Abstract
Transforming growth factor-β (TGF-β) superfamily members play an important role in growth, differentiation, adhesion, apoptosis, and development in many species from insects and worms to vertebrates. Recently, TGF-β signaling has been demonstrated to be negatively regulated by microtubules (MTs), which anchor endogenous Smad2/3 to cytosol and also directly interact with connexin43 (Cx43), and the activity of TGF-β is mediated by Cx43. However, the mechanism underlying the intracellular regulation of TGF-β activity by Cx43 remains unknown. Here, we found that the functional link between TGF-β activation and Cx43 is mediated by interactions among Smad2/3, MTs, and Cx43. We confirmed that Cx43 competes with Smad2/3 for binding to MTs, which Cx43 specifically induces release of Smad2/3 from MTs and increases phospho-Smad2 and which, as a result, Smad2/3 and Smad4 are accumulated in the nucleus, leading to activation of the transcription of target genes. Consistently, knockdown of the endogenous Cx43 activity with double-strand RNA (dsRNA) in HL1 cardiomyocytes and Cx43 knockout mice cardiomyocytes consistently show the opposite effect. Our findings demonstrate a novel mechanism for Cx43 positive regulation of TGF-β function.
INTRODUCTION
Transforming growth factor-β (TGF-β) superfamily members are multifunctional and have been shown to control various developmental and biological responses through transcriptional regulation of diverse genes via receptor-mediated activation of the Smad proteins (Patterson and Padgett, 2000; Attisano and Wrana 2002; ten Djike et al., 2002; Shi and Massagué, 2003). On binding of secreted TGF-β to the type II receptor (TβRII), type I receptors (TβRI/ALK5) are heteromerized and transphosphorylated, resulting in signal transduction through phosphorylation of the receptor-regulated Smads (R-Smads), Smad2 and Smad3. Phospho-Smad2 or phospho-Smad3 then forms a complex with a comediator Smad, i.e., Smad4, which translocates to the nucleus and regulates transcription of target genes (Heldin et al., 1997).
Connexins (Cx), not only allow direct intercellular communication but also play important roles in regulation of cell proliferation, cell differentiation, and tissue development. For example, it has been shown that the activity of TGF-β is mediated by connexin43 (Cx43; Hirschi et al., 2003). However, the mechanism underlying the intracellular regulation of TGF-β activity by Cx43 remains unknown. Furthermore, Cx43 interacts directly with β-tubulin, a major component of microtubules (MTs) through its C-terminal tail (Giepmans et al., 2001). Smad2/3 also binds to β-tubulin, which provides a negative regulatory mechanism controlling TGF-β activity (Dong et al., 2000; Zhu et al., 2004). These evidences suggest a possibility that Cx43 is involved in MTs-regulated TGF-β/Smad signaling. On the hypothesis that Cx43 regulates TGF-β activity by release of Smads from MTs, we sought to identify the molecular mechanism of Cx43 involvement in TGF-β signaling. By using biochemical approach and double-strand RNA (dsRNA) knockdown technique in HL1 cardiomyocytes and Cx43 knockout mice cardiomyocytes, we have demonstrated for the first time that Cx43 regulates TGF-β activity by triggering the release of Smads from microtubules and serves as a positive regulator in relation to the TGF-β/Smad signaling pathway.
MATERIALS AND METHODS
Plasmid Constructions
Full-length cDNAs of Smad2 and Smad3 (Remy et al., 2004) provided by S. Michnick (University of Montreal) were inserted into pGEM-3Zf(+) (Promega, Madison, WI) for in vitro translation, plasmid GST-Smad2 (Chou et al., 2003) by A. Moustakas (Ludwig Institute for Cancer Research), plasmids GST-Smad2MH1 and Smad2MH2 (Wrana et al., 1992) by J. L. Wrana (University of Toronto), and plasmids GST-Smad2ΔMH1 and GST-Smad2ΔMH2 by K. Miyazono (University of Tokyo), plasmids GST-Smad3, Smad3NL, Smad3LC, and Smad3C (Zhang et al., 1998) by R. Derynck (University of California at San Francisco), and Smad3N by Y. Zhang (National Cancer Institute, National Institutes of Health), plasmid GFPSmad2 by C. N. Hill (Cancer Research UK London Research Institute) (Nicolas et al., 2004), and various forms of GST-Cx43 (Giepmans et al., 2001) by B. N. Giepmans (University of California San Diego [UCSD]). Full-length Cx43 was inserted into pGEM-3Zf(+) for in vitro translation. Full-length cDNA of β-tubulin (Mβ3; Chang et al., 2005) provided by J. S. Chang (Daejin University) was inserted into pSPUTK (Stratagene, La Jolla, CA) for in vitro translation. Various forms of β-tubulin were inserted into pGEX vector (Pharmacia Biotech, Piscataway, NJ).
To generate dsRNA expression plasmids against rat Cx43 and mouse Cx40 and Cx45 mRNAs, the coding regions of Cx43.NT (nt 227-592), Cx43.CT (nt 593-1118), Cx40.NT (nt 121-609), Cx40.CT (nt 606-1189), Cx45.NT (nt 522-996), and Cx45.CT (nt 997-1633) were constructed by insertion into the pDECAP vector as inverted repeats with a 12-base pair spacer (GGTGCGCATATG). The dsRNA expression plasmids containing the inverted repeat were amplified in Escherichia coli GT116 strain (Invivogen, San Diego, CA), which allows the accurate replication of DNA containing inverted repeats, and purified using the Endofree Plasmid Maxi Kit (Qiagen, Chatsworth, CA). Wild-type Cx43 expression plasmid (pcDNA3.1.Cx43) was constructed as full-length Cx43 cDNA inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA). The tubulin-binding domain (TD) Cx43 mutant (pcDNA3.1.Cx43ΔTD) was constructed by deleting the amino acid residues 234–243 of pcDNA.3.1.Cx43. For construction of TD expression plasmid (pcDNA.3.1.Cx43JM228-263), the amino acid residues 228-263 were inserted into pcDNA3.1.
GST Pulldown Assays
GST pulldown assays were performed as previously described (Dai et al., 1996).
Culture of HL-1 Cardiomyocytes
HL-1 cardiomyocytes were obtained from Dr. W. C. Claycomb (Louisiana State University, New Orleans, LA). Cells were cultured on a gelatin (0.02% [wt/vol])/fibronectin (10 μg/ml) matrix and were maintained in Claycomb medium (JRH Biosciences, Lenexa, KS) supplemented with 10% (vol/vol) fetal bovine serum (FBS), glutamine (2 mM/l), norepinephrine (0.1 mM/l), penicillin (100 U/ml), and streptomycin (100 U/ml).
Coimmunoprecipitation
For coimmunoprecipitation of endogenous proteins, HL-1 cells were lysed in lysis buffer (50 mM HEPES [pH 7.5], 250 mM NaCl, 0.2 mM EDTA, 10 μM NaF, 0.5% NP-40, and cocktail of protease inhibitors). For immunoprecipitation of β-tubulin with Cx43 and Smad2/3, lysates were immunoprecipitated using anti-β-tubulin mAb (TUB 2.1, Sigma), and immune complex was analyzed by Western blotting using anti-Smad2/3 mAb (BD Biosciences, San Jose, CA) or anti-Cx43 antibodies (Sigma, St. Louis, MO) and ECL detection reagents (Amersham). For immunoprecipitation of Cx43 with β-tubulin, lysates were immunoprecipitated using anti-Cx43 antibodies, and immune complex was analyzed using anti-β-tubulin mAb. As negative controls, anti-Cx40 antibodies (C-20, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-β-galactosidase mAb (Promega) were used. For anti-phospho-Smad2 analysis, 5 μg of pDECAP, pDECAP.Cx43.NT, pDECAP.Cx43.CT, or pDECAP.Cx40.CT together with 1 μg of a puromycin-resistant expression plasmid (pact.RM2xINSPuroVer3) was cotransfected into HL-1 cells. At 24 h after transfection, the cells were incubated in puromycin (5 μg/ml)-containing Claycomb medium for 3 d. Puromycin-positive cells were harvested and lysed. Lysates were immunoprecipitated using anti-Smad2/3 mAb, and immune complex was analyzed by Western blotting using anti-Smad2/3 mAb or anti-phospho-Smad2 antibodies (Calbiochem).
Luciferase Reporter Assays
Transient transfection of cells was carried out using Lipofectamine (Invitrogen) according to the manufacturer's instructions. For the dsRNA-effected knockdown, HL-1 cells were transfected with 0.5 μg of p3TP-lux (or CAGA12-Luc), 20 ng of the internal control plasmid pRL-SV, and either 2 μg of pDECAP empty vector or distinct Cx-dsRNA expression plasmids, as indicated in the legend for Figure 4A. To examine the effect of Cx43 on TGF-β signaling, HL-1 cells were transfected with 0.5 μg of p3TP-lux, 20 ng of pRL-SV together with pcDNA3.1.Cx43, pcDNA3.1.Cx43ΔTD, or pcDNA.3.1.Cx43JM228-263 (2 or 4 μg). Similar experiments were performed in HeLa cells, with the sole difference being that the reporter vector CAGA12-luc was used. For TGF-β treatment, TGF-β1 (PeproTech, Rocky Hill, NJ) was dissolved in 1 mg/4 mM HCl/ml bovine serum albumin. HL-1 or HepG2 cells were transfected with 0.5 μg of p3TP-lux, 20 ng of pRL-SV, and 3 μg of pDECAP.Cx43.NT, pDECAP.Cx43.CT, or pDECAP.Cx40.CT, which served as a negative control. In addition, HepG2 cells were transfected with 0.5 μg of p3TP-lux, 20 ng of pRL-SV, and pcDNA3.1.Cx43 at 0, 1, 3, or 5 μg. After transfection, cells were incubated for 32 h in 10% FBS/Claycomb medium or FBS/DMEM, followed by inductions for 8 h with or without 5 ng/ml TGF-β1. For TβRI/ALK5 inhibitor treatment, SB-431542 (Tocris, Ballwin, MO) was dissolved in dimethyl sulfoxide (DMSO) and used at a final concentration of 10 μM. HL-1 cells were transfected with 20 ng of pRL-SV and the reporter vector CAGA12-luc or p3TP-lux (0, 10, 50, 200, or 500 ng). Similarly to HL-1 cells, HeLa cells were transfected with 0.5 μg of CAGA12-luc, 20 ng of the internal control plasmid pRL-CMV, and pcDNA3.1.Cx43 at 0, 1, 3, or 5 μg. Immediately after transfection, HL-1 or HeLa cells were incubated for 40 h in 10% FBS/Claycomb medium or FBS/DMEM containing either DMSO alone or 10 μM SB-431542. For luciferase assays, all samples were harvested together at 40 h after transfection. Luciferase activity in cell lysates was measured using the dual luciferase assay system (Promega) in a Berthold Lumat LB 9705 luminometer (Pforzheim, Germany). The data are expressed as means and SDs (error bars) of a representative experiment performed in triplicate.
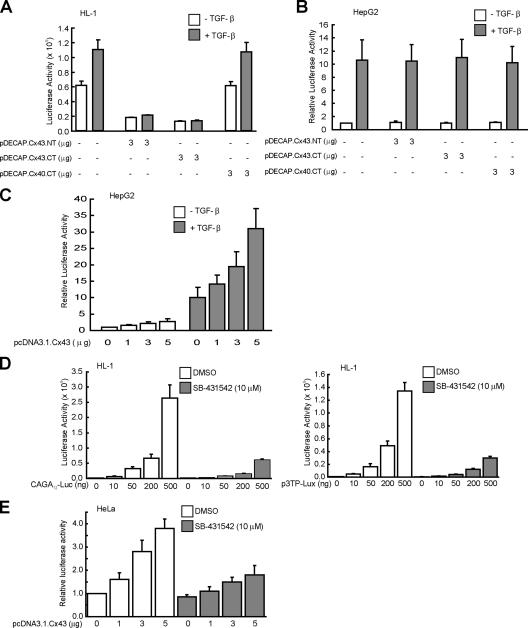
Figure 4.
Cx43 regulates TGF-β signaling. (A) Effect of Cx depletion on transcriptional activation of the TGF-β–targeted gene. HL-1 cells were cotransfected with the reporter vector p3TP-lux and internal control plasmid pRL-SV together with pDECAP empty vector, pDECAP vector expressing N-terminal Cx43 dsRNA (pDECAP.Cx43.NT), C-terminal Cx43 dsRNA (pDECAP.Cx43.CT), N-terminal Cx40 dsRNA (pDECAP.Cx40.NT), C-terminal Cx40 dsRNA (pDECAP.Cx40.CT), N-terminal Cx45 dsRNA (pDECAP.Cx45.NT), or C-terminal Cx45 dsRNA (pDECAP.Cx45.CT). (B) Cx43 is specifically required for nuclear translocation of Smad2/3 and Smad4 in cardiomyocytes. HL-1 cells were transfected with pDECAP.Cx43.CT, pDECAP.Cx43.NT, and pDECAP.Cx40.CT together with a transfectant indicator, pcDNA.3.1.mRFP1. Cells were fixed and stained with anti-Smad2/3 (left) or anti-Smad4 (right) monoclonal antibodies followed by Alexa Fluor 488 goat anti-mouse IgG and were examined by confocal fluorescence microscopy. (C) The TD of Cx43 is required for Smad-induced transcriptional activity. HL-1 cells (left) were transfected with the reporter vector p3TP-lux and internal control plasmid pRL-SV together with pcDNA3.1.Cx43, pcDNA3.1.Cx43ΔTD, or pcDNA3.1.Cx43JM228-263. HeLa cells (right) were transfected with the reporter vector CAGA12-luc, internal control plasmid pRL-CMV, and together with pcDNA3.1.Cx43, pcDNA3.1.Cx43ΔTD, or pcDNA3.1.Cx43JM228-263. All samples were harvested together at the 40-h time point, and luciferase activity was measured and normalized. The data shown in A and C are means and SDs (error bars) of a representative experiment performed in triplicate and were evaluated for statistical significance with two-tailed Student's test. *p < 0.01; **p < 0.005.
Cx43 Mutant Mice
Founder mice (C57BL/6JGjaltm1Kdr) of uniform genetic background (C57BL/6) were originally purchased from Jackson Laboratories (Bar Harbor, ME). Mice were housed in barrier facilities under standard conditions. All mice were maintained in an inbred background (C57BL/6) so that effects due to genetic variation were minimized. The genotypes of all mice were determined by PCR, using primer sequences and protocols as described (Reaume et al., 1995). All animal experiments were conducted with the approval of and in accordance with guidelines from the Committee for Animal Research, Kyoto Prefectural University of Medicine.
Primary Cell Culture
Single-cell preparations of fetal cardiomyocytes were isolated from mice deficient for Cx43 (Cx43−/− or Cx43+/−) or wild-type littermates (Cx43+/+) and cultured for 48 h in DMEM with 10% FBS. The cardiomyocytes were then used for Smad2/3 and Smad4 immunostaining.
Generation of Cell Line
The HeLa cell line stably expressing GFPSmad2 was generated by transiently transfecting HeLa cells with GFPSmad2 and then selecting transfected cells using 1 mg/ml G418.
Time-Lapse Observations
For time-lapse observations, HeLa cells stably expressing GFPSmad2 were transfected with 1 μg of Cx43 expression vector with 10 ng of pcDNA.3.1.mRFP1, which served as a transfectant indicator. After transfection, cells were incubated for 4 h and then imaged under a fluorescence microscope (IX71, Olympus, Tokyo, Japan). Images were taken with a 0.03-s exposure at 30-min intervals with 23 optical sections.
Immunocytochemistry
To express Cx-dsRNAs, 2 μg of pDECAP.Cx43.NT, pDECAP.Cx43.CT, pDECAP.Cx40.NT, pDECAP.Cx40.CT, pDECAP.Cx45.NT, or pDECAP.Cx45.CT together with 0.5 μg of pcDNA.3.1.mRFP1, which served as a transfectant indicator, was transfected into HL-1 cells. HL-1 and primary cultured cells were rinsed in ice-cold phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde in PBS for 30 min. After fixation, cells were permeabilized by incubation with 0.1% Triton X-100 in PBS for 10 min. Cells were then blocked with 3% skim milk in PBS for 1 h. Primary antibodies used were anti-Smad2/3 mAb (BD Biosciences; 1:100) and anti-Smad4 mAb (B-8, Santa Cruz; 1:100). Secondary antibodies were used with Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes). Cells were washed carefully with PBS (5 times, 10 min each time) after incubation with primary and secondary antibodies. After immunostaining, samples were mounted and analyzed by confocal microscopy (FV300, Olympus).
RESULTS
Smad2/3 and Cx43 Competitively Bind to the Same Region of β-Tubulin
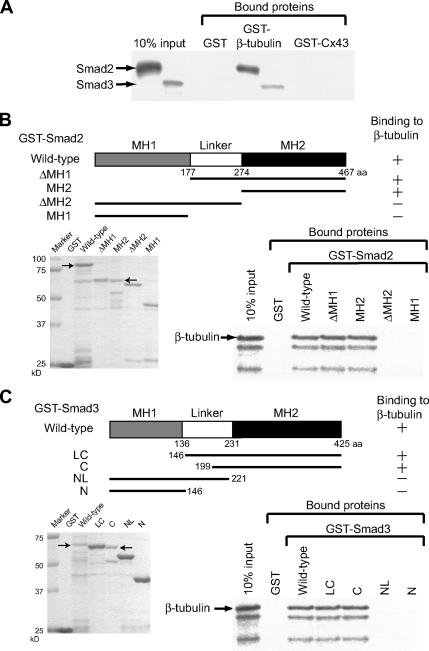
We first examined whether Smad2/3 directly interacts with mouse β-tubulin and Cx43 in vitro. The full-length forms of Smad2 and Smad3 were translated in vitro and mixed separately with each of the full-length forms of GST-β-tubulin and GST-Cx43. Smad2 and Smad3 bound to GST-β-tubulin but neither to GST-Cx43 nor to the GST resin alone (Figure 1A). To investigate which domain of Smad2 interacts with β-tubulin, we used a series of deletion mutants of GST-Smad2 in the in vitro binding assay (Figure 1B). In vitro–translated full-length β-tubulin was mixed with the various forms of GST-Smad2. The results showed that the MH2 domain of Smad2 binds to β-tubulin with almost the same efficiency as the full-length (wild-type) Smad2. Similar results were obtained for the Smad3 binding to β-tubulin (Figure 1C).
Figure 1.
Smad2/3 binds to β-tubulin but not to Cx43. (A) Smad2/3 directly binds to β-tubulin but not to Cx43. In vitro–translated Smad2 or Smad3, GST-β-tubulin, and Cx43 were used for GST pulldown assays. (B) Identification of β-tubulin–binding domain in Smad2. In vitro–translated β-tubulin and various forms of GST-Smad2 were used for GST pulldown assays. (C) Identification of the β-tubulin–binding domain in Smad3. In vitro–translated β-tubulin and various forms of GST-Smad3 were used for GST pulldown assays. Small arrows indicate the protein bands of interest in B and C. The relative binding activities of β-tubulin (B and C) are designated + and −, which indicate 5–10% and <1% binding of input protein, respectively. The purity and concentration of bacterially expressed GST-fusion proteins were analyzed on 10% SDS-PAGE followed by Coomassie blue staining in B and C.
On use of a series of GST-Cx43 proteins containing various portions of the Cx43 molecule, as reported (Giepmans et al., 2001), the C-terminal juxtamembrane region (GST-Cx43JM228-263) bound β-tubulin to the same extent as did the full-length GST-Cx43 (Figure 2A).
Figure 2.
Smad2/3 and Cx43 competitively bind to the same region of β-tubulin. (A) Identification of β-tubulin–binding domain in Cx43. In vitro–translated β-tubulin and various forms of GST-Cx43 were used for GST pulldown assays. (B) Identification of the binding domain of β-tubulin for Smad2 or Cx43. In vitro–translated Smad2 or Cx43 and various forms of GST-β-tubulin were used for GST pulldown assays. (C) Cx43 competes with Smad2 for binding to MTs. Mixtures at various ratios of in vitro–translated [35S]Smad2 and cold Cx43 (1:0, 1:1, 1:3, or 1:5) were used for binding to the affinity resin containing 10 μg of GST-β-tubulin fusion protein. Small arrows indicate the protein bands of interest in A and B. The relative binding activities of β-tubulin (A), Smad2 and Cx43 (B) are designated + and −, which indicate 5–10% and <1% binding of input protein, respectively. The purity and concentration of bacterially expressed GST-fusion proteins were analyzed on 10% SDS-PAGE followed by Coomassie blue staining in A and B.
To map the region of β-tubulin where it interacts with Smad2, we used a series of deletion mutants of GST-β-tubulin fusion for the GST pulldown assay (Figure 2B). The results revealed that the region containing amino acids 114–243 (GST-β-tubulin114/243) was required for binding to Smad2 (Figure 2B) or Smad3 (data not shown), the same region was also required for binding to Cx43 (Figure 2B).
The common binding region of β-tubulin for Smad2/3 and Cx43 (Figure 2B) indicates that there is competition between Cx43 and Smad2/3 for binding to β-tubulin. This possibility was tested by a competition assay (Figure 2C). We used in vitro–translated [35S]methionine-labeled Smad2 and a dose of cold Cx43 for competitive binding to GST-β-tubulin114/243. Cx43 interfered with the binding of Smad2 to β-tubulin.
Interactions among Cx43, β-Tubulin, and Smad2/3 In Vivo
We reasoned that Cx43-mediated TGF-β signaling could arise from competition between Cx43 and Smad2/3 on MTs. To examine interactions of MTs with Smad2/3 and Cx43 in vivo, coimmunoprecipitation assays of endogenous proteins were performed with anti-β-tubulin and anti-Cx43 using lysates prepared from HL-1 cardiomyocytes (Claycomb et al., 1998; White et al., 2004). Smad2/3 and Cx43 were coprecipitated by anti-β-tubulin (Figure 3, A and B), and β-tubulin was coprecipitated by anti-Cx43 (Figure 3D). No interaction between Cx40 and β-tubulin was detected (Figure 3C). Taken together, these results suggest that there are interactions among Cx43 and Smad2/3 at MTs.
Figure 3.
Association among Cx43, β-tubulin, and Smad2/3 in HL-1 cells. (A) Endogenous β-tubulin interacts with Smad2/3. Lysates of HL-1 cells were subjected to coimmunoprecipitation (CoIP) with anti-β-tubulin mAb, followed by immunoblot analysis (IB) with anti-Smad2/3 mAb. (B) Association of β-tubulin with Cx43. Lysates of HL-1 cells were immunoprecipitated with anti-β-tubulin mAb and subjected to immunoblot analysis using anti-Cx43 antibodies. (C) Absence of interaction between β-tubulin and Cx40. Lysates of HL-1 cells was immunoprecipitated with anti-β-tubulin mAb and subjected to immunoblot analysis using anti-Cx40 antibodies. (D) Coimmunoprecipitation of Cx43 with β-tubulin. Lysates of HL-1 cells were immunoprecipitated with anti-Cx43 antibodies and subjected to immunoblot analysis using anti-β-tubulin mAb.
Cx43 Positively Regulates TGF-β Signaling
In mammalian hearts, myocytes prominently express three major types of connexin proteins: Cx40, Cx43, and Cx45 (van Veen et al., 2001). To investigate whether Cx43 specifically contributes to TGF-β activity in vivo, we examined knockdowns of the endogenous Cx40, Cx43, and Cx45 in cardiomyocytes by producing dsRNA in HL-1 cells using the pDECAP vector (Shinagawa and Ishii, 2003). Although HL-1 cells predominantly express three types of connexin proteins (Supplementary Figure 1A), the endogenous mRNAs and the corresponding proteins of Cx40, Cx43, and Cx45 were significantly reduced by respective C-terminal dsRNAs (Supplementary Figure 1, B and C).
We examined the effect of reducing the amount of the connexin protein on Smad2/3-induced activation of transcription using a luciferase-reporter system in HL-1 cells (p3TP-lux vector; Wrana et al., 1992; Dennler et al., 1998; Stroschein et al., 1999). Luciferase levels were reduced more than fivefold by cotransfection of p3TP-lux reporter with 1 μg of pDECAP vector expressing C-terminal Cx43 dsRNA (pDECAP.Cx43.CT; Figure 4A). Cotransfection with 1 μg of pDECAP vector expressing N-terminal Cx43 dsRNA (pDECAP.Cx43.NT) resulted in reduction by about half. In contrast, cotransfection of 1 μg of pDECAP vector expressing dsRNA of Cx40 or Cx45 for the C- or N-terminal region of the gene did not significantly affect the luciferase levels of p3TP-lux reporter. Similar results were obtained in HL-1 cells with the Smad3/Smad4-dependent reporter CAGA12-luciferase (Dennler et al., 1998; data not shown). These data provide mechanistic evidence that Cx43 expression serves as a positive regulator for TGF-β signal transduction.
Next, we reasoned that the subcellular localization of Smads may be affected by the context of Cx43, i.e., whether the Cx43 functions as a positive regulator in relation to the TGF-β/Smad signaling pathway. To test this possibility, we examined whether decreased Cx43 expression results in reduction of the nuclear accumulation of Smad2/3 and Smad4 specifically. Subcellular localization of Smad2/3 and Smad4 was examined by immunocytochemical analysis of HL-1 cells transfected with the dsRNA expression vector of Cx43 or Cx40. Nuclear accumulation of Smad2/3 and Smad4 was reduced in the Cx43 dsRNA-transfected cells. In contrast to Cx43 dsRNA-expressing cells, no significant difference between the subcellular localizations of Cx40 dsRNA-expressing cells and intact cells was observed (Figure 4B). These results demonstrate that even in the absence of the TGF-β stimulation, Smad-mediated TGF-β signaling is constitutively activated by Cx43 that contributes to the basal level of Smad2/3 and Smad4 in the nucleus.
To test further whether Cx43 specifically regulates TGF-β activity among various Cxs via the TD (Giepmans et al., 2001), we constructed a Cx43 mutant, pcDNA3.1.Cx43ΔTD expression vector, by deleting a potential tubulin-binding motif (amino acids 234–243) of Cx43, and TD expression vector, pcDNA.3.1.Cx43JM228-263. In the luciferase reporter assays, we initially used HeLa cells, which express no endogenous Cx43 (Lin et al., 2004). HeLa cells showed a dose-dependent increase in TGF-β–targeted reporter activity when cotransfected with the CAGA12-luciferase reporter plasmid together with increasing amounts of wild-type Cx43 or TD expression plasmid Cx43JM228-263 (Figure 4C). Overexpression of Cx43 with wild-type Cx43 or TD expression plasmid Cx43JM228-263 at 4 μg increased the degree of TGF-β–targeted reporter transcription to about fourfold even in the absence of TGF-β. In addition, cotransfection with wild-type Cx43 or TD expression plasmid Cx43JM228-263 in HL-1 cells, which abundantly express endogenous Cx43 (Supplementary Figure 1A), revealed a similar result, albeit only a slight increase in Cx43-induced TGF-β–targeted reporter luciferase levels was observed. As expected, when the cells overexpressed Cx43ΔTD expression plasmid, no significant effect on the level of TGF-β–targeted gene luciferase activity was observed in either HeLa or HL-1 cells (Figure 4C). We also examined the effects of Cx43 or Cx43ΔTD on EGF-responsive promoter cyclin D1 (CCND1; Kasper et al., 2006), no significant effects on the level of EGF-targeted gene luciferase activity was observed by overexpressing either Cx43 or Cx43ΔTD (Supplementary Figure 2). These findings suggest that the effect of Cx43 on TGF-β signaling requires direct binding of Cx43 to MTs via its TD at the C terminus and a resultant release of Smad2/3 from MTs, which consequently readily access TβRI. Therefore Cx43 may serve as a positive regulator in relation to the TGF-β/Smad signaling pathway.
Cx43 Is Required for Smad Nuclear Translocation
To further test whether TGF-β activity is affected by the expression levels of Cx43 in vivo, we examined the subcellular localization of Smad2/3 and Smad4 in primary cultured cardiomyocytes derived from the hearts of E16 mouse littermates wild-type (Cx43+/+), heterozygous (Cx43+/−), or homozygous (Cx43−/−) for Cx43 (Figure 5, A and B). In Cx43+/+ cells, abundant nuclear localization of Smad2/3 and Smad4 was observed. Intermediate nuclear localization of these proteins was detected in Cx43+/− cells. In Cx43−/− cells, Smad2/3 or Smad4 was localized uniformly throughout the cells, i.e., it was indistinguishable from the cytoplasm and nucleus. We also examined the effects of TGF-β on subcellular localization of Smad2/3 by using primary cultured Cx43−/− cardiomyocytes. TGF-β stimulation did not affect the subcellular localization of Smad2/3 (Supplementary Figure 3). Moreover, we examined the effects of TGF-β on the protein level and subcellular localization of Cx43 by using Cx43+/+ or Cx43+/− cardiomyocytes. TGF-β stimulation affected neither the protein level nor the subcellular localization of Cx43 (Supplementary Figure 4).
Figure 5.
Cx43 is required for Smad nuclear translocation. (A) Immunofluorescence analysis of the subcellular localization of endogenous Smad2/3. Primary cultured cardiomyocytes of Cx43+/+, Cx43+/−, and Cx43−/− mice were fixed and then stained with anti-Smad2/3 mAb followed by Alexa Fluor 488 goat anti-mouse IgG and phalloidin, as an indicator of cardiomyocytes. (B) Subcellular localization of endogenous Smad4 in Cx43+/+, Cx43+/−, and Cx43−/− mouse cardiomyocytes. Similar experiments were performed in the cells as described for A with the sole difference being that the cells were stained with anti-Smad4 mAb followed by Alexa Fluor 488 goat anti-mouse IgG.
To further clarify the subcellular distribution of Smad2/3 controlled by Cx43 expression, we generated a HeLa cell line stably expressing GFPSmad2. The subcellular distribution of Smad2 was examined by time-lapse observations of GFPSmad2-expressing HeLa cells transfected with the Cx43 expression vector. GFPSmad2 localization was predominantly cytoplasmic in cells not transfected with Cx43 and predominantly nuclear after 15 h in cells transiently expressing Cx43 (Figure 6 and Supplementary Movie).
Figure 6.
Cx43 induces Smad2 for nuclear translocation. Time-lapse observation of subcellular distribution of Smad2 in HeLa cells stably expressing GFPSmad2 were transfected with pcDNA3.1.Cx43 together with a transfectant indicator, pcDNA.3.1.mRFP1.
Thus, these results suggest that TGF-β activity depends on Cx43 expression levels, which control the subcellular localization of Smads.
Cx43 Cooperatively Interacts with TGF-β in Regulating TGF-β/Smad Signaling
To examine whether the expression levels of Cx43 affect TGF-β–induced activation, we initially cotransfected HL-1 cells with the TGF-β–targeted reporter together with Cx43 dsRNA or Cx40 dsRNA expression plasmid with or without TGF-β stimulation (Figure 7A). TGF-β–induced an increase in targeted reporter luciferase activity. This increase was reduced or completely abrogated by expression of Cx43 dsRNA at basal levels comparable to that in the absence of TGF-β stimulation in HL-1 cells. In contrast, Cx40 dsRNA had no significant effect on TGF-β–induced transcription of the targeted reporter. In addition, cotransfection with the TGF-β–targeted reporter together with Cx43 dsRNA or Cx40 dsRNA expression plasmid with or without TGF-β stimulation in HepG2 cells, which express no endogenous Cx43 (data not shown). As expected, neither Cx43 dsRNA nor Cx40 dsRNA reduced TGF-β–induced targeted reporter luciferase activity (Figure 7B). We also cotransfected HepG2 cells with the TGF-β–targeted reporter together with Cx43 expression plasmid with or without TGF-β stimulation (Figure 7C). Cx43 cooperatively potentiated the effect in the presence of TGF-β on TGF-β–induced transcription of the targeted reporter. Moreover, a similar result, albeit only a slight increase in TGF-β–induced targeted reporter luciferase levels was observed in HeLa cells, (Supplementary Figure 5). These data indicate that Cx43 cooperatively interacts with TGF-β in regulating TGF-β/Smad signaling.
Figure 7.
Cx43 cooperatively interacts with TGF-β in regulating TGF-β/Smad signaling. (A and B) Cx43 depletion abrogates TGF-β–induced Smad-mediated transcription. HL-1 (A) or HepG2 (B) cells were transfected with p3TP-lux and pRL-SV and together with pDECAP.Cx43.NT, pDECAP.Cx43.CT, or pDECAP.Cx40.CT, which served as a negative control. (C) Cx43 cooperatively potentiates TGF-β–induced transcriptional activity. HepG2 cells transfected with p3TP-lux, pRL-SV and a dose of pcDNA.3.1.Cx43. After transfection, cells were incubated for 32 h in 10% FBS/Claycomb medium or FBS/DMEM, followed by inductions for 8 h with or without 5 ng/ml TGF-β1 in A–C. (D) Activated TGF-β receptor TβRI/ALK5 is required for Cx43-mediated TGF-β/Smad activity in HL-1 cardiomyocytes. HL-1 cells were transfected with internal control plasmid pRL-SV and the indicated dose of the reporter vector CAGA12-luc (left) or p3TP-lux (right). (E) TβRI/ALK5 kinase activity is required for Cx43-mediated TGF-β/Smad activity in HeLa cells. HeLa cells were transfected with the reporter vector CAGA12-luc, internal control plasmid pRL-CMV, and the indicated dose of pcDNA3.1.Cx43. Immediately after transfection, cells were incubated for 40 h in 10% FBS/DMEM containing DMSO alone or 10 μM SB-431542 in D and E. For luciferase assays, all samples were harvested together at the 40-h time point after transfection and luciferase activity was then measured and normalized. All data shown in A–E are means and SDs (error bars) of a representative experiment performed in triplicate.
To elucidate whether Cx43-mediated TGF-β activity is mediated by continuous stimulation of the TGF-β receptors, we used a highly specific and potent TβRI/ALK5 inhibitor, SB-431542, which instantaneously inhibits ALK5 kinase activity (Inman et al. 2002a,b; Laping et al., 2002). HL-1 cells transfected with either p3TP-lux or CAGA12-luciferase reporter plasmid were treated with or without SB-431542 for 40 h before reporter assays. A significant inhibition of luciferase reporter transcription was observed in the presence of SB-431542. In contrast, treatment with DMSO produced no remarkable effect on TGF-β–targeted reporter activity (Figure 7D). Concomitantly, we observed similar inhibitory effects of SB-431542 on Cx43-induced TGF-β–targeted reporter activity after cotransfection of HeLa cells with pcDNA3.1.Cx43 together with CAGA12-luciferase reporter. Under treatment with SB-431542, Cx43-induced TGF-β–targeted reporter activity was abrogated (Figure 7E). Taken together, these data suggest that Cx43-mediated TGF-β signaling acts via type I receptor, TβRI/ALK5, and phosphorylates Smad2 and Smad3 that form complexes with Smad4, which accumulate in the nucleus and directly regulate transcription of target genes.
Cx43 Regulates TGF-β/Smad Signaling via the Phosphorylation of Smad2
TGF-β signal transduction involves phosphorylation of serine residues located at the C termini of Smad2 and Smad3 by the activated TβRI (Abdollah et al., 1997; Souchelnytskyi et al., 1997). To determine whether the Cx43-mediated TGF-β transcriptional activity is accompanied by a change in the phosphorylation state of Smad2, we performed Western blotting analysis using a phospho-Smad2-specific antibody (Dong et al., 2000). We used similar total levels of endogenous Smad2/3 proteins extracted from intact HL-1 cells (untransfected) and HL-1 cells transfected with empty vector pDECAP, dsRNA expression plasmid of Cx43 or Cx40 (Figure 8A). In untransfected intact HL-1 cells, a high level of phospho-Smad2 was detected, even in the absence of TGF-β stimulation. Although Smad2 phosphorylation was not markedly altered by transfection with empty vector or Cx40 dsRNA expression plasmid, transfection with Cx43 dsRNA expression plasmid remarkably reduced phosphorylation of Smad2 (Figure 8A). These data provide mechanistic evidence that intact Cx43 expression may serve as a cytoplasmic regulator for Smad2, mediating Smad2 access to and phosphorylation by TβRI. Thus, Cx43 functions as a positive regulator of TGF-β signaling affecting transcription of TGF-β/Smad target genes.
Figure 8.
Cx43 regulates TGF-β/Smad signaling via the phosphorylation of Smad2 in vivo. (A) Cx43 enhances the phosphorylation of Smad2. HL-1 cells were transfected with a puromycin-resistant expression plasmid (pact.RM2xINSPuroVer3), which served as a resistant transfectant, and the empty vector pDECAP, pDECAP.Cx43.NT pDECAP.Cx43.CT, or pDECAP.Cx40.CT. The cells were incubated with puromycin-containing Claycomb medium (puromycin, 5 μg/ml) for 3 d. TGF-β stimulation was performed as that HL1 cells were by inductions for 8 h with or without 5 ng/ml TGF-β1. Lysates of puromycin-positive cells were immunoprecipitated using anti-Smad2/3 mAb. Immune complex was analyzed by Western blotting using anti-Smad2/3 mAb (top), and then the same blotting membrane was analyzed by using anti-phospho-Smad2 antibodies (bottom). (B) A proposed model for the role of Cx43 in TGF-β signaling.
DISCUSSION
Nuclear accumulation of Smad2, Smad3, and Smad4 transcription factors is a key event heralding the effective initiation and maintenance of the TGF-β signaling pathway (Heldin et al., 1997). TGF-β has pivotal and reciprocal roles in normal and pathological developmental-homeostatic processes (Shi and Massagué, 2003), and gap junction protein Cx43 has been shown to mediate TGF-β activity (Hirschi et al., 2003). However, the detailed molecular mechanisms by which these activities control TGF-β signaling are not clear. Our results for the first time suggest a simple mechanism by which the translocation of Smad2/3 and Smad4 from the cytoplasm to the nucleus is mediated by Cx43 competition with Smad2/3 for binding to MTs (Figure 8B). We found that the Cx43 binding to MTs induces TGF-β signaling through interference with Smad2/3 binding to MTs at the same region named the “tubulin signature motif” (GGGTGSG). Interference by Cx43 induces release of Smad2/3 from MTs, facilitating the association of Smad2/3 with the receptor kinase TβRI and subsequent phosphorylation/activation of Smad2/3, leading to translocation of phosphorylated Smad2/3 with Smad4 to the nucleus. Thus, Cx43 may affect the basal level of R-Smads in the nucleus by competitive R-Smads binding to MTs.
To date, TGF-β signaling in endothelial cells has been shown to occur via two pathways, the classical pathway involving type I receptor, TβRI/ALK-5 and phosphorylation of Smad2/3, and another pathway involving ALK-1 for transduction of signals and phosphorylation of Smad1 or Smad5 (Macias-Silva et al., 1998; Chen and Massagué, 1999; Oh et al., 2000). In the present study, Cx43 was shown to regulate TGF-β activity by triggering the release of Smad2/3 from MTs and increase the amount of phosphor-Smad2, with resulting Smad2/3 and Smad4 accumulation in the nucleus (Figures 2C, 5, 6, and 8A and Supplementary Movie). It is likely that Cx43-mediated TGF-β signaling occurs via the classical pathway.
TGF-β stimulation did not affect the subcellular localization of Smad2/3 in primary cultured Cx43−/− cardiomyocytes (Supplementary Figure 3). Moreover, TGF-β–induced transcriptional activity was abolished by knockdown of the endogenous Cx43 activity with Cx43 dsRNA at basal levels comparable to that in the absence of TGF-β stimulation in HL-1 cells (Figure 7A). These results suggest that in the nondividing, terminally differentiated cardiomyocytes, which express abundant endogenous Cx43, Cx43 contribution to Smad transcriptional activity mainly depends on Cx43 expression and not on TGF-β stimulation.
Recent genetic studies suggest that Cx43 expression is essential for normal heart formation and function. Homozygous Cx43 knockout mice die neonatally from cono-truncal heart malformation and right ventricular outflow obstruction (Reaume et al., 1995; Waldo et al., 1999). Moreover, Cx gene products have come to be regarded as tumor suppressors, in that loss of intercellular communication frequently results in transformed cells, whereas restoration of coupling by transfection with Cx genes may serve to slow cell growth (Mehta et al., 1991; Hotz-Wagenblatt and Shalloway, 1993). Cx43 expression is important for the maintenance of lung homeostasis, and Cx43+/− mice show a greater susceptibility to urethane-induced lung carcinogenesis (Avanzo et al., 2004). TGF-β signaling is well known to have a negative effect on cell growth, and inactivation of this pathway contributes to tumorigenesis. Tumor-driven mutations have been observed in both TGF-β family receptors and the Smad proteins (Massagué et al., 2000). The present study for the first time demonstrates that Cx43 specifically regulates endogenous Smad2/3 and Smad4 nuclear translocation without TGF-β stimulation. The capacity of Cx43 to positively mediate TGF-β signaling suggests a novel mechanism through which Cx gene products may regulate cell growth in a manner quite distinct from their channel-forming properties.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. C. Claycomb for HL-1 cardiomyocytes, J. Massagué (Memorial Sloan-Kettering Cancer Center) for plasmid of p3TP-Lux, P. ten Dijke (Leiden University Medical Center) for plasmids of CAGA12-MLP-Luc and MLP-Luc, R. Y. Tsien (University of California, San Diego) for mRFP1 cDNA, S. Michnick for full-length cDNAs of Smad2 and Smad3, J. L. Wrana for GST-Smad2MH1 and Smad2MH2, A. Moustakas for GST-Smad2, K. Miyazono for GST-Smad2ΔMH1 and Smad2ΔMH2, R. Derynck for GST-Smad3, Smad3NL, Smad3LC and Smad3C, Y. Zhang for GST-Smad3N, C. S. Hill for GFPSmad2, B. N. Giepmans for various forms of GST-Cx43, J. S. Chang for full-length cDNA of β-tubulin (Mβ3), F. Aberger (University of Salzburg) for CCND1 and S. Ishii (RIKEN Tsukuba Institute) and T. Shinagawa (RIKEN Tsukuba Institute) for valuable discussion and plasmids of pDECAP and pact.RM2xINSPuroVer3. This work was supported by funds for Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology (JST) and by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Abbreviations used:
- Cx43
connexin 43
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-12-1064) on April 11, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abdollah S., Macías-Silva M., Tsukazaki T., Hayashi H., Attisano L., Wrana J. L. TGFβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J. Biol. Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Attisano L., Wrana J. L. Signal transduction by the TGF-β superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Avanzo J. L., et al. Increased susceptibility to urethane-induced lung tumors in mice with decreased expression of connexin43. Carcinogenesis. 2004;25:1973–1982. doi: 10.1093/carcin/bgh193. [DOI] [PubMed] [Google Scholar]
- Chang J. S., Kim S. K., Kwon T. K., Bae S. S., Min do S., Lee Y. H., Kim S. O., Seo J. K., Choi J. H., Suh P. G. Pleckstrin homology domains of phospholipase C-γ1 directly interact with β-tubulin for activation of phospholipase C-γ1 and reciprocal modulation of β-tubulin function in microtubule assembly. J. Biol. Chem. 2005;280:6897–6905. doi: 10.1074/jbc.M406350200. [DOI] [PubMed] [Google Scholar]
- Chen Y. G., Massagué J. Smad1 recognition and activation by the ALK1 group of transforming growth factor-b family receptors. J. Biol. Chem. 1999;274:3672–3677. doi: 10.1074/jbc.274.6.3672. [DOI] [PubMed] [Google Scholar]
- Chou W. C., Prokova V., Shiraishi K., Valcourt U., Moustakas A., Hadzopoulou-Cladaras M., Zannis V. I., Kardassis D. Mechanism of a transcriptional cross talk between transforming growth factor-β-regulated Smad3 and Smad4 proteins and α nuclear receptor hepatocyte nuclear factor-4. Mol. Biol. Cell. 2003;14:1279–1294. doi: 10.1091/mbc.E02-07-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P., Akimaru H., Tanaka Y., Hou D. X., Yasukawa T., Kanei-Ishii C., Takahashi T., Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Li Z., Alvarez R., Jr, Feng X. H., Goldschmidt-Clermont P. J. Microtubule binding to Smads may regulate TGFβ activity. Mol. Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- Giepmans B. N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M. M., Moolenaar W. H. Gap junction protein connexin-43 interacts directly with microtubules. Curr. Biol. 2001;11:1364–1368. doi: 10.1016/s0960-9822(01)00424-9. [DOI] [PubMed] [Google Scholar]
- Heldin C. H., Miyazono K., ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hirschi K. K., Burt J. M., Hirschi K. D., Dai C. Gap junction communication mediates transforming growth factor-β activation and endothelial-induced mural cell differentiation. Circ. Res. 2003;93:429–437. doi: 10.1161/01.RES.0000091259.84556.D5. [DOI] [PubMed] [Google Scholar]
- Hotz-Wagenblatt A., Shalloway D. Gap junctional communication and neoplastic transformation. Crit. Rev. Oncog. 1993;4:541–558. [PubMed] [Google Scholar]
- Inman G. J., Nicolas F. J., Callahan J. F., Harling J. D., Gaster L. M., Reith A. D., Laping N. J., Hill C. S. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002a;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Inman G. J., Nicolas F. J., Hill C. S. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell. 2002b;10:283–294. doi: 10.1016/s1097-2765(02)00585-3. [DOI] [PubMed] [Google Scholar]
- Kasper M., et al. Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 2006;26:6283–6298. doi: 10.1128/MCB.02317-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laping N. J., et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542. Mol. Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- Lin G. C., Rurangirwa L. K., Koval M., Steinberg T. H. Gap junctional communication modulates agonist-induced calcium oscillations in transfected HeLa cells. J. Cell Sci. 2004;117:881–887. doi: 10.1242/jcs.00942. [DOI] [PubMed] [Google Scholar]
- Macias-Silva M., Hoodless P. A., Tang S. J., Buchwald M., Wrana J. L. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J. Biol. Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- Massagué J., Blain S. W., Lo R. S. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;13:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Mehta P. P., Hotz-Wagenblatt A., Rose B., Shalloway D., Loewenstein W. R. Incorporation of the gene for a cell-cell channel protein into transformed cells leads to normalization of growth. J. Membr. Biol. 1991;124:207–225. doi: 10.1007/BF01994355. [DOI] [PubMed] [Google Scholar]
- Nicolas F. J., De Bosscher K., Schmierer B., Hill C. S. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 2004;117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- Oh S. P., et al. Activin receptor like kinase 1 (ALK-1) modulates TGFβ1 signalling in regulation of angiogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:2626–2631. doi: 10.1073/pnas.97.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. I., Padgett R. W. TGF-β-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/s0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- Reaume A. G., de Sousa P. A., Kulkarni S., Langille B. L., Zhu D., Davies T. C., Juneja S. C., Kidder G. M., Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- Remy I., Montmarquette A., Michnick S. W. PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nat. Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- Shi Y., Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shinagawa T., Ishii S. Generation of Ski-knockdown mice by expressing along double-strand RNA from an RNA polymerase II promoter. Genes Dev. 2003;17:1340–1345. doi: 10.1101/gad.1073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S., Tamaki K., Engstrom U., Wernstedt C., ten Dijke P., Heldin C. H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J. Biol. Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Stroschein S. L., Wang W., Luo K. Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates transforming growth factor β-induced Smad-dependent transcriptional activation. J. Biol. Chem. 1999;274:9431–9441. doi: 10.1074/jbc.274.14.9431. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Goumans M. J., Itoh F., Itoh S. Regulation of cell proliferation by Smad proteins. Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- van Veen A. A., van Rijen H. V., Opthof T. Cardiac gap junction channels: modulation of expression and channel properties. Cardiovasc. Res. 2001;51:217–229. doi: 10.1016/s0008-6363(01)00324-8. [DOI] [PubMed] [Google Scholar]
- Waldo K. L., Lo C. W., Kirby M. L. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev. Biol. 1999;208:307–323. doi: 10.1006/dbio.1999.9219. [DOI] [PubMed] [Google Scholar]
- White S. M., Constantin P. E., Claycomb W. C. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- Wrana J. L., Attisano L., Carcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng X. H., Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhu S., Goldschmidt-Clermont P. J., Dong C. Transforming growth factor-β-induced inhibition of myogenesis is mediated through Smad pathway and is modulated by microtubule dynamic stability. Circ. Res. 2004;19:617–625. doi: 10.1161/01.RES.0000118599.25944.D5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.