E., Maxfield F. R. Iterative fractionation of recycling receptors from lysosomally destined ligands in an early sorting endosome. J. Cell Biol. 1989;109:3303–3314. doi: 10.1083/jcb.109.6.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elangovan M., Wallrabe H., Chen Y., Day R. N., Barroso M., Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- Ford M. G., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J., Evans P. R., McMahon H. T. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Futter C. E., Gibson A., Allchin E. H., Maxwell S., Ruddock L. J., Odorizzi G., Domingo D., Trowbridge I. S., Hopkins C. R. In polarized MDCK cells basolateral vesicles arise from clathrin-gamma-adaptin-coated domains on endosomal tubules. J. Cell Biol. 1998;141:611–623. doi: 10.1083/jcb.141.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze H. J., Slot J. W., Schwartz A. L. Membranes of sorting organelles display lateral heterogeneity in receptor distribution. J. Cell Biol. 1987;104:1715–1723. doi: 10.1083/jcb.104.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze H. J., Slot J. W., Strous G. J., Peppard J., von Figura K., Hasilik A., Schwartz A. L. Intracellular receptor sorting during endocytosis: comparative immunoelectron microscopy of multiple receptors in rat liver. Cell. 1984;37:195–204. doi: 10.1016/0092-8674(84)90315-5. [DOI] [PubMed] [Google Scholar]
- Gibson A., Futter C. E., Maxwell S., Allchin E. H., Shipman M., Kraehenbuhl J. P., Domingo D., Odorizzi G., Trowbridge I. S., Hopkins C. R. Sorting mechanisms regulating membrane protein traffic in the apical transcytotic pathway of polarized MDCK cells. J. Cell Biol. 1998;143:81–94. doi: 10.1083/jcb.143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffroy D., Courtoy P. J., Vaerman J. P. Polymeric IgA binding to the human pIgR elicits intracellular signalling, but fails to stimulate pIgR-transcytosis. Scand. J. Immunol. 2001;53:56–64. doi: 10.1046/j.1365-3083.2001.00843.x. [DOI] [PubMed] [Google Scholar]
- Hannan L. A., Lisanti M. P., Rodriguez-Boulan E., Edidin M. Correctly sorted molecules of a GPI-anchored protein are clustered and immobile when they arrive at the apical surface of MDCK cells. J. Cell Biol. 1993;120:353–358. doi: 10.1083/jcb.120.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra D., Tyteca D., Van IJzendoorn S. C. The subapical compartment: a traffic center in membrane polarity development. J. Cell Sci. 2004;117:2183–2192. doi: 10.1242/jcs.01217. [DOI] [PubMed] [Google Scholar]
- Hunziker W., Whitney J. A., Mellman I. Selective inhibition of transcytosis by brefeldin A in MDCK cells. Cell. 1991;67:617–627. doi: 10.1016/0092-8674(91)90535-7. [DOI] [PubMed] [Google Scholar]
- Kenworthy A. K., Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of < 100 A using imaging fluorescence resonance energy transfer. J. Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy A. K., Petranova N., Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R., Gryczynski I., Gryczynski Z., Dattelbaum J. D. Anisotropy-based sensing with reference fluorophores. Anal. Biochem. 1999;267:397–405. doi: 10.1006/abio.1998.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. M., Ray S., Babyonyshev M., Galluser R., Borhani D. W., Harrison S. C. Crystal structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- Legendre-Guillemin V., Wasiak S., Hussain N. K., Angers A., McPherson P. S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Leung S. M., Ruiz W. G., Apodaca G. Sorting of membrane and fluid at the apical pole of polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 2000;11:2131–2150. doi: 10.1091/mbc.11.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luton F., Mostov K. E. Transduction of basolateral-to-apical signals across epithelial cells: ligand-stimulated transcytosis of the polymeric immunoglobulin receptor requires two signals. Mol. Biol. Cell. 1999;10:1409–1427. doi: 10.1091/mbc.10.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luton F., Verges M., Vaerman J. P., Sudol M., Mostov K. E. The SRC family protein tyrosine kinase p62yes controls polymeric IgA transcytosis in vivo. Mol. Cell. 1999;4:627–632. doi: 10.1016/s1097-2765(00)80213-0. [DOI] [PubMed] [Google Scholar]
- Maxfield F. R., McGraw T. E. Endocytic recycling. Nat Rev. Mol. Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- Mayor S., Presley J. F., Maxfield F. R. Sorting of membrane components from endosomes and subsequent recycling to the cell surface occurs by a bulk flow process. J. Cell Biol. 1993;121:1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H. T., Gallop J. L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Mostov K., Su T., ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat. Cell Biol. 2003;5:287–293. doi: 10.1038/ncb0403-287. [DOI] [PubMed] [Google Scholar]
- Mu F. T., Callaghan J. M., Steele-Mortimer O., Stenmark H., Parton R. G., Campbell P. L., McCluskey J., Yeo J. P., Tock E. P., Toh B. H. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Pearse A., Domingo D., Trowbridge I. S., Hopkins C. R. Apical and basolateral endosomes of MDCK cells are interconnected and contain a polarized sorting mechanism. J. Cell Biol. 1996;135:139–152. doi: 10.1083/jcb.135.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino S., Sarnataro D., Pillich R., Tivodar S., Nitsch L., Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentcheva T., Edidin M. Clustering of peptide-loaded MHC class I molecules for endoplasmic reticulum export imaged by fluorescence resonance energy transfer. J Immunol. 2001;166:6625–6632. doi: 10.4049/jimmunol.166.11.6625. [DOI] [PubMed] [Google Scholar]
- Prydz K., Hansen S. H., Sandvig K., van Deurs B. Effects of brefeldin A on endocytosis, transcytosis and transport to the Golgi complex in polarized MDCK cells. J. Cell Biol. 1992;119:259–272. doi: 10.1083/jcb.119.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Musch A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- Rojas R., Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat. Rev. Mol. Cell Biol. 2002;3:944–955. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- Sachse M., Urbe S., Oorschot V., Strous G. J., Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol. Biol. Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff D. R., Daro E. A., Hull M., Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Lippe R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998a;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Lippe R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998b;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Singer K. L., Mostov K. E. Dimerization of the polymeric immunoglobulin receptor controls its transcytotic trafficking. Mol. Biol. Cell. 1998;9:901–915. doi: 10.1091/mbc.9.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Bomsel M., Casanova J., Vaerman J. P., Mostov K. Stimulation of transcytosis of the polymeric immunoglobulin receptor by dimeric IgA. Proc. Natl. Acad. Sci. USA. 1994;91:163–166. doi: 10.1073/pnas.91.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E. T., Pentcheva T., Edidin M. Probing for membrane domains in the endoplasmic reticulum: retention and degradation of unassembled MHC class I molecules. Mol. Biol. Cell. 2002;13:1566–1581. doi: 10.1091/mbc.01-07-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W., Geuze H. J., Griffith J. M., Schwartz A. L., Strous G. J. Relations between the intracellular pathways of the receptors for transferrin, asialoglycoprotein, and mannose 6-phosphate in human hepatoma cells. J. Cell Biol. 1989;108:2137–2148. doi: 10.1083/jcb.108.6.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkasskaya O., Klushin L., Gronenborn A. M. Effective lattice behavior of fluorescence energy transfer at lamellar macromolecular interfaces. Biophys. J. 2002;82:988–995. doi: 10.1016/S0006-3495(02)75458-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Van IJzendoorn S. C., Tuvim M. J., Weimbs T., Dickey B. F., Mostov K. E. Direct interaction between Rab3b and the polymeric immunoglobulin receptor controls ligand-stimulated transcytosis in epithelial cells. Dev. Cell. 2002;2:219–228. doi: 10.1016/s1534-5807(02)00115-6. [DOI] [PubMed] [Google Scholar]
- Varma R., Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Wallrabe H., Barroso M. Confocal FRET microscopy: study of clustered distribution of receptor-ligand complexes in endocytic membranes. In: Periasamy A., Day R. N., editors. Molecular Imaging: FRET Microscopy and Spectroscopy. New York: Oxford University Press; 2005. pp. 95–111. [Google Scholar]
- Wallrabe H., Chen Y., Periasamy A., Barroso M. Issues in confocal microscopy for quantitative FRET analysis. Microsc. Res. Tech. 2006;69:196–206. doi: 10.1002/jemt.20281. [DOI] [PubMed] [Google Scholar]
- Wallrabe H., Elangovan M., Burchard A., Periasamy A., Barroso M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophys. J. 2003a;85:559–571. doi: 10.1016/S0006-3495(03)74500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H., Stanley M., Periasamy A., Barroso M. One- and two-photon fluorescence resonance energy transfer microscopy to establish a clustered distribution of receptor-ligand complexes in endocytic membranes. J. Biomed. Opt. 2003b;8:339–346. doi: 10.1117/1.1584444. [DOI] [PubMed] [Google Scholar]
- Wan J., Taub M. E., Shah D., Shen W. C. Brefeldin A enhances receptor-mediated transcytosis of transferrin in filter-grown Madin-Darby canine kidney cells. J. Biol. Chem. 1992;267:13446–13450. [PubMed] [Google Scholar]
- Wang E., Brown P. S., Aroeti B., Chapin S. J., Mostov K. E., Dunn K. W. Apical and basolateral endocytic pathways of MDCK cells meet in acidic common endosomes distinct from a nearly-neutral apical recycling endosome. Traffic. 2000;1:480–493. doi: 10.1034/j.1600-0854.2000.010606.x. [DOI] [PubMed] [Google Scholar]
- Wang E., Pennington J. G., Goldenring J. R., Hunziker W., Dunn K. W. Brefeldin A rapidly disrupts plasma membrane polarity by blocking polar sorting in common endosomes of MDCK cells. J. Cell Sci. 2001;114:3309–3321. doi: 10.1242/jcs.114.18.3309. [DOI] [PubMed] [Google Scholar]
- Weissman A. M., Klausner R. D., Rao K., Harford J. B. Exposure of K562 cells to anti-receptor monoclonal antibody OKT9 results in rapid redistribution and enhanced degradation of the transferrin receptor. J. Cell Biol. 1986;102:951–958. doi: 10.1083/jcb.102.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters F. S., Bastiaens P. I., Wirtz K. W., Jovin T. M. FRET microscopy demonstrates molecular association of non-specific lipid transfer protein (nsL-TP) with fatty acid oxidation enzymes in peroxisomes. EMBO J. 1998;17:7179–7189. doi: 10.1093/emboj/17.24.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Sun W., Kujala P., Lotfi Y., Vida T. A., Bean A. J. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol. Biol. Cell. 2005;16:2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zimet D. B., Thevenin B. J., Verkman A. S., Shohet S. B., Abney J. R. Calculation of resonance energy transfer in crowded biological membranes. Biophys. J. 1995;68:1592–1603. doi: 10.1016/S0006-3495(95)80332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]