Abstract
CENP-C is a conserved inner kinetochore component. To understand the precise roles of CENP-C in the kinetochore, we created a cell line with a conditional knockout of CENP-C with the tetracycline-inducible system in which the target protein is inactivated at the level of transcription. We found that CENP-C inactivation causes mitotic delay. However, observations of living cells showed that CENP-C-knockout cells progressed to the next cell cycle without normal cell division after mitotic delay. Interphase cells with two nuclei before subsequent cell death were sometimes observed. We also found that ∼60% of CENP-C–deficient cells had no Mad2 signals even after treatment with nocodazole, suggesting that lack of CENP-C impairs the Mad2 spindle checkpoint pathway. We also observed significant reductions in the signal intensities of Mis12 complex proteins at centromeres in CENP-C–deficient cells. CENP-C signals were also weak in interphase nuclei but not in mitotic chromosomes of cells with a knockout of CENP-K, a member of CENP-H complex proteins. These results suggest that centromere localization of CENP-C in interphase nuclei occurs upstream of localization of the Mis12 complex and downstream of localization of the CENP-H complex.
INTRODUCTION
Faithful chromosome segregation during mitosis is essential for accurate transmission of genetic material. A kinetochore is assembled at the centromere of each chromatid of a replicated chromosome, and it forms a dynamic interface with microtubules of the mitotic spindle (Pluta et al., 1995; Cleveland et al., 2003). To promote alignment and proper segregation of mitotic chromosomes, sister kinetochores must attach to microtubules and regulate cell cycle progression. This process requires the integrated activities of multiple kinetochore proteins. However, the mechanisms of kinetochore assembly in vertebrate cells for promotion of accurate chromosome segregation remain unclear. To understand the molecular mechanism of kinetochore assembly in vertebrate cells, components of vertebrate kinetochores must be identified, and each component must be characterized. In budding yeasts, >60 kinetochore proteins have been identified by means of genetic and biochemical approaches (Cheeseman et al., 2002; De Wulf et al., 2003; Westermann et al., 2003). Because the vertebrate kinetochore may be more complicated than that of budding yeast and because <60 kinetochore components have been identified in higher vertebrate cells to date, it is likely that other components of the vertebrate kinetochore remain to be isolated. Moreover, many kinetochore components of higher vertebrate cells, including the Mis12 complex proteins (Cheeseman et al., 2004; Obuse et al., 2004a; Kline et al., 2006) and the CENP-H/I complex proteins (Obuse et al., 2004b; Minoshima et al., 2005; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006), have been isolated in recent years.
CENP-C, a kinetochore component in higher vertebrate cells, was originally identified as an antigen of anti-centromere antibodies from patients with various autoimmune diseases (Moroi et al., 1980; Earnshaw and Rothfield, 1985). CENP-C is localized to the inner kinetochore plates adjacent to the centromeric DNA (Saitoh et al., 1992), and it is known to have DNA binding ability (Yang et al., 1996). Conditional knockout of CENP-C in chicken DT40 cells (Fukagawa and Brown, 1997) and disruption of the CENP-C gene in mice (Kalitsis et al., 1998) revealed that CENP-C is essential for cell proliferation. Analysis of a conditional knockout of CENP-C in DT40 cells revealed that the inactivation of CENP-C caused mitotic delay, chromosome missegregation, and apoptosis (Fukagawa and Brown, 1997; Fukagawa et al., 1999). Mitotic arrest has been observed after microinjection of anti-CENP-C antibodies into human HeLa cells (Tomkiel et al., 1994). Antibody microinjection experiments revealed that CENP-C or an associated protein is involved in determination of kinetochore size. CENP-C homologues were identified from several species, including yeast, nematode, and fly (Brown et al., 1993; Moore and Roth, 2001; Oegema et al., 2001; Heeger et al., 2005; Holland et al., 2005). The Caenorhabditis elegans homologue of CENP-C, HCP4, is involved in sister kinetochore resolution (Moore et al., 2005). Recently, a Drosophila CENP-C homologue was isolated as a genetic interactant with Separase, which is essential for sister chromatid separation (Heeger et al., 2005). However, the role of CENP-C in sister chromatid separation and resolution in higher vertebrate cells remains unclear.
We previously reported creation of a cell line with a conditional knockout of CENP-C with an estrogen receptor (ER) system (Fukagawa and Brown, 1997). In this conditional knockout system, the target protein is inactivated by posttranslational regulation, and it is possible that other proteins associated with CENP-C may be inactivated. To investigate the precise phenotype of knockout cells, it is better to use a knockout system, such as a tetracycline (tet)-inducible system, in which the target protein is inactivated at the level of transcription. We have used the tet system to create several knockout cell lines for centromere proteins (Fukagawa, 2004). Many kinetochore components have been identified in recent years, and it is necessary to use conditional knockout cells in which expression of CENP-C is controlled at the transcriptional level to investigate interactions between kinetochore proteins. In the present study, we created a CENP-C-conditional knockout cell line by using the tet system, and we examined the phenotype of the knockout cells, including analyses of the relation of CENP-C with several kinetochore components. We observed a significant reduction in the intensities of immunofluorescent signals of Mis12 complex proteins and of Nuf2 complex proteins at centromeres in CENP-C–deficient cells. In addition, CENP-C signals were weak and diffuse in interphase nuclei of cells with a knockout of CENP-K, a member of the CENP-H complex proteins. These results suggest that localization of CENP-C to the centromere occurs upstream of Mis12 complex localization and that it occurs downstream of CENP-H complex localization in interphase nuclei. We also observed that CENP-C–deficient cells progress to the next cell cycle after mitotic delay. Consistent with this observation, we found that CENP-C–deficient cells had no or weak signals for checkpoint proteins, suggesting that CENP-C–deficient cells cannot maintain spindle checkpoint function.
MATERIALS AND METHODS
Molecular Biology, Cell Culture, and Transfection
All plasmids were constructed by standard methods. For the CENP-C disruption constructs, a histidinol- (hisD) or puromycin-resistance cassette under control of the β-actin promoter was inserted between the two arms. Target constructs were transfected into DT40 cells with a Gene Pulser II electroporator (Bio-Rad, Tokyo, Japan). All molecular biology experiments, including Southern blot analysis and Western blot analysis, followed standard methods.
DT40 cells were cultured and transfected as described previously (Fukagawa et al., 2004). All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin. To suppress expression of the tetracycline-responsive CENP-C transgenes, tet (Sigma, Tokyo, Japan) was added to the culture medium at a final concentration of 2 μg/ml. To examine localization of the Mis12 complex proteins, Dsn1/Q9H410-, Nnf1/PMF1-, and Nsl1/DC31-green fluorescent protein (GFP) plasmids (Kline et al., 2006) were transfected into CENP-C–deficient cells. For transfection of these plasmids into CENP-C–deficient cells, we used a Gene Pulser II electroporator (Bio-Rad) to generate cell lines stably expressing GFP fusion proteins.
Immunocytochemistry and Fluorescence In Situ Hybridization (FISH)
Immunofluorescence staining of whole cells was performed as described previously (Fukagawa et al., 2001, 2004). Cells were collected onto slides with a cyto-centrifuge and fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature or in 100% methanol for 15 min at −20°C, permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature, rinsed three times in 0.5% bovine serum albumin (BSA) and incubated for 1 h at 37°C with primary antibody. Binding of primary antibody was then detected with Cy3- or fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted to an appropriate concentration in phosphate-buffered saline (PBS)/0.5% BSA. Anti-CENP-A, -C, -H, -K, -I, and -O and anti-Mis12, -Hec1, -Nuf2, -BubR1, and -Mad2 antibodies were described previously (Fukagawa et al., 2001; Nishihashi et al., 2002; Hori et al., 2003; Regnier et al., 2005; Kline et al., 2006; Okada et al., 2006). FISH was performed as described previously (Fukagawa et al., 2001, 2004). For chromosome stability assay, the numbers of chromosomes 1 and 2 (5 chromosomes in normal DT40 cells) were scored in ∼100 metaphase cells at the indicated time points. Chromosomes and nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) at 0.2 μg/ml in Vectashield Antifade (Vector Laboratories, Burlingame, CA). All immunofluorescence and FISH images were obtained with a cooled charge-coupled device camera (CoolSNAP HQ; Photometrics Image Point, Tucson, AZ) mounted on an Olympus IX71 inverted microscope (Olympus, Tokyo, Japan) with a 60× objective lens (PlanApo 60×, numerical aperture [NA] 1.40) together with a filter wheel. All subsequent analyses and processing of images were performed with IPLab software (Signal Analytics, Fairfax, VA).
Observations of Living Cells
For live cell imaging, a histone H2B-RFP plasmid was transfected into CENP-C-conditional knockout cells to visualize chromosomes. Fluorescently stained living cells were observed with an Olympus inverted microscope IX71 with an oil immersion objective lens (PlanApo 60×, NA 1.40) in temperature-controlled box to keep temperature at 38°C. Time-lapse images were recorded at 5-min intervals with an exposure time of 0.2–0.3 s.
RESULTS
Generation of Conditional Knockout Cells of CENP-C in DT40 Cells with the tet System
We previously created a cell line with a conditional knockout of CENP-C by using a steroid agonist 4-hydroxytamoxifen (4-OHT)–ER system (Fukagawa and Brown, 1997). We disrupted the first CENP-C allele and replaced the second CENP-C allele with a cDNA encoding a fusion protein in which the CENP-C gene was fused to the ER gene. Although the CENP-C–ER fusion protein associates with the centromere and functions like CENP-C in the presence of 4-OHT, the CENP-C–ER fusion protein translocates to cytoplasm, and it does not function properly in the absence of 4-OHT. Because CENP-C function was inactivated by delocalization of CENP-C in the ER-based knockout system, proteins that bind to or interact with CENP-C may also be inactivated. Therefore, to investigate CENP-C–dependent protein localization, ER-based CENP-C-knockout cells are not appropriate. To characterize the role of CENP-C in kinetochore assembly, we generated a tet-inducible system-based conditional knockout of CENP-C. The CENP-C disruption construct was generated such that genomic fragments encoding several exons were replaced with a histidinol-resistance (hisD) cassette (1st allele construct), and we isolated CENP-C+/− clones (Figure 1A). One CENP-C+/− clone was cotransfected with a plasmid construct encoding a chicken CENP-C transgene under the control of a tet-repressible promoter and a plasmid construct encoding a tet-repressible transactivator containing a zeocin (zeo)-resistance cassette. We selected zeo-resistant colonies and identified several clones carrying these constructs integrated at random sites in the genome (CENP-C+/−CENP-Ctransgene). Two clones with the CENP-C+/−CENP-Ctransgene genotype were transfected with the puromycin CENP-C disruption construct (2nd allele construct) to disrupt the remaining CENP-C allele (Figure 1B). We obtained five clones with the CENP-C−/−CENP-Ctransgene genotype, and one clone, CC6-121-13, was chosen for further analysis. CENP-C protein in CC6-121-13 cells was reduced at 6 h after addition of tet, and it was not detected by 48 h by Western blot analysis (Figure 1C). We did not observe localization of CENP-C protein to kinetochores by immunofluorescence analysis (Figure 1C). The proliferative properties of CC6-121-13 cells were monitored by growth curves. Viable and dead cells were assessed by trypan blue exclusion. The growth curve of CC6-121-13 cells (tet−; CENP-C+) was indistinguishable from that of wild-type DT40 cells (Figure 1D). In the presence of tet, CC6-121-13 cells stopped proliferating at 48 h after tet addition (Figure 1D), and most cells had died by 96 h.
Figure 1.
Generation of a tet-based conditional knockout cell line for CENP-C. (A) Restriction maps of the chicken CENP-C locus, gene disruption constructs, and targeted loci. Black boxes indicate the positions of exons. BamHI restriction sites are shown. There is a BamHI polymorphism in this locus; a BamHI site is absent from the 1st round targeted allele. The position of the probe used for Southern hybridization is indicated. Novel 16.2- and 16.9-kb BamHI fragments hybridize to the probe if targeted integration of the construct occurs. (B) Restriction analysis of genomic DNAs from cells with targeted integration of a CENP-C disruption construct. Genomic DNAs from wild-type DT40 cells (Cl18), a clone after first-round targeting (+/−, 1st), a clone after first-round targeting and random integration of the CENP-C transgene (+/− CENP-C+, 1st + cDNA), and a clone after second-round targeting (−/− CENP-C+, 2nd + cDNA, clone CC6-121-13) were analyzed by Southern hybridization with the probe indicated in A. (C) Western blot analysis of CC6-121-13 cell extracts (3 × 105 cells) with anti-CENP-C antibody. Wild-type DT40 cell extracts are also shown on the SDS-PAGE gel (lane a, 3 × 105 cells; lane b, 1 × 105 cells; and lane c, 3 × 104 cells). Immunofluorescence staining with anti-CENP-C antibody (green) is also shown. CENP-C protein was not detected by 48 h after tet addition. Bars, 5 μm. (D) Representative growth curves for the indicated cell cultures (Cl18 and CC6-121-13 cells). tet was added at time 0 to cultures of CC6-121-13 CENP-C–deficient cells (tet+), and the number of cells not stained with trypan blue was counted.
CENP-C-deficient Cells Exhibit Mitotic Delay with Progression to the Next Cell Cycle before Subsequent Cell Death
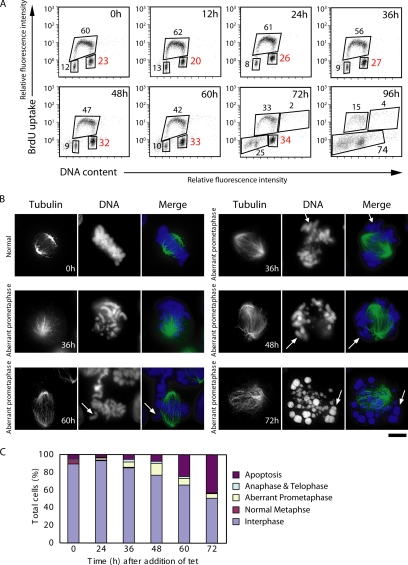
We then examined the delayed growth and cell death after CENP-C depletion in CC6-121-13 cells. We measured both cellular DNA content and DNA synthesis by fluorescence-activated cell sorting (FACS) after pulse-5-bromo-2′-deoxyuridine (BrdU) labeling (Figure 2A). Asynchronous wild-type DT40 and CC6-121-13 cells (tet−; CENP-C+) showed similar cell cycle patterns (data not shown), with S phase accounting for two thirds of the entire cell cycle time (Figure 2A, 0 h). In contrast, when CC6-121-13 cells were cultured with tet, cells began to accumulate in G2/M phase, and the proportion of cells in G2/M phase reached 32% of the total cells by 48 h (Figure 2A). Degradation of chromosomal DNA due to massive cell death was observed between 72 and 96 h (data not shown).
Figure 2.
CENP-C–deficient cells show mitotic defects. (A) Cell cycle distribution of CC6-121-13 cells after inhibition of CENP-C transgene expression due to addition of tet at time 0. Cells were stained with FITC-anti-BrdU (y-axis, log scale) to detect BrdU incorporation (DNA replication) and with propidium iodide to detect total DNA (x-axis, linear scale). The bottom left box represents G1 phase cells, the upper box represents S phase cells, and the bottom right box represents G2/M phase cells. The numbers given in the boxes indicate the percentage of gated events. (B) Abnormal mitotic morphologies of CENP-C–deficient cells. Cells were stained with α-tubulin (green), and DNA was counterstained with DAPI (blue). Several chromosomes are not aligned at the metaphase plate (arrows). Bars, 10 μm. (C) Quantitation of aberrant CC6-121-13 cells following inhibition of CENP-C transgene expression by addition of tet at time 0. We scored the number of interphase cells, normal metaphase cells, aberrant metaphase cells described in B, anaphase and telophase cells, and apoptotic cells.
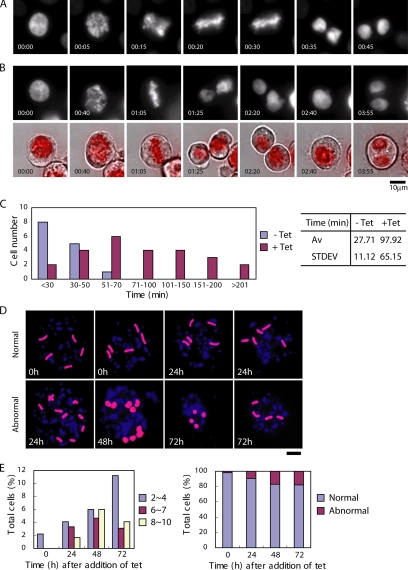
Because we could not distinguish the exact stage at which tet-treated CC6-121-13 cells accumulated in G2/M phase by FACS analysis, we used DNA staining and immunocytochemical staining of microtubules to examine the time course of the kinetics of the mitotic index (Figure 2, B and C). In the presence of tet, we observed aberrant mitotic CC6-121-13 cells in which some chromosomes seemed misaligned on the metaphase plate (Figure 2B), whereas chromosomes seemed ordered and aligned on the metaphase plate in untreated CC6-121-13 cells (Figure 2B, 0 h). At 48 h after tet addition, the percentage of aberrant prometaphase cells was 13% of total cells, which is ∼3 times higher than that of control cells (Figure 2C). Although we detected a significant increase in the number of aberrant mitotic cells, only 13% of the total cells were mitotic, suggesting that the phenotype of CENP-C–deficient cells is not simple mitotic arrest. Therefore, it was necessary to observe the behavior of individual living cells after inactivation of CENP-C. We transfected the histone H2B-RFP plasmid into CC6-121-13 cells to visualize nuclei and chromosomes. We then observed the behavior of individual CENP-C-conditional knockout cells expressing histone H2B-RFP by microscopy at 38°C. Microscope images were typically obtained at intervals of 5 min, and chromosomes in individual cells were observed through an entire mitotic cell cycle. An example of the time-lapse observation of CC6-121-13 cells in the absence of tet (control) is shown in Figure 3A. Selected frames of the dynamics of chromosomes from prophase to telophase are shown. We were able to observe well-ordered metaphase plates (20 min) and normal cell division. It took control cells ∼27 min (n = 14) to progress from prophase to telophase. After addition of tet for 48 h, we performed the time-lapse imaging for CC6-121-13 cells and observed mitotic delay. It took ∼98 min (n = 25) to complete mitosis from prophase (Figure 3, B and C). This time was 3 times longer than that of control cells (CC6-121-13 cells in the absence of tet), and it was consistent with the mitotic index data (Figure 2). We continued the live cell observations after mitotic delay and found that CENP-C–deficient cells progressed to the next cell cycle, but some cells failed to complete cytokinesis and contained two nuclei due to fusion of daughter cells before subsequent cell death (Figure 3B and Supplemental Material 1). These observations suggest that CENP-C deficiency compromises mitotic checkpoint function.
Figure 3.
CENP-C–deficient cells proceed to the next cell cycle with abnormal chromosome segregation after mitotic delay. (A) Dynamics of chromosomes in control cells by time-lapse observations of living cells. Selected images of chromosomes from prophase to telophase in CC6-121-13 (−tet) are shown. (B) Dynamics of chromosomes in CENP-C–deficient cells by time-lapse observations of living cells. In the superimposed images (bottom), chromosomes (red) and bright-field images are displayed. Bars, 10 μm. (C) Evaluation of time to progress from prophase to anaphase for wild-type (n = 14) and CENP-C–deficient (n = 25) cells by observation of living cells. Average time to progress from prophase to anaphase was ∼27 min for wild-type cells and 98 min for CENP-C–deficient cells. Bars, 10 μm. (D) To examine chromosome loss, FISH analysis was performed with chromosome-specific painting probes. We used painting probes for chicken chromosomes 1 and 2. Because DT40 cells have three copies of chromosome 2, five painted chromosomes are visible in normal cells (top left). CC6-121-13 cells showed gains and losses of chromosomes (bottom) after addition of tet. Time (hours) after addition of tet is indicated on each image. (E) Distribution of the number of painted chromosomes per cell. CC6-121-13 cells were examined after culture with tet.
We observed that mitotically delayed CENP-C–deficient cells progress to the next cell cycle without normal chromosome segregation, indicating that aneuploidy may be induced by CENP-C depletion. We performed FISH analysis of metaphase spreads with chromosome painting probes specific for chicken chromosomes 1 and 2. Because DT40 cells contain three copies of chromosome 2, five fluorescent chromosomes can be observed in each wild-type cell (Figure 3D, top). As shown in Figure 3D, we observed abnormal numbers of painted chromosomes in CENP-C–deficient cells in the presence of tet. The proportion of these aneuploid cells gradually increased after 24 h (Figure 3E), suggesting that CENP-C deficiency induces aneuploidy accompanied by chromosome missegregation.
CENP-C-deficient Cells Impaired Kinetochore Localization of Mad2
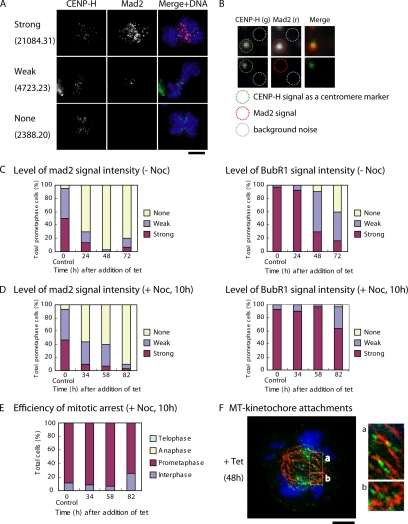
In eukaryotic cells, chromosome segregation is regulated by a spindle checkpoint that prevents anaphase onset until all chromosomes have aligned at the metaphase plate (Musacchio and Hardwick, 2002). Because our living cell analysis suggested that CENP-C–deficient cells have impaired spindle checkpoint function, we examined kinetochore localization of mitotic checkpoint proteins Mad2 and BubR1 (Figure 4). Because some prometaphase CENP-C–deficient cells showed no or weak Mad2 and BubR1 signals, we performed quantitative immunofluorescence analysis with anti-Mad2 and anti-BubR1 antibodies. To quantify Mad2 and BubR1 signals at each kinetochore, IPLab software was used, and the signal intensity at each kinetochore was quantified in arbitrary units. We introduced the CENP-H-GFP knockin construct into CC6-121-13 cells, and we used the CENP-H-GFP signal as a marker of the kinetochore region (Figure 4B). We measured signal intensity of Mad2 and BubR1 at >30 kinetochores in each cell, and we obtained the average value per individual cell. Cells were classified into three groups on the basis of unit values of signal intensities: strong (unit value of signal intensity >7001), weak (unit value of signal intensity, 3001–7000), and none (unit value of signal intensity <3000). Examples of signal intensity values per individual cell are shown in Figure 4A. All data for signal intensities and representative images are presented in Supplemental Material 2. More than 90% of prometaphase cells showed no Mad2 signal at 48 h after tet addition (Figure 4C). We also observed that 60% of prometaphase cells were classified into the weak signal group for BubR1 staining at 48 h after tet addition. These signal reductions for Mad2 and BubR1 in CENP-C–deficient cells are consistent with the weak mitotic delay and subsequent cell cycle progression observed in CENP-C–deficient cells (Figure 3).
Figure 4.
Localization of mitotic checkpoint proteins in CENP-C–deficient cells. (A) Immunofluorescence analysis of Mad2 in CENP-C–deficient cells. Because cells with various intensities of Mad2 signals are observed, we classified cells into three groups on the basis of arbitrary unit values of Mad2 signal intensity: strong (unit value of signal intensity >7001), weak (unit value of signal intensity, 3001–7000), and none (unit value of signal intensity <3000). The signal intensities of Mad2 and BubR1 for >30 kinetochores in each cell were measured, and the average value per individual cell was calculated. Unit value of signal intensity per cell is shown. Typical examples for measurements are shown in B. Bars, 10 μm. (B) Examples of quantification of Mad2 level. CENP-H was used as a marker of the centromere region. Mad2 signal intensities for each kinetochore were measured, and background noise was subtracted. (C) Quantification of cells with various Mad2 and BubR1 signal intensities in CC6-121-13 cells after inhibition of CENP-C transgene expression after addition of tet at time 0. Because we confirmed that amount of CENP-C at time 0 is similar to that of wild-type cells, CC6-121-13 cells at time 0 are used as control cells. (D) Quantification of Mad2 and BubR1 signals in CC6-121-13 cells after inhibition of CENP-C transgene expression after addition of tet at time 0. In these experiments, cells were treated with nocodazole for 10 h before immunofluorescence analysis. (E) Mitotic index after treatment of CENP-C–deficient cells with the spindle poison nocodazole. (F) Immunofluorescence analysis of CC6-121-13 cells expressing CENP-H-GFP (green) with anti-α-tubulin (red) at 48 h after tet addition (+tet). Many chromosomes seem to have formed kinetochore–microtubule attachments. Bars, 5 μm.
The ability of CENP-C–deficient cells to exit mitosis as well as the defects in localization of the checkpoint proteins Mad2 and BubR1 led us to examine whether CENP-C–deficient cells could still activate the spindle checkpoint in response to a spindle poison. We treated CENP-C-conditional knockout cells with nocodazole, which depolymerizes microtubules, after tet addition. Although ∼90% of prometaphase cells still showed no or weak Mad2 signals after incubation of tet-treated CC6-121-13 cells with nocodazole for 10 h, >90% of cells showed strong BubR1 signals at the same time point (Figure 4D). We observed accumulation of mitotic CENP-C–deficient cells after nocodazole treatment for 10 h (Figure 4E), suggesting that loss of CENP-C did not impair overall activation of the checkpoint pathway in response to nocodazole. Our data indicate that BubR1 but not Mad2 is involved in activation of the checkpoint pathway in CENP-C–deficient cells in response to a spindle poison. In the vertebrate spindle checkpoint pathway, Mad2 and BubR1 operate as elements of distinct pathways sensing tension and attachment (Skoufias et al., 2001).
In CENP-C–deficient cells, the spindle checkpoint function was partially defective. Therefore, we were interested in whether kinetochore–microtubule attachment occurred in CENP-C–deficient cells. We investigated kinetochore–microtubule attachment by staining microtubules with anti-α-tubulin antibodies in CC6-121-13 cells expressing CENP-H-GFP (Figure 4F). Although we cannot conclude that kinetochores attached to microtubules without electron microscopy, our light microscopy data suggest that most kinetochores of prometaphase chromosomes, including chromosomes not aligned at the metaphase plate, in CENP-C–deficient cells were attached to microtubules.
Centromere Localization of the Mis12 Complex Proteins Is Abolished in CENP-C–deficient Cells
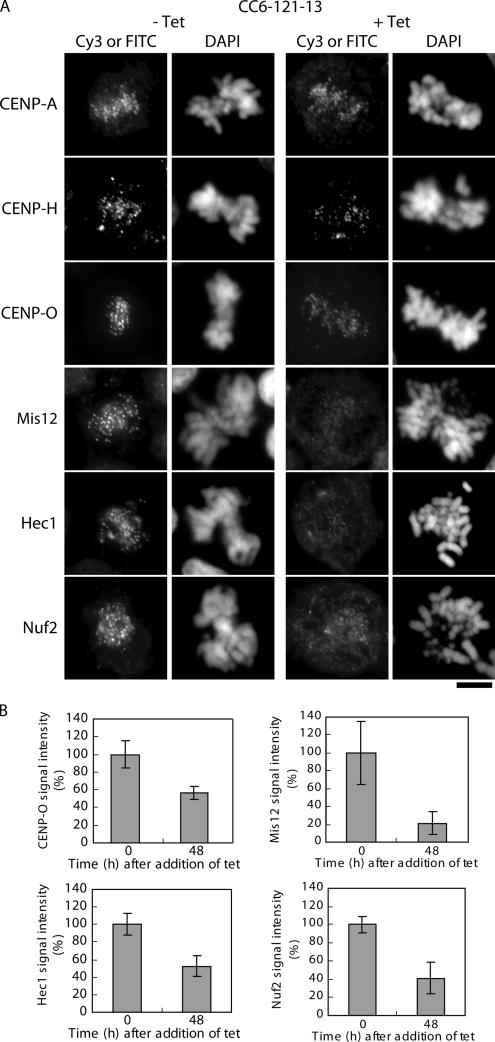
CENP-C is recognized as an important component of the inner kinetochore (Saitoh et al., 1992), and we were interested in how CENP-C is involved in centromere assembly. We performed immunocytochemical analyses of CENP-C–deficient cells (both interphase and mitotic cells) with antibodies against CENP-A, CENP-H complex proteins, Mis12, and Nuf2 complex proteins (Figure 5). As shown in Figure 5A, all antibodies tested gave typical centromere signals in wild-type cells. In CENP-C–deficient cells, anti-CENP-A antibodies gave centromere signals that were of similar intensities to those observed in control cells. Antibodies against the CENP-H complex proteins (CENP-H and CENP-O) gave clear centromere signals in CENP-C-deficient cells (Figure 5A), although the signal intensities were slightly reduced (see CENP-O in Figure 5B, ∼60% of the level of control cells). We previously reported that CENP-H showed centromere signals in the ER-based CENP-C-knockout cells (Fukagawa et al., 2001). Immunofluorescence results with CENP-H and CENP-O in this study are consistent with those of our previous report. Antibodies against Mis12 showed weak centromere signals in CENP-C–deficient cells. The signal intensities of Mis12 in CENP-C–deficient cells were reduced to <20% of the level observed in control cells (Figure 5B). Signal intensities of the Nuf2 complex proteins, including Nuf2 and Hec1, in CENP-C–deficient cells were also reduced to <40% of the levels (Nuf2) observed in control cells (Figure 5, A and B). Our present results are consistent with reports that the Nuf2 complex proteins are associated with the Mis12 complex proteins (Cheeseman et al., 2004; Obuse et al., 2004a).
Figure 5.
Localization of centromere proteins in CENP-C–deficient cells. (A) Immunofluorescence analysis of CC6-121-13 cells at 0 h (−tet) or 48 h (+tet) after tet addition. Cells were stained with anti-CENP-A, anti-CENP-H, anti-CENP-O, anti-Mis12, anti-Hec1, and anti-Nuf2 antibodies. Antibody signals were detected with Cy3- or FITC-conjugated secondary antibodies. DNA was counterstained with DAPI. Bars, 5 μm. (B) Quantification of signal intensities for CENP-O, Mis12, Hec1, and Nuf2 in CC6-121-13 cells at 0 h (−tet) or 48 h (+tet) after tet addition.
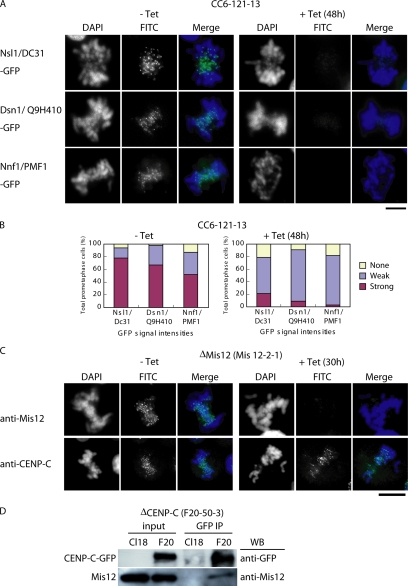
Mis12 forms a complex that includes PMF1/Nnf1, Q9H410/Dsn1, and DC31/Nsl1 (Cheeseman et al., 2004; Obuse et al., 2004a; Kline et al., 2006). If Mis12 protein is mislocalized in CENP-C–deficient cells, other Mis12 complex proteins also would be mislocalized. Therefore, we created GFP-fusion constructs for PMF1/Nnf1, Q9H410/Dsn1, and DC31/Nsl1, and we introduced them into CENP-C-conditional knockout cells. We analyzed the GFP signals of the fusion proteins in these cells in both the presence and absence of tet. Because expression varied between individual cells, we counted the numbers of cells with strong, weak, and no signals on the basis of signal intensity. Average intensity in the weak group was ∼20% of the level in the strong group. In the absence of tet, 60–80% of cells showed strong kinetochore signals for the Mis12 complex proteins (Figure 6, A and B). In the presence of tet, >80% of these cells (both interphase and mitotic cells) showed weak or no Mis12 complex signals (Figure 6, A and B). Typical mitotic figures are shown in Figure 6A (interphase figures not shown). In contrast, centromere signals for CENP-C were observed in Mis12-deficient cells, and the signal intensities were similar to those in control cells (Figure 6C). These results indicate that localization of Mis12 complex proteins to the centromere of both interphase and mitotic cells is dependent on CENP-C, but CENP-C localization is not dependent on the Mis12 complex proteins.
Figure 6.
Centromere localization of the Mis12 complex proteins is abolished in CENP-C–deficient cells. (A) To investigate localization of Mis12 complex proteins, plasmid constructs for Nsl1/DC31-GFP, Dsn1/Q9H410-GFP, and Nnf1/PMF1-GFP were transfected into CC6-121-13 cells. Localization of each GFP fusion protein (green) was observed in the absence (−tet) or presence (+tet) of tetracycline. DNA was counterstained with DAPI (blue). Bars, 10 μm. (B) Quantification of cells with various intensities of Nsl1-GFP, Dsn1-GFP, and Nnf1-GFP signals in CC6-121-13 cells at 0 h (−tet) or 48 h (+tet) after addition of tet. Average of intensity in the weak group is ∼20% of the level in the strong group. (C) Immunofluorescence analysis of Mis12-2-1 cells, which carry a conditional knockout of Mis12, with anti-Mis12 and anti-CENP-C antibodies. Antibody signals were detected with FITC-conjugated secondary antibodies (green). DNA was counterstained with DAPI (blue). Bars, 10 μm. (D) Coimmunoprecipitation of Mis12 protein with CENP-C-GFP. A cell line (F20-50-3) in which expression of CENP-C was replaced with that of CENP-C-GFP was created. The chromatin fractions of wild-type (Cl18) and F20-50-3 cells were immunoprecipitated with anti-GFP antibody. Immunoprecipitates were separated by SDS-PAGE and analyzed by Western blotting with anti-GFP and anti-Mis12 antibodies.
Because Mis12 localization is dependent on CENP-C, we examined the interaction between CENP-C and Mis12 in DT40 cells by coimmunoprecipitation assay. We prepared a cell line in which expression of CENP-C was replaced by expression of CENP-C-GFP (F20-50-3 cells). We then isolated the chromatin fraction from the F20-50-3 cells and performed immunoprecipitation with anti-GFP antibody. Immunoprecipitates were then separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting with anti-GFP or anti-Mis12 antibody. Mis12 was detected in immunoprecipitates with anti-GFP antibody (Figure 6D), suggesting that Mis12 is closely associated with CENP-C. This association was observed in both interphase and mitotic extracts (data not shown).
Centromere Localization of CENP-C and Mis12 Is Dependent on CENP-H Complex Proteins in Interphase Nuclei but Not in Mitotic Chromosomes
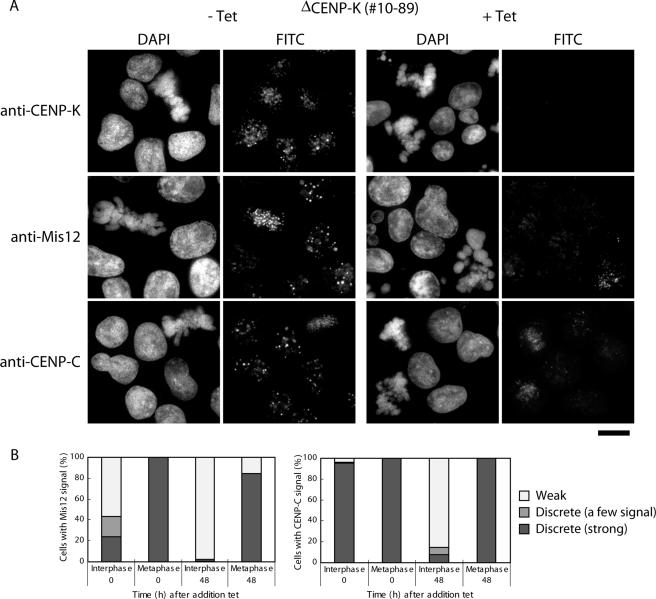
We previously reported that CENP-C shows weak centromeric localization in CENP-H–deficient interphase nuclei (Fukagawa et al., 2001). Recently, we and other groups identified several CENP-H–associated proteins and created conditional knockout cell lines for these proteins (Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006). We examined localization of CENP-C in cells with a conditional knockout of CENP-K, a member of the CENP-H complex proteins. Similar to CENP-H–deficient cells, CENP-C showed weak (and diffuse) centromeric localization in ∼90% of CENP-K–deficient interphase nuclei (Figure 7, A and B). The weak intensity at each kinetochore was ∼20% of the strong intensity level observed in control cells (Figure 7B). When we expressed CENP-C-GFP in CENP-K–deficient cells, diffuse CENP-C-GFP signals were also observed in CENP-K–deficient interphase cells (data not shown). Although we observed clear mislocalization of CENP-C in CENP-K–deficient interphase nuclei, we observed CENP-C signals in kinetochores in mitotic cells. We also detected CENP-C kinetochore signals in CENP-H–deficient mitotic cells (data not shown). These data suggest that localization of CENP-C to the centromere is dependent on the CENP-H complex proteins in interphase cells but not in mitotic cells.
Figure 7.
Centromere localization of CENP-C and Mis12 at interphase nuclei is dependent on the CENP-H complex proteins. (A) Immunofluorescence analysis of #10–89 cells, which carry a conditional knockout of CENP-K, with anti-CENP-K, anti-Mis12, and anti-CENP-C antibodies. Antibody signals were detected with FITC-conjugated secondary antibodies. DNA was counterstained with DAPI. Bars, 10 μm. (B) Quantification of cells with various intensities of Mis12 and CENP-C signals in #10–89 cells after inhibition of CENP-K transgene expression after addition of tet at time 0. Both Mis12 and CENP-C signals were weak in interphase nuclei but not in mitotic cells at 48 h after tet addition. Average of intensity in the weak group is ∼20% of the level in the strong group.
Because centromere localization of Mis12 is dependent on CENP-C in both interphase (data not shown) and mitotic cells (Figure 5), we hypothesized that centromeric localization of Mis12 may be abolished in CENP-K–deficient interphase nuclei. To test this hypothesis, we performed immunofluorescence analysis with anti-Mis12 antibody in CENP-K-conditional knockout cells. Centromeric localization of Mis12 was diminished in CENP-K–deficient interphase nuclei (Figure 7B). Mis12 signal in 90% of CENP-K–deficient interphase cells was <20% level of control cells (weak group in Figure 7B). We still observed clear centromeric localization of Mis12 in CENP-K–deficient mitotic cells. These data suggest that localization of CENP-C and Mis12 to centromeres in interphase nuclei is dependent on the CENP-H complex proteins.
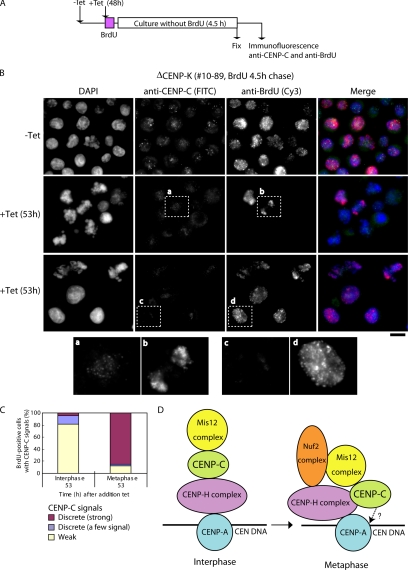
Our present results raise the question as to why centromere localization of CENP-C and Mis12 is dependent on CENP-H complex proteins only in interphase cells and not in mitotic cells. One possibility is the proteins that interact with CENP-C may differ between interphase cells and mitotic cells, which would suggest that organization of inner kinetochore is dynamically changed through cell cycle progression. To strengthen this hypothesis, we designed a pulse-chase experiment with BrdU in CENP-K–deficient cells (Figure 8). CENP-K–deficient cells were incubated with BrdU for 20 min, washed, and then cultured for 4.5 h. Cells were then fixed, and immunofluorescence experiments were performed with anti-CENP-C or anti-BrdU antibodies (Figure 8A). We were interested in CENP-C localization in BrdU-positive mitotic cells, which incorporated BrdU in interphase cells that lack CENP-C at centromeres. We detected clear CENP-C signals in >80% of BrdU-positive mitotic cells (Figure 8, B and C), suggesting that CENP-C can be targeted to kinetochores during mitosis even if CENP-C and CENP-H are not present at centromeres during interphase.
Figure 8.
CENP-C can be targeted into kinetochores in the absence of the CENP-H complex proteins during mitosis. (A) Schematic presentation of a pulse-chase experiment with BrdU in CENP-K–deficient cells. #10–89 cells (CENP-K-conditional knockout cells) were cultured for 48 h after tet addition. BrdU was added to the culture for 20 min (pulse). Cells were cultured without BrdU for 4.5 h (chase) and then fixed. Cells were then subjected to immunofluorescence experiments with anti-CENP-C or anti-BrdU antibody. (B) Typical images of immunofluorescence experiments. CENP-C signals were detected in BrdU-positive mitotic cells (a and b). CENP-C signals were weak in interphase nuclei (c and d). (C) Quantification of BrdU-positive cells with various intensities of CENP-C signals in #10–89 cells after the pulse-chase experiments. CENP-C signals were detected in >80% of BrdU-positive mitotic cells. (D) Current model for kinetochore assembly in interphase nuclei and mitotic chromosomes in chicken DT40 cells. Centromere-specific histone CENP-A is upstream of other centromere proteins for centromere localization. Our previous data showed that the CENP-H complex is closely associated with CENP-A (Okada et al., 2006). In interphase nuclei, centromere localization of CENP-C occurs downstream of that of the CENP-H complex but occurs upstream of localization of the Mis12 complex. During mitosis, CENP-C dependence on CENP-H is altered. CENP-C can be targeted into kinetochores even in the absence of CENP-H. Localization of Nuf2 complex is dependent on both the Mis12 complex and the CENP-H complex (Cheeseman et al., 2004; Obuse et al., 2004b; Mikami et al., 2005). Localization of CENP-H complex to centromeres is somewhat dependent on CENP-C. CENP-C also may be involved directly in DNA binding (Yang et al., 1996) or may interact with CENP-B (Suzuki et al., 2004).
DISCUSSION
CENP-C-conditional Knockout Cells with the tet System
Loss-of-function analyses of vertebrate CENP-C were performed in several ways, including microinjection of anti-CENP-C into HeLa cells (Tomkiel et al., 1994), creation of an ER-based DT40 knockout (Fukagawa and Brown, 1997; Fukagawa et al., 1999), and creation of CENP-C-knockout mice (Kalitsis et al., 1998). The results of these analyses led us to conclude that CENP-C is essential for cell viability and completion of mitosis. However, it is difficult to conduct further analyses of CENP-C function with these systems due to technical problems. Each kinetochore protein associates with other kinetochore proteins for full centromere function. Therefore, inactivation of CENP-C protein by antibody microinjection or the ER-based DT40 knockout may lead to unpredicted inactivation of other kinetochore proteins. Analysis of CENP-C-knockout mice is complicated by the fact that CENP-C-knockout mice die during early embryonic development (Kalitsis et al., 1998). Therefore, to further our understanding of the function of CENP-C, cells with a conditional knockout of CENP-C at the level of transcription are needed. In the present study, we created and characterized a tet-based CENP-C-conditional knockout cell line. Characterization of this cell line will further our understanding of the roles of CENP-C in vertebrate cells.
Our phenotypic analysis of the tet-based CENP-C-conditional knockout cells revealed defects in mitosis, as indicated by the accumulation of prometaphase cells and the data from live cell observations. This mitotic phenotype is consistent with previous observations (Tomkiel et al., 1994; Fukagawa and Brown, 1997). Use of the tet-based conditional knockout system allowed us to show that CENP-C–deficient cells were only transiently delayed in prometaphase and progressed to the next cell cycle without normal chromosome segregation. This mitotic phenotype is different from that observed in mutants for the CENP-H complex proteins, which are also components of the inner kinetochore. In comparison with the percentage of mitotic-delayed cells in mutants for the CENP-H complex proteins, which was ∼50% at 48 h after tet addition (Okada et al., 2006), the percentage of CENP-C–deficient cells showing mitotic delay was low (13% of CC6-121-13 cells at 48 h after tet addition) (Figure 2C). In addition, live cell observations revealed that the average time to complete mitosis is ∼98 min in CENP-C–deficient cells, whereas mitosis requires ∼800 min in mutants of the CENP-H complex proteins (Nishihashi et al., 2002; Okada et al., 2006). These differences in the mitotic phenotype of CENP-C–deficient cells from those of mutants for the CENP-H complex proteins indicate that the function of CENP-C is distinct from the function(s) of the CENP-H complex proteins.
Mitotic Checkpoint Pathway and Inner Kinetochore Proteins
The observation that CENP-C–deficient cells undergo a transient mitotic delay before exit from mitosis with chromosome missegregation indicates that the mitotic checkpoint is activated in these cells but that the checkpoint response is subsequently overcome. We also observed that localization of checkpoint components Mad2 and BubR1 to the kinetochore is altered in CENP-C–deficient cells. Mad2 signals are nearly absent in CENP-C–deficient mitotic cells, and Mad2 localization did not recover even after nocodazole treatment. We also observed a reduction in the BubR1 signal in CENP-C–deficient cells. However, BubR1 localization recovered after nocodazole treatment. Because both checkpoint proteins detached from kinetochores in CENP-C–deficient mitotic cells after transient mitotic delay, our data suggest that CENP-C is involved in maintenance of the mitotic checkpoint. This phenotype is also observed in CENP-A–deficient cells (Regnier et al., 2005). In CENP-H– and CENP-I–deficient cells, we found that a subset of cells progressed to the next cell cycle even after strong mitotic delay (Fukagawa et al., 2001; Nishihashi et al., 2002). Defects in the mitotic checkpoint are common in cells with knockouts of inner kinetochore proteins (Nishihashi et al., 2002; Regnier et al., 2005). We think that depletion of individual inner kinetochore proteins does not disrupt the entire kinetochore structure (Nishihashi et al., 2002). Although the mechanism of how mitosis progresses in cells with knockouts of inner kinetochore proteins is not clear, our present data suggest that checkpoint proteins associate with the kinetochores but that this association is not maintained in CENP-C–deficient cells due to partial disruption of the inner kinetochore structure.
Another question is why a spindle checkpoint is activated first, although our data suggest that many kinetochores seem to bind microtubules in CENP-C–deficient cells (Figure 4F). One possible explanation is that there may not be sufficient tension to complete chromosome segregation even if kinetochores bind microtubules in CENP-C–deficient cells. Therefore, a mitotic checkpoint pathway can be activated through a BubR1-dependent mechanism. However, the checkpoint response cannot be maintained and is subsequently overcome in CENP-C–deficient cells, as mentioned above.
It is important that CENP-C–deficient cells were able to activate and maintain the checkpoint after treatment with a spindle poison, nocodazole. After exposure to spindle poison, BubR1 signals were efficiently relocalized to kinetochores; however, Mad2 signals were not, suggesting that CENP-C–deficient cells have impaired Mad2-dependent signaling from kinetochores of mitotically arrested cells but that the checkpoint activation can occur through a BubR1-dependent pathway. In CENP-A–depleted cells, only the BubR1-dependent pathway is activated (Regnier et al., 2005). In the vertebrate spindle checkpoint system, Mad2 and BubR1 operate as elements of distinct pathways sensing microtubule attachment and tension, respectively (Skoufias et al., 2001). The Mad2-dependent pathway seems to be easily disrupted by knockout of inner kinetochore proteins.
When treated with the spindle poison nocodazole, CENP-C–deficient cells maintained prolonged mitotic arrest (>10 h). CENP-H– and CENP-I–deficient cells have the ability to maintain the checkpoint for >10 h even in the absence of the spindle poison, although these cells subsequently progress to the next cell cycle (Nishihashi et al., 2002). Like previous reports in cells depleted of some kinetochore proteins (Waters et al., 1998; Nishihashi et al., 2002; Liu et al., 2003; Weaver et al., 2003; Tanudji et al., 2004; Regnier et al., 2005), it is possible that, in the absence of a spindle poison, CENP-C–deficient cells are only able to generate a weak checkpoint signal with a level that is below a necessary threshold. The mitotic kinetochores that lack CENP-C contain small amounts of the checkpoint proteins, and they can generate a weak inhibitory signal that might not be strong enough to sustain a strong mitotic block. In the presence of spindle poison, the inhibitory signals from the kinetochores would reach the threshold for checkpoint activation and maintenance. The response to the mitotic checkpoint pathway differs between the various inner kinetochore protein mutants (Fukagawa et al., 2001; Nishihashi et al., 2002; Regnier et al., 2005; Okada et al., 2006). Signal transmission may require phosphorylation of inner kinetochore proteins, and as part of a preliminary study, we found that several inner kinetochore proteins are phosphorylated (our unpublished data).
Role of CENP-C in Kinetochore Assembly
To understand kinetochore assembly, we used a tet-based DT40 conditional knockout system to generate knockouts of several inner kinetochore proteins (Fukagawa et al., 2001; Nishihashi et al., 2002; Regnier et al., 2005; Okada et al., 2006). Although we found that CENP-H complex proteins function as a marker for incorporation of newly synthesized CENP-A into the centromere (Okada et al., 2006), we showed that CENP-A is upstream of CENP-H complex proteins in centromere targeting because endogenous CENP-A localization is not altered in CENP-H–deficient cells, whereas CENP-H localization is abolished in CENP-A–deficient cells (Fukagawa et al., 2001; Regnier et al., 2005; Okada et al., 2006). We also reported that localization of CENP-C to the centromere is abolished in CENP-H–deficient interphase nuclei, indicating that CENP-H is required for centromere targeting of CENP-C in interphase nuclei (Fukagawa et al., 2001). To further our understanding of kinetochore assembly, we analyzed CENP-C–deficient cells in the present study. Mis12 complex proteins were reduced in both interphase and mitotic cells in the absence of CENP-C. Recent RNA interference (RNAi) experiments in human HeLa cells support our data (Liu et al., 2006). Because CENP-C localization was not altered in Mis12-deficient DT40 cells, we concluded that localization of Mis12 complex proteins to the centromere occurs downstream of localization of CENP-C (Figure 8D). In C. elegans, the Mis12 protein coprecipitates with the CENP-C homologue HCP4 (Cheeseman et al., 2004), and our experiments indicate that CENP-C coprecipitates with Mis12 (Figure 6D), suggesting that Mis12 complex proteins may interact directly with CENP-C in vertebrate cells.
Because localization of CENP-C to the centromere is abolished in interphase nuclei of CENP-H and CENP-I mutants (Fukagawa et al., 2001; Nishihashi et al., 2002), we examined CENP-C localization in cells deficient for another CENP-H complex protein, CENP-K. Centromeric localization of CENP-C in interphase nuclei was abolished in CENP-K–deficient cells (intensities of CENP-C signals were ∼20% of the control signal intensity in >90% of CENP-K–deficient interphase cells), indicating that localization of CENP-C to the centromere in interphase nuclei is dependent on the CENP-H complex proteins CENP-H, CENP-I, and CENP-K. We also investigated centromere localization of CENP-H complex proteins in CENP-C–deficient cells. Although we observed a weak reduction in the CENP-H complex proteins (60–70% signal intensity of the control cells), we clearly detected discrete signals of these proteins at centromeres in CENP-C–deficient cells. These data suggest that localization of CENP-C to centromeres in interphase cells occurs downstream of localization of the CENP-H complex proteins, although localization of the CENP-H complex is dependent in part on CENP-C (Figure 8D). If our model is correct, Mis12 should not be localized to the centromere in interphase cells deficient for CENP-H complex proteins. Indeed, we observed mislocalization of Mis12 in CENP-K–deficient interphase nuclei. Previous RNAi experiments revealed that centromere localization of Mis12 occurs upstream of that of CENP-H in human cells (Goshima et al., 2003; Kline et al., 2006). This discrepancy between our present results and those of previous reports may reflect differences in the functional interactions among kinetochore components between humans and chickens. The previous reports (Goshima et al., 2003; Kline et al., 2006) also focused mainly on dependency of proteins at mitotic chromosomes. We emphasize that the dependency of Mis12 and CENP-C on the CENP-H complex for centromere localization is restricted to interphase nuclei of DT40 cells. Mis12 and CENP-C localization to centromeres is dependent on CENP-H in human interphase cells but not mitotic cells (Figure 8D). Our pulse-chase experiments with BrdU suggest that organization of the inner kinetochore changes throughout cell cycle progression. These results caused us to consider the organization of C. elegans kinetochore. HCP-4 (CENP-C in C. elegans) is distributed in the cytoplasm during interphase, but it is targeted into kinetochores during mitosis (Moore and Roth, 2001; Oegema et al., 2001). At present, homologues of the CENP-H complex proteins have not been identified in C. elegans. Lack of CENP-H complex proteins may cause deficiency of kinetochore targeting of CENP-C (HCP4) in interphase cells of C. elegans. During mitosis, CENP-C can be targeted into kinetochores in the absence of CENP-H complex proteins in both C. elegans and chicken DT40 cells. To understand kinechore assembly, it is important to observe dynamic changes in inner kinetochore organization during the cell cycle.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to K. Suzuki, M. Takahashi, and K. Kita for technical assistance. We also thank H. Kimura for the H2B-RFP plasmid. M.K. thanks T. Motohashi for encouragement. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Science and Technology of Japan.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-01-0045) on March 28, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Brown M. T., Goetsch L., Hartwell L. H. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., III, Chan C. S., Drubin D. G., Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., III, Oegema K., Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Mao Y., Sullivan K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., III, Cleveland D. W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Brown W.R.A. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 1997;6:2301–2308. doi: 10.1093/hmg/6.13.2301. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., Brown W. CENP-C is necessary but not sufficient to induce formation of functional centromere. EMBO J. 1999;18:4196–4209. doi: 10.1093/emboj/18.15.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T. Assembly of kinetochore in vertebrate cells. Exp. Cell Res. 2004;296:21–27. doi: 10.1016/j.yexcr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., Lehner C. F. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 2005;19:2041–2053. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S., Ioannou D., Haines S., Brown W. R. Comparison of Dam tagging and chromatin immunoprecipitation as tools for the identification of the binding sites for S. pombe CENP-C. Chromosome Res. 2005;13:73–83. doi: 10.1007/s10577-005-7062-z. [DOI] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 2003;116:3347–3362. doi: 10.1242/jcs.00645. [DOI] [PubMed] [Google Scholar]
- Izuta H., et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Fowler K. J., Earle E., Hill J., Choo K.H.A. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. USA. 1998;95:576–582. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S. L., Cheeseman I. M., Hori T., Fukagawa T., Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T., Hittle J. C., Jablonski S. A., Campbell M. S., Yoda K., Yen T. J. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 2003;5:341–345. doi: 10.1038/ncb953. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y., Hori T., Kimura H., Fukagawa T. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 2005;25:1958–1970. doi: 10.1128/MCB.25.5.1958-1970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y. C., Kawashima T., Kitamura T., Fukagawa T. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol. Cell. Biol. 2005;25:10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. L., Roth M. B. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. L., Stanvitch G., Roth M. B., Rosen D. HCP-4/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans. Mol. Cell. Biol. 2005;25:2583–2592. doi: 10.1128/MCB.25.7.2583-2592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y., Peebles C., Fritzler M. J., Steigerwald J., Tan E. M. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Hardwick K. G. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W. C., Fukagawa T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004a;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004b;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., III, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Pluta A. F., Mackay A. M., Ainsztein A. M., Goldberg I. G., Earnshaw W. C. The centromere: Hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C. A., Ratrie H., Maure M., Rothfield N. F., Earnshaw W. C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Skoufias D. A., Andreassen P. R., Lacroix F. B., Wilson L., Margolis R. L. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 2001;98:4492–4497. doi: 10.1073/pnas.081076898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Nakano M., Nozaki N., Egashira S., Okazaki T., Masumoto H. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J. Biol. Chem. 2004;279:5934–5946. doi: 10.1074/jbc.M306477200. [DOI] [PubMed] [Google Scholar]
- Tanudji M., Shoemaker J., L'Italien L., Russell L., Chin G., Schebye X. M. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J., Cooke C. A., Saitoh H., Bernat R. L., Earnshaw W. C. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B. A., Bonday Z. Q., Putkey F. R., Kops G. J., Silk A. D., Cleveland D. W. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., III, Drubin D. G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Tomkiel J., Saitoh H., Johnson D. H., Earnshaw W. C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.