Abstract
A variety of spindle and kinetochore defects have been shown to induce a mitotic delay through activation of the spindle checkpoint. With the aim of identifying novel mitotic defects we carried out a mad1 synthetic lethal screen in budding yeast. In this screen, four novel alleles of sfi1 were isolated. SFI1 is an essential gene, previously identified through its interaction with centrin/CDC31 and shown to be required for spindle pole body (SPB) duplication. The new mutations were all found in the C-terminal domain of Sfi1p, which has no known function, but it is well conserved among budding yeasts. Analysis of the novel sfi1 mutants, through a combination of light and electron microscopy, revealed duplicated SPBs <0.3 μm apart. Importantly, these SPBs have completed duplication, but they are not separated, suggesting a possible defect in splitting of the bridge. We discuss possible roles for Sfi1p in this step in bipolar spindle assembly.
INTRODUCTION
The spindle checkpoint acts as a surveillance system and monitors kinetochore–microtubule interactions during mitosis (Musacchio and Hardwick, 2002; Cleveland et al., 2003; Taylor et al., 2004). It helps ensure that all sister chromatids are bioriented, with sisters attached to opposite spindle poles, before anaphase onset. This checkpoint is not essential in yeasts; indeed, deletion of the mitotic arrest defective (MAD) genes leads to a rather subtle phenotype in the absence of additional perturbation of the mitotic machinery (Warren et al., 2002). However, mouse knockouts of Mad2 and Bub3 are early embryonic lethal (Dobles et al., 2000; Kalitsis et al., 2000), and it has been argued that this is because the spindle checkpoint plays a critical role in the timing of mitosis in vertebrates (Meraldi et al., 2004). Efficient RNA interference (RNAi) of Mad and Bub components in tissue culture shortens mitosis so much that it is catastrophic, because there simply is not enough time to build a spindle and attach all kinetochores properly before anaphase onset (Meraldi et al., 2004; Michel et al., 2004).
The nonessential nature of the yeast MAD genes makes them extremely tractable genetically, and allowed us to perform a synthetic lethal screen with the aim of identifying novel mitotic defects that activate the spindle checkpoint. Neither the mitotic mutation nor a mad1Δ on their own kill cells, but when combined they become lethal. Synthetic lethal screens with mad mutants have been carried out using the viable haploid gene deletion set, and many interactions found, including a range of nonessential spindle and kinetochore functions (Lee and Spencer, 2004; Daniel et al., 2006). Our screen is unbiased: any essential or nonessential component of the mitotic machinery that can be mutated to cause a defect that is recognized by the spindle checkpoint can be identified. A pilot screen that identified 25 mitotic mutants was conducted (Hardwick, unpublished data), but among them were four novel alleles of sfi1.
SFI1 was initially identified as a suppressor of the fil1 mutation (Ma et al., 1999), and it has since been demonstrated to be an essential and widely conserved component of centrosomes, which are known in yeast as spindle pole bodies (SPBs) (Kilmartin, 2003).
The budding yeast spindle pole body is a layered structure embedded in the nuclear envelope and is the sole microtubule-organizing center, nucleating both nuclear and cytoplasmic microtubules from its inner and outer plaques, respectively. Associated with one side of the SPB is a structure called the half-bridge, a specialization of nuclear membrane with a layer of material on the cytoplasmic face (O'Toole et al., 1999). The half-bridge serves several important roles during spindle pole body duplication. On initiation of SPB duplication, the half-bridge extends and a small satellite body is formed on the distal cytoplasmic tip (Byers, 1981). The satellite expands to form a duplication plaque, which is then inserted in the membrane by an unknown process, possibly involving contraction of the half-bridge (Adams and Kilmartin, 1999). After the inner plaque assembles on the nuclear face of duplication plaque, the result is two side-by-side SPBs joined by the bridge. The bridge then cleaves or dissociates, allowing the two SPBs to separate and organize opposite ends of the spindle.
One important component of the half-bridge is Cdc31p, the single budding yeast member of the conserved centrin family. Members of the centrin family have important roles in microtubule-organizing centers such as the SPB, centrosome, and basal bodies; and in higher organisms, they are often the major component of Ca2+-sensitive contractile fibers (Schiebel and Bornens, 1995). Sfi1p biochemically interacts with Cdc31p, is localized to the half-bridge of the SPB, and like Cdc31p is required for early steps in SPB duplication (Kilmartin, 2003). Sfi1p contains a central domain composed of multiple, evolutionarily conserved repeats, each of which can bind to Cdc31p (Kilmartin, 2003). It was suggested that Sfi1p–centrin fibers could act as elastic connections between various elements of the centrosome and that they might mediate a licensing step necessary for centrosome duplication (Salisbury, 2004). Recently, the crystal structures of two Sfi1p–centrin complexes were reported, and it was confirmed that multiple molecules of Cdc31p bind a single Sfi1p, resulting in a filamentous structure (Li et al., 2006). On the basis of electron microscopy (EM) evidence placing the N terminus of Sfi1p at the SPB and the C terminus at the center of the bridge, it was proposed that an Sfi1p/Cdc31p filament could span the length of the half-bridge. Kilmartin and coworkers suggested that during SPB duplication, the half-bridge could double in length through association of two, end-to-end Sfi1 C termini, providing a new Sfi1 N terminus as an assembly site for the new SPB (Li et al., 2006). However, these important functions for the N- and C-terminal domains of Sfi1p have yet to be investigated experimentally.
Here, we describe new sfi1 mutants that arrest in mitosis with duplicated, side-by-side SPBs, indicating that Sfi1p does have further roles to play in addition to its previously described function in centrin-mediated SPB duplication. These mutations all map to the C terminus of Sfi1p, and they are entirely consistent with models in which the Sfi1p C terminus has an important role in separation or splitting of the duplicated SPBs during bipolar spindle assembly.
MATERIALS AND METHODS
General yeast methods and media were used except where stated (Guthrie et al., 2004). Standard stocks were 10 mg/ml α factor, 10 mg/ml nocodazole, 100 mg/ml hydroxyurea, and 30 mg/ml benomyl. Yeast transformation and polymerase chain reaction (PCR)-mediated epitope tagging were carried out using standard procedures (Ito et al., 1983; Longtine et al., 1998), and for mCherry (Shaner et al., 2004; Snaith et al., 2005). DNA sequencing was carried out by PCR according to manufacturer's instructions (ABI Big Dye; Applied Biosystems, Foster City, CA). Table 1 lists the yeast strains used, all of which are derivatives of W303.
Table 1.
Yeast strains used, all of which are derivatives of W303
| Yeast strains | |
|---|---|
| slm65 MATa | sfi1-65, mad1Δ::HIS3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, p(MAD1, URA3, ADE3) |
| slm120 MATa | sfi1-120, mad1Δ::HIS3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, p(MAD1, URA3, ADE3) |
| slm229 MATa | sfi1-229, mad1Δ::HIS3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, p(MAD1, URA3, ADE3) |
| slm273 MATa | sfi1-273, mad1Δ::HIS3, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, p(MAD1, URA3, ADE3) |
| IAY18 MATa | TRP1::SPC42-GFP, spc42Δ1::LEU2, ade2-1, trp1-1, can1-100, leu2,-3, his3-11, ura3 |
| VAY3 MATa | SFI1-YFP::G418, SPC42-CFP::TRP1 |
| VAY20 MATa | sfi1-65, TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| VAY21 MATa | sfi1-120, TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| VAY22 MATa | sfi1-229, TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| VAY23 MATa | sfi1-273, TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| VAY24 MATa | TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
| KH321 MATa | ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100, CFIII URA3 SUP11 |
| VAY25 MATa | sfi1-65, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100, CFIII URA3 SUP11 |
| VAY26 MATa | sfi1-120, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100, CFIII URA3 SUP11 |
| VAY27 MATa | sfi1-229, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100, CFIII URA3 SUP11 |
| VAY28 MATa | sfi1-273, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100, CFIII URA3 SUP11 |
| VAY33 MATa | cdc4, TRP1::SPC42-GFP |
| VAY55 MATa | sfi1-3, TRP1::SPC42-GFP |
| VAY78 MATa | sfi1-120-mCherry:NAT, TRP1::SPC42-GFP, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1 |
| VAY87 MATa | SFI1-mCherry:NAT, ura3-1, leu2,3-112, his3-11, trp1-1, ade2-1, can1-100 |
Yeast Colony Sectoring Assays
The quantitative chromosome loss assay (CFIII) was performed as described previously (Warren et al., 2002).
Linkage Analysis of the Novel sfi1 Alleles
To test genetically whether the synthetic lethal mutations were tightly linked to the sfi1 locus, we constructed a wild-type SFI1, mad1Δ strain in which the URA3 marker was integrated just upstream of SFI1. This strain was then crossed with the synthetic lethal mutants, diploids were sporulated, and tetrads were dissected. If the synthetic lethal mutation was linked to the SFI1 locus, no spores would be recovered that were URA3+ and that contained the synthetic lethal mutation. The latter was assessed visually, using the MAD1, ADE3 plasmid and colony sectoring assay. We dissected at least 24 tetrads for each sfi1 allele, and we never saw recombination between the mutation and URA3. This showed that each mutation was tightly linked to, and most likely lay within, the SFI1 gene. This was later confirmed by sequencing these sfi1 alleles (Figure 1B).
Figure 1.
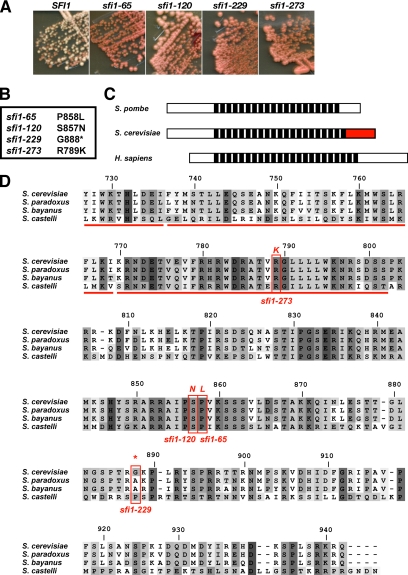
The C terminus of Sfi1p is highly conserved among budding yeast, and it is mutated in the novel mad1-synthetic lethal sfi1 alleles. (A) The four mutants (slm65, 120, 229, and 273) are synthetically lethal with a mad1Δ; therefore, they are unable to grow without the MAD1, ADE3 plasmid. Colonies containing this ADE3 plasmid were red (as they are ade2−), whereas those that lost it were white (ade2−, ade3−). Only the strain containing wild-type SFI1 was able to lose the plasmid and form white sectors and colonies. (B) Three of the sfi1 alleles have mutations in conserved residues, and the fourth encodes a truncated protein. (C) Schematic diagram of the Sfi1 protein indicating the three domains. The central domain consist of ∼20 repeats that are conserved from yeast to human. The red region highlights the domain of Sfi1p containing the synthetic lethal mutations. (D) Sequence alignment of the C terminus of Sfi1p from four different Saccharomyces species (modified from http://db.yeastgenome.org/fungi/YLL003W.html). The four mutations identified in this study are indicated. Red bars underneath the sequence indicate the last three repeats (19–21) from the central domain.
Synthetic Lethality of the sfi1-C-Terminal Mutations with mad3Δ and bub1Δ
The sfi1 mutations were all crossed with strains containing either a mad3 or bub1 deletion that was complemented by a plasmid carrying MAD3 or BUB1 and the URA3 marker. The double mutants isolated from these crosses were able to grow on −URA media but not on plates containing 5-fluoroorotic acid, indicating that the MAD3 or BUB1 gene were needed for viability, and that the sfi1 alleles are also synthetic lethal with other checkpoint mutations.
Sfi1p Antibodies
Nucleotides 1-1655 of SFI1 were PCR amplified and cloned into pGEX-6P. Bacterial cultures were induced with isopropyl β-d-thiogalactoside overnight at 18°C, pelleted, frozen, and then ground in a pestle and mortar under liquid nitrogen. Extracts were thawed in lysis buffer (phosphate-buffered saline, 1 M NaCl, 1 mM phenylmethylsulfonyl fluoride, and 0.5% Tween 20), sonicated for 1 min, and clarified. Sfi1–glutathione S-transferase (GST) fusion protein was then purified on glutathione agarose by using standard protocols, dialyzed overnight (50 mM HEPES, pH 7.6, 100 mM KCl, and 30% glycerol), and used as antigen. Anti-Sfi1p serum was affinity purified on a column of Sfi1-GST coupled to Affigel 10 (Bio-Rad, Hercules, CA) as described previously (Hardwick and Murray, 1995). Immunoblotting was carried out as described previously (Hardwick and Murray, 1995).
Light Microscopy
Strains were grown in complete synthetic media (CSM) or the appropriate selective CSM media (Qbiogene, Carlsbad, CA) made up to double recommended concentration. For GFP/YFP/CFP/mCherry imaging, cells were analyzed live, or they fixed briefly with formaldehyde (typically 3.7% for 5–10 min). Microscopy was performed with a Zeiss Axiovert 200 Marianas inverted microscope system from Intelligent Imaging Innovations (Denver, CO), with a CoolSNAP HQ charge-coupled device camera. Images were taken and analyzed using Slidebook software (Intelligent Imaging Innovations).
Electron Microscopy
Cells were fixed, processed, and imaged as described previously (Giddings et al., 2001). Twenty-four sfi1 cells were analyzed very thoroughly by serial section.
RESULTS
Isolation of Novel Alleles of sfi1
A mad1 synthetic lethal screen (Hardwick, unpublished data) isolated four novel sfi1 alleles. The screen used an ADE3, MAD1 plasmid sectoring assay and ade2, ade3, mad1Δ mutants. These strains are red if they maintain the ADE3 plasmid, but they are white if they lose it. The synthetic lethal mutants die if they lose the MAD1, ADE3 plasmid and therefore grow to form entirely red colonies (Figure 1A). These four mutants were all rescued with an SFI1 clone from a YCP50-based genomic library (Hardwick and Murray, 1995), enabling them to lose the MAD1, ADE3 plasmid and form sectored colonies (data not shown, but see Figure 6). Linkage experiments suggested that the synthetic lethal mutations were likely to be sfi1 alleles rather than being suppressed by the SFI1 gene (see Materials and Methods for details). The sfi1 locus was then PCR amplified using genomic DNA from all four strains, sequenced, and found to contain mutations in, or very close to, the C-terminal domain of Sfi1p (Figure 1B). This domain lies after the central repeat domain that has been shown to interact with Cdc31p. The sfi1-273 allele (R789K) actually lies in the last of the 21 repeats thought to interact with centrin (Li et al., 2006), but it could perturb the overall structure of the C terminus. Although the C terminus is not conserved in other model organisms or human Sfi1p, database analysis showed that it, if the last repeat is included, is the section of Sfi1p with the highest levels of homology among budding yeast (ranging from 40% identity for Saccharomyces cerevisiae and Saccharomyces castelli to 98% identity for S. cerevisiae and S. paradoxus; Figure 1C). This suggested that the C terminus of Sfi1p has an important conserved function, at least in budding yeast.
Figure 6.
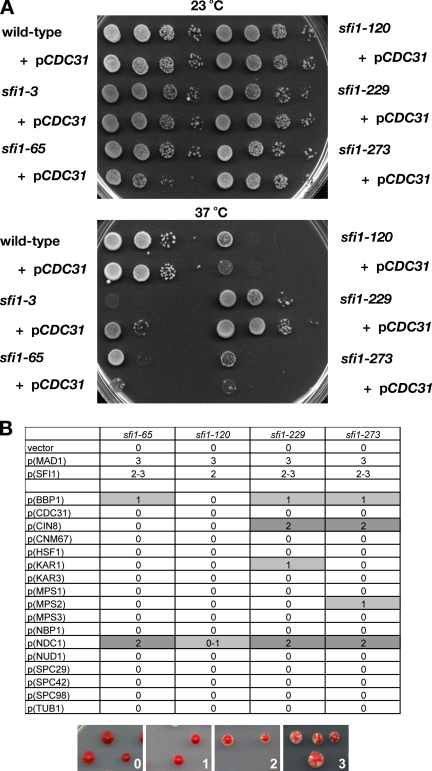
The sfi1 C-terminal mutants are not suppressed by overexpression of CDC31, but they do display genetic interactions with other SPB components. (A) The strains indicated, either containing or lacking extra plasmid-borne copies of the CDC31 gene, were diluted and spotted onto YPD plates. Images were taken after 3-d growth at either 23 or 37°C. Only sfi1-3 was suppressed, and sfi1-65, sfi1-120, and sfi1-273 all grew slower in the presence of extra CDC31. (B) The four sfi1, mad1Δ alleles were all transformed with the bank of plasmids listed in the left column. The ability of the transformant to sector was scored on the scale 0 (no sectoring, red colonies) to 3 (considerable sectoring with large white sectors). Sectoring indicates that the strain had been able to lose a MAD1, ADE3 plasmid. This could be due to either rescue of the mad1 or sfi1 mutation (as in rows 2 and 3) or suppression of the sfi1 mutant phenotype by the other plasmids.
Their isolation as mad1 synthetic lethal mutations suggested that these sfi1 alleles have mitotic defects, even at 23°C, because they are dependent on the spindle checkpoint for viability. To further our understanding of their phenotypes, we analyzed the growth of these sfi1 alleles in different conditions, and we compared them with the tight temperature-sensitive sfi1-3 allele (this contains mutations H207Q, F208C, Y247C; Kilmartin, 2003). The new sfi1 mutants grew significantly slower than wild type at 18 and 36°C, although their cold-sensitive phenotype was somewhat variable. We analyzed their temperature sensitivity further by shifting log phase cultures to 36°C for 4 h and then analyzing cells by fluorescence-activated cell sorting and microscopy. No clear DNA replication defect was observed by FACS (Figure 2B), but a significant increase in the number of large-budded cells was observed in all four strains (Figure 2C). Such an accumulation of large-budded cells with 2C DNA is consistent with a mitotic delay due to spindle checkpoint activation. Next, we tested whether these sfi1 alleles were sensitive to antimicrotubule drugs, and we found them to be benomyl resistant (Figure 2D). This is a relatively unusual phenotype and suggests the possibility that these mutants somehow stabilize microtubules, thereby counteracting the effects of antimicrotubule drugs.
Figure 2.
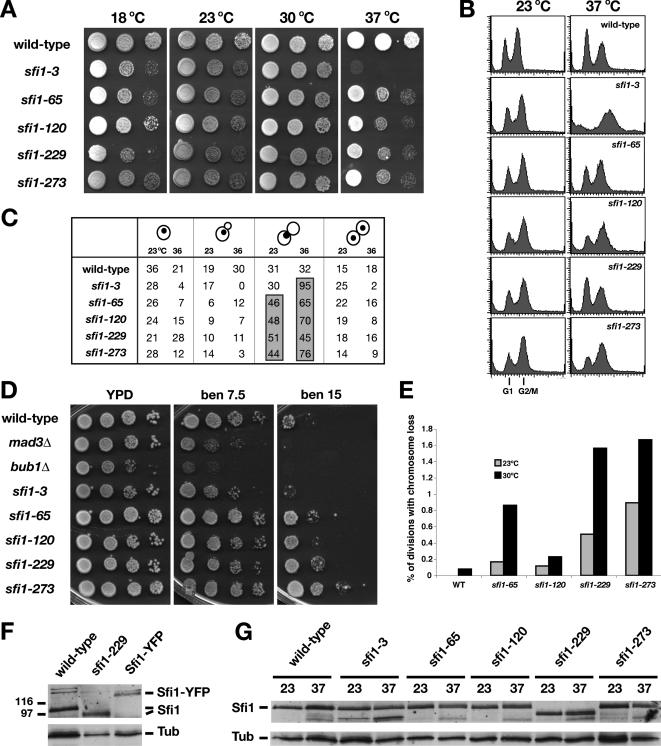
The novel sfi1 alleles are temperature sensitive, resistant to benomyl, and display increased chromosome loss. (A) sfi1-65, sfi1-120, sfi1-229, and sfi1-273 are temperature and cold sensitive, but far less so than sfi1-3. Yeast strains were diluted then spotted onto rich media (YPD) plates. Images were taken after 3-d growth at the indicated temperatures. (B) Wild-type, sfi1-3, sfi1-65, sfi1-120, sfi1-229, and sfi1-273 cultures were shifted to 37°C for 4 h, fixed, and processed for FACS analysis (n = 20,000 cells). Although sfi1-3 clearly arrested with a G2/M DNA content (74% of cells), this was less apparent for the other sfi1 alleles (62, 63, 53, and 66% respectively), which is consistent with them having a less penetrant arrest. (C) Cells from B were analyzed by light and fluorescence microscopy (DNA was stained with propidium iodide during the FACS protocol), and the budding index of the various cultures was scored. The four novel sfi1 alleles all displayed an accumulation of large-budded cells, and with the exception of sfi1-229, this was enhanced at 37°C. (D) The novel sfi1 alleles are benomyl resistant. The yeast strains were diluted and spotted onto YPD plates containing the indicated concentration of the antimicrotubule drug benomyl (micrograms per milliliter). Images were taken after 3-d growth at 24°C. (E) A colony sectoring assay (CFIII) was used to measure the chromosome loss rates of the novel sfi1 alleles at 23 and 30°C. Numbers indicate the percentage of divisions leading to loss of the marker chromosome during the first division on the solid media (half-sectored). (F) Immunoblots of total protein extracts using anti-Sfi1p antibody. Specificity of the antibody is shown by the gel shifts in Sfi1p found in the sfi1-229 truncation allele and a strain where the wild-type protein had been tagged with YFP. (G) All mutant alleles of sfi1 express stable proteins. Yeast cultures were grown at 23 or 37°C for 3 h, total protein extracts were made, and they were immunoblotted for Sfi1p.
Mitotic defects in budding yeast tend to lead to an increased chromosome loss rate. To quantitate this in these sfi1 alleles, we used another colony sectoring assay (see Materials and Methods for details). All four sfi1 alleles demonstrated increased chromosome loss at semipermissive temperatures (30°C), and they all showed subtle defects at 23°C (Figure 2C). The latter result was to be expected, because even at 23°C these mutations require the spindle checkpoint for viability. Note, these sfi1 mutations are also synthetic lethal with mad3Δ and bub1Δ (see Materials and Methods for details), indicating that this is not a genetic interaction that is specific to the loss of Mad1p function.
Protein Stability and Localization of Sfi1p Are Unaffected
Next, we analyzed the mutant Sfi1 proteins, to determine whether their stability was affected upon temperature shift. To do this, we raised polyclonal antibodies to the N terminus of Sfi1p (see Materials and Methods), and we used these on immunoblots of total protein extracts. These antibodies recognized wild-type Sfi1p, which ran at the predicted size of 112 kDa. As expected, a truncated protein was detected in sfi1-229 (97 kDa), and a larger fusion protein of ∼140 kDa was recognized in the strain expressing Sfi1-YFP (Figure 2F). We immunoblotted extracts made from wild-type and sfi1 mutant cells grown at 23 and 37°C. This showed that all of the mutant Sfi1 proteins were stable, even at 37°C (Figure 2G). We conclude that the C-terminal sfi1 mutations do not significantly affect the stability of the Sfi1 protein.
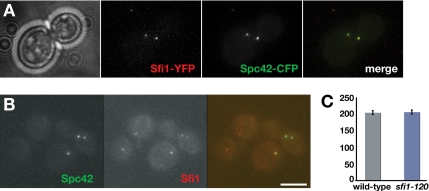
We used double-label fluorescence experiments to confirm that Sfi1p localizes to budding yeast SPBs (Figure 3A; Kilmartin, 2003). To determine whether our mad1 synthetic lethal sfi1 alleles were defective in SPB targeting, we fused the monomeric red fluorescent protein mCherry (Shaner et al., 2004) to sfi1-120. To ensure identical exposure times and image processing, we imaged a mixture of mCherry-tagged sfi1-120p mutant cells, which also expressed Spc42-green fluorescent protein (GFP), and mCherry-tagged wild-type Sfi1p cells that did not express Spc42-GFP. The sfi1-120 mutant protein was still detected at SPBs (Figure 3B), and quantitative analysis showed that it was there at wild-type levels (Figure 3C). Thus, the sfi1 C-terminal point mutants do not seem to affect the stability of Sfi1p or its targeting to the SPB.
Figure 3.
The C-terminal mutation of Sfi1p does not affect its targeting to the yeast SPB. (A) Wild-type Sfi1-YFP colocalizes with the spindle pole body marker Spc42-CFP. Cells were fixed briefly for 5 min in 3.7% formaldehyde, and then they were imaged using a CFP/YFP filter set. (B) Cells containing Sfi1-mCherry, or sfi1-120-mCherry and Spc42-GFP, were fixed briefly for 5 min in 3.7% formaldehyde, and then they were mixed and imaged together. Spc42-GFP was used to distinguish the two cell types. The mutant Sfi1 protein was found at SPBs. Bar, 5 μm. (C) Quantitation of images from these same mixed cultures showing that both wild-type and the sfi1-120 mutant protein were found at SPBs at very similar levels. The y-axis shows mean pixel intensity at the SPBs, in arbitrary units, as measured in Slidebook. We analyzed 149 sfi1-120 cells and 270 wild-type cells. Bars indicate SD.
Analysis of SPBs and Spindles in sfi1 Mutants
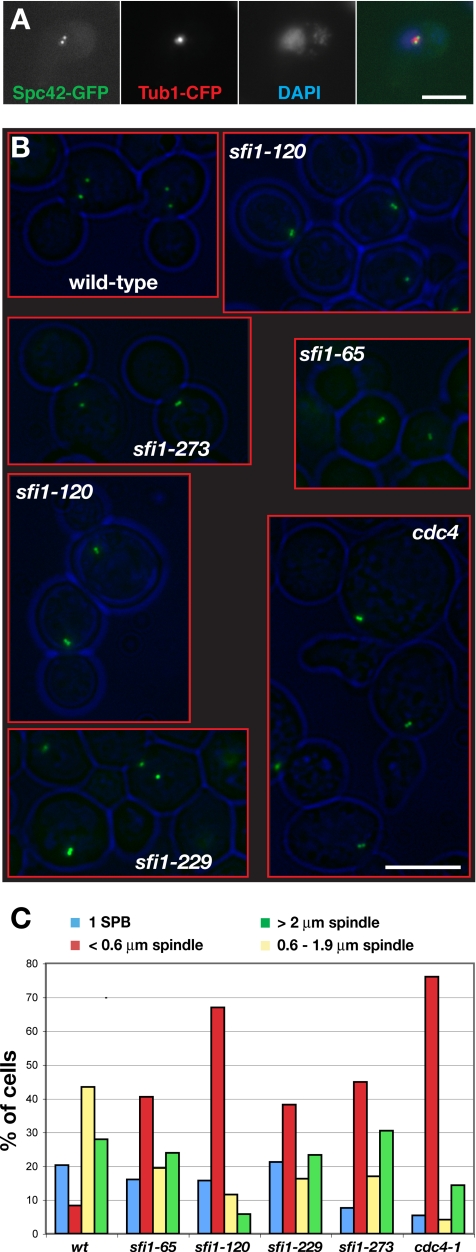
sfi1-3 (H207Q, F208C, Y247C) and sfi1-7 (N218D, D242A) are tight conditional (ts) alleles that arrest with an elongated but unduplicated SPB (Kilmartin, 2003). Analysis of spindles in sfi1-65 and sfi1-120 alleles revealed a very distinct phenotype (Figure 4). We presynchronized cells in G1 with α-factor, and then we released them to 37°C for 3 h, before fixing them briefly with formaldehyde. When analyzed by light microscopy, using cyan fluorescent protein (CFP)-tubulin and Spc42-GFP, we observed a significant enrichment of cells with paired GFP spots (Figure 4, A and B). The two Spc42-GFP spots were typically ∼0.3 μm apart. These spindle poles were much closer together than in a normal short mitotic spindle (∼1.2 μm in early wild-type mitosis), and they had a very similar separation to the spindles observed in cdc4 mutants (also 0.3 μm), which are known to arrest with duplicated but unseparated SPBs (Figure 4C; Byers and Goetsch, 1974). This suggested that the new sfi1 alleles had arrested with side-by-side SPBs or with very short bipolar spindles.
Figure 4.
The sfi1 C-terminal mutants have duplicated SPBs but abnormally short spindles. (A) sfi1-65 cells containing Spc42-GFP and Tub1-CFP were shifted to 37°C for 3 h, fixed briefly for 5 min in 3.7% formaldehyde, and then they were imaged to analyze SPB separation and spindle structures. (B) Cells were presynchronized cells in G1 with α-factor, released at 37°C for 3 h, and then fixed briefly with formaldehyde. These strains contain Spc42-GFP, and they were imaged to look at their SPB number and position. Bar, 5 μm. (C) Quantitation of the distances separating the SPBs in the aforementioned strains. sfi1-65, sfi1-120, sfi1-229, and sfi1-273 all displayed increased numbers of short (<0.6-μm) spindles, which were very similar in appearance to the cdc4 control strain. A minimum of 118 cells were counted per sample (n = 444 for wild type and 380 for sfi1-65, 312 for sfi1-120, 283 for sfi1-229, 118 for sfi1-273, and 167 for cdc4).
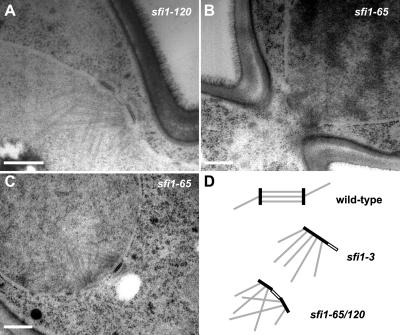
To look at these spindle defects in more detail, we used thin-section electron microscopy, and we analyzed serial sections. sfi1-65 and sfi1-120 cells were α-factor arrested and then released at 37°C for 3 h before high-pressure freezing. Although some large-budded cells had bipolar spindles that looked normal, 50% had duplicated side-by-side SPBs (Figure 5). These SPBs were of equal and normal size, had a clear half-bridge connecting them, and had nuclear microtubules emanating from them. This demonstrates that the sfi1-65 and sfi1-120 defect is postduplication and that it does not affect the ability of the SPBs to nucleate microtubules. Thus, these sfi1 mutants have a defect in SPB separation. This analysis by electron microscopy demonstrates that Sfi1p has an additional role to play in bipolar spindle formation after SPB duplication.
Figure 5.
Electron microscopy reveals duplicated but unseparated SPBs in the sfi1 C-terminal mutants. Cells were presynchronized cells in G1 with α-factor and released at 37°C for 3 h. Cells were high-pressure frozen, and then they were freeze-substituted in 2% osmium tetroxide and 0.1% uranyl acetate (see Giddings et al., 2001 for further details). (A) EM image of side-by-side SPBs in sfi1-120. (B and C) EM images of SPBs in sfi1-65. Bars are 0.3 μm. (D) Schematic representation of the SPB phenotype observed by EM, in different sfi1 alleles. Note, this is our interpretation of the sfi1-3 phenotype from data in Kilmartin (2003).
Genetic Interactions of sfi1 C-Terminal Mutants
The spindle defect in the sfi1 C-terminal mutants is quite distinct from that of the previously described sfi1-3 and sfi1-7 alleles. The sfi1-3 and sfi1-7 defect is probably due to impaired interaction with the budding yeast centrin protein Cdc31p, and their temperature sensitivity could be suppressed by overexpression of CDC31 (Kilmartin, 2003). To confirm that the sfi1 C-terminal defect(s) of mutants was mechanistically distinct from that of sfi1-3 and sfi1-7, we tested whether CDC31 overexpression could also suppress their temperature sensitivity. This was not the case (Figure 6A). We next tested a bank of high copy (2-μm) plasmids encoding other SPB components to see whether any of them could suppress the sfi1 C-terminal alleles. To do this, we tested whether their overexpression could suppress the synthetic lethality of the sfi1 C-terminal mutant alleles with mad1Δ, by using the MAD1, ADE3 plasmid and colony sectoring assay. These sfi1 mutants require MAD1 for viability, and by maintaining the MAD1, ADE3 plasmid, they form red colonies (because they are chromosomally ade2, ade3 mutants). If the 2-μm plasmid suppresses the sfi1 mutation, they are then able to lose the ADE3 plasmid and form red/white sectored colonies (Figure 6B). As expected, plasmids encoding either SFI1 or MAD1 allowed frequent colony sectoring. In addition, we found that BBP1, NDC1, MPS2, KAR1, and CIN8 plasmids could all suppress the synthetic lethality to different extents (Figure 6C). Bbp1, Mps2, and Ndc1 are all implicated in the process of inserting the newly formed SPB duplication plaque into the nuclear membrane, and they possibly have roles in anchoring the SPB once inserted (Chial et al., 1998; Schramm et al., 2000; Park et al., 2004; Araki et al., 2006). This suggests that at least part of the sfi1 C-terminal mutant phenotype could reflect a defective interaction between the SPB and the nuclear envelope. However, our EM analysis does not support an insertion defect, because the duplicated SPBs seem to be sitting normally in the nuclear envelope, and they are able to nucleate microtubules. It is not known what other roles the Mps2/Bbp1 membrane protein complex has to play in spindle biogenesis.
DISCUSSION
We have isolated several novel sfi1 alleles and shown by light and electron microscopy that they have phenotypes distinct from those described previously for other ts alleles of sfi1. This was confirmed through analysis of genetic interactions with CDC31. These findings demonstrate that Sfi1p is involved in aspects of bipolar spindle biogenesis after the SPBs have been duplicated. The electron microscopy suggests that the novel sfi1 defect is in separation of the duplicated SPBs, possibly defective splitting of the bridge.
Multiple Functions for Sfi1p
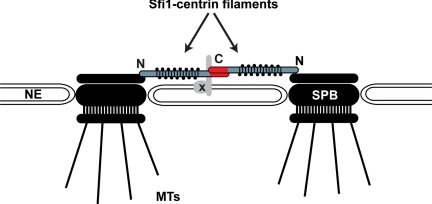
Sfi1p had already been shown to have a role in the initial stage of SPB duplication. The sfi1-3 and sfi1-7 alleles, which have mutations in the Cdc31p binding repeats, arrest with only one spindle pole body before satellite assembly (Kilmartin, 2003). Recent work from the Kilmartin laboratory has described the crystal structure of Sfi1p–Cdc31p complexes (Li et al., 2006). In addition, it was shown by EM shadowing that Sfi1p–Cdc31p complexes form filaments in isolation. The authors proposed an elegant model describing the role of Sfi1p in SPB duplication. The model has Sfi1p molecules spanning the half-bridge with their N termini anchored in the spindle pole body and their C termini at the distal end. They suggested that upon initiation of SPB duplication additional Sfi1p molecules are recruited to the half-bridge and interact with the existing half-bridge Sfi1p molecules, end on, via their C termini. This end-on interaction results in a doubling of the bridge length, and the Sfi1 C termini are now positioned at the center of the bridge (Figure 7). This model was strongly supported by immuno-EM analysis of strains that expressed Sfi1p with either N- or C-terminal GFP tags: the N terminus was positioned next to the junction between the SPB and the proximal end of the half-bridge, whereas the C-terminal tag was found to be close to the distal end, or in the center of the bridge in paired SPBs (Li et al., 2006). Thus, the Sfi1p C termini are ideally placed to mediate interactions necessary for bridge separation. It was suggested that an important role for the Sfi1p C terminus might be in mediating the cleavage process, either alone or in combination with other proteins.
Figure 7.
Model of Sfi1 function during SPB duplication and splitting. Schematic representation of the Sfi1–centrin filament spanning the bridge of the duplicated SPBs. Factor X is predicted to interact with the C terminus of Sfi1p and to be instrumental in the cleavage of the bridge upon bipolar spindle formation.
Our findings provide very strong support for such a model. In this study, we have isolated novel alleles of sfi1 that arrest late in spindle pole body duplication, with duplicated but unseparated SPBs. Our EM analysis of these sfi1 alleles is consistent with a defect in bridge cleavage, and the mutations conferring this phenotype all mapped to the C terminus of Sfi1p. To develop this model further, it will be important to find proteins that interact with the C terminus of Sfi1p. In an attempt to do this, we carried out two-hybrid screens and Sfi1p affinity chromatography (with C-terminal Sfi1p–GST fusion proteins), but as yet we have failed to find any interactors (data not shown). One possible explanation for this failure is that the interacting Sfi1p partners could be integral membrane proteins, which would be difficult to identify with either of the approaches taken.
Our genetic studies confirm that the sfi1 C-terminal mutant phenotype is novel and shows that it is not due to a defective interaction with Cdc31p, as was found for the sfi1-3 and sfi1-7 alleles. In addition, our multicopy suppression analysis identified other components of the spindle pole body that can partially rescue the defect of sfi1 C-terminal mutations. One of these components was the kinesin motor protein Cin8p. This is a plus-end–directed motor required to separate duplicated SPBs by producing an outward force on the bipolar spindle (Saunders and Hoyt, 1992; Saunders et al., 1997). We suggest that overexpression of the Cin8 motor protein might increase this outwards force to such an extent that it is capable of splitting and driving apart the duplicated SPBs in the sfi1-229 and sfi1-273 mutants.
Some of the sfi1 suppressors encode integral membrane proteins (Ndc1p, Mps2p) of the nuclear envelope, and others are known to form a complex with them (Mps2-Bbp1-Nbp1). The terminal phenotype of conditional alleles of these SPB components is typically a “dead” SPB, which fails to nucleate nuclear microtubules (Chial et al., 1998; Schramm et al., 2000; Park et al., 2004; Araki et al., 2006). This is due to a failure to insert the new SPB into the nuclear envelope. Could these complexes interact with the Sfi1p C termini? It was recently shown that bridge components, such as Sfi1p, Cdc31p, Mps3p, and Kar1p, are all present on these dead SPBs (Araki et al., 2006), so it is unlikely that these factors are involved in targeting or maintaining Sfi1p at the bridge. Whether the “insertion” factors have later roles to play in spindle biogenesis is unclear, although it is thought that Nbp1 is required to maintain Mps2p at the SPB (Araki et al., 2006). We propose that the C termini of Sfi1p do have an important role to play in bridge splitting, as proposed originally (Li et al., 2006), and that this separation event might require interaction with integral components of the nuclear envelope, such as the Mps2–Bbp1 complex (see model in Figure 7).
Almost nothing is known about separation of the SPB bridge. Another mutant that arrests at a similar stage is cdc4, an F-box component of the ubiquitin ligase known as the SCF (Mathias et al., 1996; Cardozo and Pagano, 2004). This suggests the possibility that an SCF substrate needs to be degraded, by ubiquitin-mediated proteolysis, for bridge separation to occur. It is tempting to think that a half-bridge linker molecule is itself the SCF substrate, but it is also possible that the SCF substrate regulates bridge separation. Indeed, SCF activity may simply be needed to get to the correct stage of the cell cycle for SPB separation to occur, perhaps through the activation of microtubule motors and changes in microtubule dynamics after S phase onset. That increased Cin8 motor protein expression can suppress two of the sfi1 alleles is interesting and suggests that increased motor activity can be sufficient to drive defective Sfi1p C termini apart leading to SPB separation.
It will be of interest to discover whether Sfi1 homologues have roles to play in splitting and separation of vertebrate centrosomes, and if so, whether the Sfi1 proteins are perturbed in cancers. Many human tumors display centrosome aberrations, but it is unclear as to whether these are typically the cause or consequence of cancer progression (Nigg, 2002). Either way, it is possible that the spindle checkpoint could make a useful drug target for anticancer therapy, because we have shown here that the combined loss of Sfi1p and Mad1p functions can be completely lethal to cells.
ACKNOWLEDGMENTS
We thank John Kilmartin (MRC, LMB, Cambridge) for yeast strains, plasmids, advice, and communication of unpublished observations; Mark Winey (University of Colorado, Boulder) and Sue Jasperson (Stowers Institute, Kansas City) for yeast strains, plasmids, and advice; Julie Blyth (University of Edinburgh) for Sfi1p antibodies; Julie Luquet (University of Edinburgh) for sequence analysis of the sfi1 mutant alleles; Antje Geissenhoener (University of Edinburgh) for back-crossing and complementation analysis of the mad1 synthetic lethal mutants; Hilary Snaith (University of Edinburgh) and Ken Sawin (University of Edinburgh) for the mCherry tagging cassette; Oliver Kerscher (John Hopkins University School of Medicine), Elmar Schiebel (ZMBH, Heidelberg), Sue Biggins (Fred Hutchinson Cancer Research Center, Seattle), and the NCRR Yeast Resource Center for CFP and YFP reagents and yeast strains; and Karen May (University of Edinburgh) for Slidebook assistance and image quantitation. This work was supported by the Wellcome Trust of which K.G.H. is a Wellcome Trust Senior Research Fellow in Biomedical Science, and V.A. was a Wellcome Trust Prize Student. J.P. and S.P. were supported by a grant from the Fred Hutchison Cancer Center (Seattle, WA).
Abbreviations used:
- EM
electron microscopy
- MAD
mitotic arrest defective
- NAT
nourseothricin
- SPB
spindle pole body.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Lau C. K., Maekawa H., Jaspersen S. L., Giddings T. H., Jr, Schiebel E., Winey M. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol. Biol. Cell. 2006;17:1959–1970. doi: 10.1091/mbc.E05-07-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. Mol. Genet. Yeast. 1981;16:119–133. [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chial H. J., Rout M. P., Giddings T. H., Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Daniel J. A., Keyes B. E., Ng Y. P., Freeman C. O., Burke D. J. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 2006;172:53–65. doi: 10.1534/genetics.105.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M. L., Benezra R., Sorger P. K. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Giddings T. H., Jr, O'Toole E. T., Morphew M., Mastronarde D. N., McIntosh J. R., Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–42. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G., Abelson J, Simon M. Guide to Yeast Genetics and Molecular Biology, Part A. Vol. 194. San Diego, CA: Elsevier Academic Press; 2004. [Google Scholar]
- Hardwick K. G., Murray A. W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Fowler K. J., Choo K. H. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Spencer F. A. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA. 2004;101:10655–10660. doi: 10.1073/pnas.0404102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Sandercock A. M., Conduit P., Robinson C. V., Williams R. L., Kilmartin J. V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ma P., Winderickx J., Nauwelaers D., Dumortier F., De Doncker A., Thevelein J. M., Van Dijck P. Deletion of SFI1, a novel suppressor of partial Ras-cAMP pathway deficiency in the yeast Saccharomyces cerevisiae, causes G(2) arrest. Yeast. 1999;15:1097–1109. doi: 10.1002/(SICI)1097-0061(199908)15:11<1097::AID-YEA437>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mathias N., Johnson S. L., Winey M., Adams A. E., Goetsch L., Pringle J. R., Byers B., Goebl M. G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Draviam V. M., Sorger P. K. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Michel L., Diaz-Rodriguez E., Narayan G., Hernando E., Murty V. V., Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc. Natl. Acad. Sci. USA. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nigg E. A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M., McIntosh J. R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C. J., Song S., Giddings T. H., Jr, Ro H. S., Sakchaisri K., Park J. E., Seong Y. S., Winey M., Lee K. S. Requirement for Bbp1p in the proper mitotic functions of Cdc5p in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1711–1723. doi: 10.1091/mbc.E03-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J. L. Centrosomes: Sfi1p and centrin unravel a structural riddle. Curr. Biol. 2004;14:R27–R29. doi: 10.1016/j.cub.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Saunders W. S., Hoyt M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M. A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E., Bornens M. In search of a function for centrins. Trends Cell Biol. 1995;5:197–201. doi: 10.1016/s0962-8924(00)88999-0. [DOI] [PubMed] [Google Scholar]
- Schramm C., Elliott S., Shevchenko A., Schiebel E. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 2000;19:421–433. doi: 10.1093/emboj/19.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Snaith H. A., Samejima I., Sawin K. E. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 2005;24:3690–3699. doi: 10.1038/sj.emboj.7600838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Scott M. I., Holland A. J. The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12:599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- Warren C. D., Brady D. M., Johnston R. C., Hanna J. S., Hardwick K. G., Spencer F. A. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]