Abstract
In Arabidopsis thaliana, the SOS1 (Salt Overly Sensitive 1) locus is essential for Na+ and K+ homeostasis, and sos1 mutations render plants more sensitive to growth inhibition by high Na+ and low K+ environments. SOS1 is cloned and predicted to encode a 127-kDa protein with 12 transmembrane domains in the N-terminal part and a long hydrophilic cytoplasmic tail in the C-terminal part. The transmembrane region of SOS1 has significant sequence similarities to plasma membrane Na+/H+ antiporters from bacteria and fungi. Sequence analysis of various sos1 mutant alleles reveals several residues and regions in the transmembrane as well as the tail parts that are critical for SOS1 function in plant salt tolerance. SOS1 gene expression in plants is up-regulated in response to NaCl stress. This up-regulation is abated in sos3 or sos2 mutant plants, suggesting that it is controlled by the SOS3/SOS2 regulatory pathway.
Soil salinity is a major abiotic stress for plant agriculture. Sodium ions in saline soils are toxic to plants because of their adverse effects on K+ nutrition, cytosolic enzyme activities, photosynthesis, and metabolism (1, 2). Three mechanisms function cooperatively to prevent the accumulation of Na+ in the cytoplasm, i.e., restriction of Na+ influx, active Na+ efflux, and compartmentalization of Na+ in the vacuole (1). The wheat high-affinity K+ transporter HKT1 functions as a Na+-K+ cotransporter, which confers low-affinity Na+ uptake at toxic Na+ concentrations (3). Thus, HKT1 could represent one of the Na+ uptake pathways in plant roots. The low-affinity cation transporter LCT1 from wheat may also mediate Na+ influx into plant cells (4). In addition, patch-clamp studies have shown that nonselective cation channels play important roles in mediating Na+ entry into plants (5). The Arabidopsis thaliana AtNHX1 gene encodes a tonoplast Na+/H+ antiporter and functions in compartmentalizing Na+ into the vacuole (6). Overexpression of AtNHX1 enhances the salt tolerance of Arabidopsis plants (7).
No Na+ efflux transporter has been cloned from plants. Plants do not seem to have a Na+-ATPase at the plasma membrane (1). It is expected that proton motive force created by H+-ATPases would drive Na+ efflux from plant cells through plasma membrane Na+/H+ antiporters (8). Fungal cells contain both Na+-ATPases and Na+/H+ antiporters at the plasma membrane. In the yeast Saccharomyces cerevisiae, plasma membrane Na+-ATPases play a predominant role in Na+ efflux and salt tolerance (9). In contrast, Na+/H+ antiporters are more important for Na+ efflux and salt tolerance in the fungus Schizosaccharomyces pombe (10).
Recently, several Arabidopsis sos (for salt overly sensitive) mutants defective in salt tolerance were characterized (11–13). The sos mutants are specifically hypersensitive to high external Na+ or Li+ and also unable to grow under very low external K+ concentrations (13). Allelic tests indicated that the sos mutants define three SOS loci, i.e., SOS1, SOS2, and SOS3 (13). The SOS3 gene encodes an EF-hand type calcium-binding protein with similarities to animal neuronal calcium sensors and the yeast calcineurin B subunit (14). In yeast, calcineurin plays a central role in the regulation of Na+ and K+ transport. Mutations in calcineurin B lead to increased sensitivity of yeast cells to growth inhibition by Na+ and Li+ stresses (15). The SOS2 gene was cloned recently and shown to encode a serine/threonine type protein kinase (16). Interestingly, SOS2 physically interacts with and is activated by SOS3 (17). Therefore, SOS2 and SOS3 define a previously uncharacterized regulatory pathway for Na+ and K+ homeostasis and salt tolerance in plants. The SOS3/SOS2 pathway has been predicted to control the expression and/or activity of ion transporters (17).
Among the three SOS loci, SOS1 plays the greatest role in plant salt tolerance. Compared with sos2 and sos3 mutant plants, sos1 mutant plants are even more sensitive to Na+ and Li+ stresses (13). Double-mutant analysis indicated that SOS1 functions in the same pathway as SOS2 and SOS3 (12, 13). Thus, SOS1 may be a target for regulation by the SOS3/SOS2 pathway.
We have now isolated the SOS1 locus through positional cloning. It is predicted to encode a transmembrane protein with similarities to plasma membrane Na+/H+ antiporters from bacteria and fungi. The results suggest that a plasma membrane-type Na+/H+ antiporter is essential for plant salt tolerance. The steady-state level of SOS1 transcript is up-regulated by NaCl stress. The sos2 mutation abolishes SOS1 up-regulation in the shoot. In the sos3 mutant, no SOS1 up-regulation is found in the shoot or root. Therefore, SOS1 gene expression under NaCl stress is controlled by the SOS3/SOS2 regulatory pathway.
Materials and Methods
Genetic Mapping.
sos1 mutant plants in the Columbia background were crossed to wild-type plants of the Landsberg ecotype. sos mutants were selected from the segregating F2 population by the root-bending assay (11). Genomic DNA from 1,663 individual mutant F2 plants was analyzed for cosegregation with simple sequence length polymorphism (SSLP) markers. For the fine mapping of SOS1, seven SSLP markers were developed based on genomic sequences of the bacterial artificial chromosome (BAC) clones at the top of chromosome 2. The primer pairs for the SSLP markers that are polymorphic between Columbia and Landsberg are as follows: T20F6-1-F, 5′-GGATGATGATCGATTCGGAT-3′; T20F6-1-R, 5′-ATCTGACTCATAGGATATCG-3′; nga1145-F, 5′-CCTTCACATCCAAAACCCAC-3′; nga1145-R, 5′-GCACATACCCACAACCAGAA-3′; F5O4-3-F, 5′-GAATGTTTTGAAGGATATCTCAG-3′; F5O4-3-R, 5′-GAAAAATGGAGCACGAAATAAGC-3′; F14H20-3-F, 5′-CCCGAGATTAATACACAATC-3′; F14H20-3-R, 5′-GCAGATTATGTAATTGTGACC-3′; T23K3-1-F, 5′-TCGTGTTTACCGGGTCGGAT-3′; T23K3-1-R, 5′-TGATGAGAATCTTAGCGAGC-3′; CCC-1-F, 5′-TGGTAAGACCAAATTACACTC-3′; CCC-1-R, 5′-CGTAATTAAAATGTGTTAAACCG-3′; F10A8-1-F, 5′-AACCGCATAGTACAATGCAG-3′; F10A8-1-R, 5′-CGGTAAAGATCAACTAATAACG-3′; F23H14-3-F, 5′-AACGGAAACGGCAACTAGAC-3′; and F23H14-3-R, 5′-ACCCTAAATGTTTCGATTCG-3′.
DNA Sequencing.
To determine the nucleotide sequence of SOS1 gene in sos1 mutant alleles, synthetic oligonucleotide primers were made that would enable sequencing of the entire gene. Overlapping fragments encompassing the entire SOS1 gene were PCR amplified by using these primers. The amplified products were sequenced on both strands. To avoid errors caused by PCR, three independent PCR samples were mixed and batch sequenced.
Isolation of cDNA.
cDNA containing the complete SOS1 ORF was obtained by reverse transcription–PCR amplification. RNA from salt-treated Columbia wild-type plants was used as template for the reverse transcription–PCR. Three overlapping cDNA fragments obtained from reverse transcription–PCR were mixed as the template to amplify a full-length cDNA, which was then cloned into pCR-Blunt II-TOPO Vector (Invitrogen).
Plant Transformation and Complementation Test.
SOS1 cDNA containing the entire ORF was cloned into the XbaI and SacI sites of pBI121. The construct was introduced into the Agrobacterium GV3101 strain, and the resulting bacteria were used to transform sos1-1 mutant plants by vacuum infiltration (18). Kanamycin-resistant T2 transgenic plants were selected and subjected to complementation tests on Murashige and Skoog (MS) agar medium supplemented with 100 mM NaCl.
RNA Analysis.
Arabidopsis seedlings were grown on MS agar medium under continuous light (11), and 10-day-old seedlings were used for different treatments. For salt treatment, the seedlings were transferred onto Whatman filter paper soaked with 300 mM NaCl and treated for 5 h. For abscisic acid (ABA) treatment, the seedlings were sprayed with 100 μM ABA and kept for 3 h. For cold treatment, the seedlings on MS agar medium were incubated at 0°C for 24 h. To determine gene expression in root and shoot separately, seedlings were grown on an agar surface in vertical plates for 10 days and treated with NaCl by immersing the roots in MS nutrient solution supplemented with 200 mM NaCl for 6 h. RNA extraction and Northern analysis were carried out as described (13).
Results
Positional Cloning of SOS1.
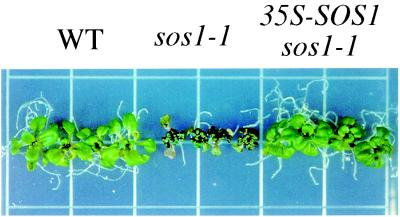
By examining several PCR-based molecular markers, we found that the SSLP marker nga1145 near the top of chromosome 2 is closely linked to the sos1 mutation. Seven previously unidentified SSLP markers were then developed based on the genomic sequence of BAC clones at the top of chromosome 2. Fine mapping with these markers delimited SOS1 to about a 70-kilobase region between the molecular markers T23K3-1 and F14H20-3 (Fig. 1A). Candidate genes in this region were amplified from sos1 mutants and sequenced. The sequence analysis revealed that a putative gene, F14H20.5, contains a 2-bp deletion in the sos1-13 mutant allele generated by fast neutron bombardment. Further analyses showed that all sos1 alleles contain mutations in this putative gene and that each mutation causes a change in the amino acid sequence (Table 1). Furthermore, expression of this candidate gene under control of the cauliflower mosaic virus 35S promoter complemented the salt-hypersensitive phenotype of sos1-1 mutant plants (Fig. 2). When sos1-1 mutant seedlings were treated with 100 mM NaCl, their growth was arrested. In these mutant plants, older leaves became chlorotic, and young leaves became dark in color. In contrast, sos1-1 mutant plants containing the 35S-SOS1 transgene could grow and remained green under 100 mM NaCl treatment, as did the wild-type plants. Based on these results, we conclude that this putative gene is SOS1.
Figure 1.
Positional cloning of the SOS1 gene. (A) Genetic and physical mapping of SOS1. All of the SSLP markers shown except nga1145 were developed in this study based on sequence information of the BACs. The BAC contig was assembled based on information available at http://www.Arabidopsis.org/cgi-bin/maps. cM, centimorgan. (B) Structure of the SOS1 gene. Positions are relative to the initiation codon. Filled boxes indicate the ORF, and lines between boxes indicate introns.
Table 1.
Molecular basis of sos1 mutations
| Mutant line | Allele | Mutagen | Nucleotide change | Protein change |
|---|---|---|---|---|
| ssr1, Icss1-3, Icss1-18 | sos1-1 | EMS | Δ14 bp, 1,330–1,343 | Frameshift |
| ss1-6, ss3-13 | sos1-2 | EMS | C5,410T | Stop |
| ss1-16, Icss1-24 | sos1-3 | EMS | C2,520T | Arg-365-Cys |
| IIcss1-13, IIcss1-22 | sos1-4 | EMS | G2,480A | Stop |
| Icss1-10 | sos1-5 | EMS | G2,766A | Splicing junction |
| Icss1-25 | sos1-6 | EMS | G3,652A | Stop |
| IIcss1-59, css1-61 | sos1-7 | EMS | Δ1 bp, 4,539 | Frameshift |
| Icss2-21 | sos1-8 | EMS | G4,594A | Gly-777-Glu |
| Icss2-7 | sos1-9 | EMS | G4,615A | Gly-784-Asp |
| tss2-1, p2901-3503 2-1 | sos1-10 | T-DNA | Δ63 bp, 2,792–2,854 | Splicing junction |
| P800 1-2, p800 1-3 | sos1-11 | T-DNA | Δ7 bp, 5,953–5,959 | Frameshift |
| FN50css2-3, FN50css3-22, FN75css1-24, FN75css1-14, FN75css3-18 | sos1-12 | Fast neutron | G668A | Gly-136-Glu |
| FN50css2-9, FN75css1-22, FN75css1-23 | sos1-13 | Fast neutron | Δ2 bp, 5,149–5,150 | Frameshift |
| FN50css1-8, FN50css3-3, FN75css1-17, B46, B47 | Fast neutron | Whole gene deletions | Whole gene deletions |
EMS, ethylmethane sulfonate.
Figure 2.
Complementation of sos1 by 35S-SOS1: 7-day-old seedlings grown on MS agar medium were transferred to MS medium supplemented with 100 mM NaCl. The picture was taken after 10 days of treatment on the NaCl medium. (Left) Wild-type plants (WT). (Center) sos1-1 mutant plants. (Right) Transgenic sos1-1 plants containing the wild-type SOS1 gene under control of the cauliflower mosaic virus 35S promoter. These plants did not show any difference when grown on MS medium without supplementation of NaCl (not shown).
SOS1 Encodes a Putative Na+/H+ Antiporter.
The SOS1 ORF was determined by sequencing several overlapping cDNAs obtained from young Arabidopsis seedlings by reverse transcription–PCR. Comparison with the genomic sequence revealed that SOS1 has 22 introns and 23 exons (Fig. 1B). SOS1 is predicted to encode a polypeptide of 1,146 amino acid residues (Fig. 3A) with a theoretical molecular mass of 127 kDa. Hydrophobicity plot analysis showed that the N-terminal portion of SOS1 is highly hydrophobic and has 12 predicted transmembrane domains (Fig. 3B). Database searches revealed substantial similarities between the transmembrane region of SOS1 and Na+/H+ antiporters of animal or microbial origins (Fig. 4A). Over a stretch of 342 amino acid residues (113–443), SOS1 has 26% identity and 45% similarity with NHE1 from Chinese hamster (19). The highest sequence similarities for SOS1 are with the “eukaryotic” type Na+/H+ antiporters from bacteria, of which only NhaP from P. aeruginosa has been characterized functionally (20). SOS1 exhibits 31% identity and 48% similarity with the NhaP sequence over a stretch of 289 amino acids (131–408 in SOS1). The C-terminal portion of SOS1 is hydrophilic and predicted to reside in the cytoplasm (Fig. 5). The long hydrophilic C-terminal tail makes SOS1 the largest Na+/H+ antiporter sequence known to date. No similarities were found between the SOS1 tail region and other amino acid sequences in the GenBank database.
Figure 3.
SOS1 is predicted to encode a transmembrane protein. (A) The deduced amino acid sequence of SOS1. The 12 putative transmembrane domains (TM) are underlined. (B) Hydrophobicity plot of SOS1. The hydrophobicity values were calculated by the program tmpred available at http://www.ch.embnet.org/software/TMPRED_form.html.
Figure 4.
SOS1 is similar to Na+/H+ antiporters. (A) Alignment of SOS1 (accession number AF256224) with Na+/H+ antiporters NHE1 from Chinese hamster (P48761) and NhaP from Pseudomonas aeruginosa (BAA31695). The sequences were aligned by the program clustalw (http://dot.imgen.bcm.tmc.edu:9331/multialign/Options/clustalw.html). Amino acids identical in at least two proteins are highlighted in black, and conservative substitutions are highlighted in gray. Asterisks indicate conserved residues that were substituted in sos1 mutant alleles. (B) Phylogenetic analysis of SOS1 and other representative Na+/H+ antiporters. Multiple sequence alignment was performed with clustalw. The alignment is based on the N-terminal 450 amino acids of SOS1. Evolutionary distances were calculated by the neighbor joining method, and the phylogenetic tree was drawn by the program drawgram (http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html). The accession numbers and sources of each of the other representative Na+/H+ antiporters are as follows: NHE1 (P19634), Homo sapiens; NHE2 (AAD41635), H. sapiens; NHE3 (P48764), H. sapiens; NHE4 (P26434), Rattus norvegicus; NHE5 (AAC98696.1), H. sapiens; NHE6 (NP_006350), H. sapiens; NHA1 (NP_013239), S. cerevisiae; NHX1 (NP_010744), S. cerevisiae; AtNHX1 (AAD16946.1), A. thaliana; SOD2 (CAA77796.1), S. pombe; NhaA (P13738), Escherichia coli; and NhaP (BAA31695.1), P. aeruginosa.
Figure 5.
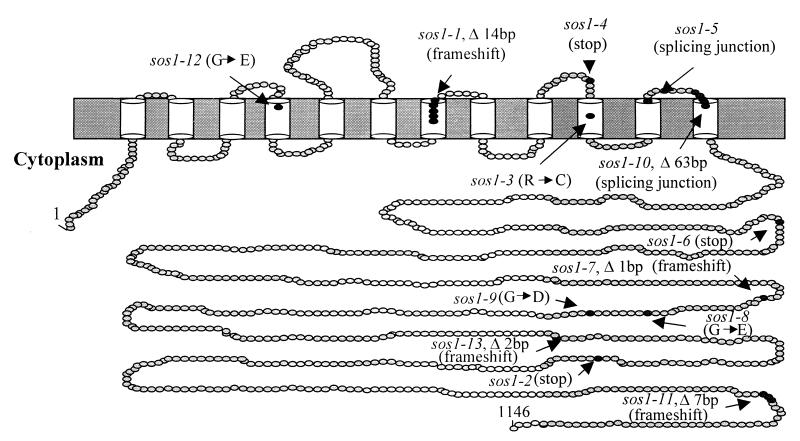
Diagramatic representation of SOS1 structure. The diagram was drawn based on the prediction of hydrophobicity profile of SOS1. Putative transmembrane helices are shown as cylinders. The positions of mutations in sos1 alleles are indicated.
Phylogenetic analysis showed that SOS1 clusters with plasma membrane Na+/H+ antiporters such as SOD2, NHA1, NhaA, and NhaP (Fig. 4B). SOD2 and NHA1 function on the plasma membrane of S. pombe and S. cerevisiae, respectively, to export Na+ from cytosol to the extracellular space (21–23). NhaA and NhaP are Na+/H+ antiporters that function in Na+ efflux in E. coli and P. aeruginosa, respectively (20, 24). SOS1 is more distantly related to a cluster of organellar Na+/H+ antiporters such as AtNHX1, NHX1, or NHE6 (Fig. 4B). AtNHX1 functions on the tonoplast to compartmentalize Na+ into the vacuole of Arabidopsis cells (6, 7). NHX1 plays a role in transporting Na+ to the yeast prevacuolar compartment (25, 26). The animal Na+/H+ antiporter NHE6 has been reported to have a mitochondrial localization (27). SOS1 does not cluster with plasma membrane Na+/H+ antiporters from animals, which function in mediating Na+ influx (28). These results suggest that SOS1 is distinct from vacuolar Na+/H+ antiporters and may function at the plant-cell plasma membrane to mediate Na+ efflux.
Analysis of sos1 Mutant Alleles Reveals Several Residues and Regions Essential for SOS1 Function.
The SOS1 gene was amplified from 32 independent sos1 mutant lines (13) and sequenced to determine the molecular basis of each mutation. Several mutant lines were found to harbor identical mutations (Table 1). Five of the fast neutron alleles result in relatively large deletions and were not assigned specific allele designations, because the boundaries of the deletions are not known. Analysis of the various sos1 mutations reveals several amino acid residues and regions essential for SOS1 function. The sos1-3 and sos1-12 alleles contain single amino acid substitutions in the membrane-spanning region (Fig. 5). Both mutations affect residues that are conserved in all antiporters (Fig. 4A) and presumably abolish SOS1 antiporter activity. Two other single amino acid substitution mutations (i.e., sos1-8 and sos1-9) are found in the hydrophilic tail region (Fig. 5). The sos1-10 allele was obtained from T-DNA mutagenesis and contains a 7-bp deletion that causes a frameshift that truncates the last 40 amino acids from the C terminus of SOS1 (Fig. 5). Similarly, sos1-2 and sos1-6 mutations truncate the cytoplasmic tail of SOS1 (Fig. 5). These and other mutations that do not affect the transmembrane region reveal an essential role of the tail region for SOS1 function. Like the hydrophilic tail of animal NHE1 antiporters (29), the tail of SOS1 may interact with various regulators of antiporter activity. As such, these mutations likely disrupt interaction between SOS1 and its regulators.
SOS1 Expression Is Up-Regulated Specifically by Salt Stress.
To examine the expression of SOS1 gene under stress, RNA gel blot analysis was performed. SOS1 mRNA was detected without stress treatment but was up-regulated significantly by salt stress (Fig. 6A). Consistent with its specific role in Na+ tolerance, SOS1 gene expression was not up-regulated by cold stress or ABA (Fig. 6A). In comparison, the RD29A gene was induced by ABA, cold, as well as salt stresses. SOS1 mRNA was more abundant in roots than in shoots. In both roots and shoots, SOS1 expression was up-regulated by NaCl stress (Fig. 6B).
Figure 6.
SOS1 expression is up-regulated by NaCl stress and is under the control of the SOS3/SOS2 regulatory pathway. (A) SOS1 expression is specifically up-regulated by NaCl stress in wild-type Arabidopsis seedlings. (B) Up-regulation of SOS1 expression in roots and shoots of wild-type plants. (C) SOS1 expression in sos2-1 mutant seedlings. (D) SOS1 expression in sos3-1 mutant seedlings. The same RNA blots were hybridized successively with SOS1, RD29A, and actin cDNA probes. Actin was used as loading control, and RD29A was used as control for the stress treatments. C, unstressed control.
SOS1 Up-Regulation Is Controlled by the SOS3/SOS2 Pathway.
To determine whether NaCl up-regulation of SOS1 is under control of the SOS3/SOS2 regulatory pathway, SOS1 expression in sos2-1 and sos3-1 mutant plants was analyzed. In the sos2 mutant, SOS1 was up-regulated by NaCl stress in the root but not in the shoot (Fig. 6C). In sos3 plants, no SOS1 up-regulation was seen in either the root or shoot (Fig. 6D). These results show that SOS1 expression is regulated at least in part by the SOS3/SOS2 pathway.
Discussion
Plant salt tolerance is a complex trait that is still not well understood. Very few genes have been shown to be required for plant salt tolerance. SOS1 is a genetic locus that was previously identified as essential for plant salt tolerance (11). Mutations in SOS1 render Arabidopsis plants extremely sensitive to high Na+ or low K+ environment (11, 13). To understand how the SOS1 gene functions in salt tolerance, it is necessary to clone this gene. Even though several sos1 mutant lines were recovered from a T-DNA insertion population, the T-DNA did not cosegregate with the sos1 mutant phenotype (13). Therefore, a map-based strategy had to be used to clone the SOS1 gene. Fine genetic mapping narrowed the search of SOS1 to a very short region of chromosome 2. The fine mapping of SOS1 was made possible by several molecular markers that we have developed and by the large number of recombinant chromosomes examined. Several candidate genes in the region where SOS1 is mapped were sequenced to identify the sos1 mutation. One of the candidate genes was found to contain a mutation in every sos1 mutant allele. Further confirmation that this candidate is indeed SOS1 came from a genetic complementation test.
The SOS1 protein is predicted to have 12 transmembrane domains in its N-terminal part. Throughout this transmembrane region, SOS1 shows substantial sequence similarities with Na+/H+ antiporters from microbes and animals. The sequence similarities combined with the Na+ hypersensitive phenotype of sos1 mutant plants strongly indicate that SOS1 is a Na+/H+ antiporter. Phylogenetic analysis showed that SOS1 is more closely related to plasma membrane Na+/H+ antiporters from microorganisms than to the vacuolar antiporters from either plants or fungi. This finding suggests that SOS1 is probably a plasma membrane Na+/H+ antiporter in Arabidopsis. As such, SOS1 is expected to function in exporting Na+ from the cytosol to the extracellular space to prevent rapid accumulation of Na+ in the cytoplasm.
SOS1 is predicted to have a cytoplasmic tail approximately 700 amino acids in length. Our sequence analysis of the multitude of sos1 mutant alleles revealed that both the tail and transmembrane regions of SOS1 are necessary for its function in plant salt tolerance. The sos1-3, sos1-8, sos1-9, and sos1-12 mutations each cause a single amino acid substitution in the SOS1 protein. Two of these substitutions occur in the transmembrane region, and the other two occur in the tail. These four residues are clearly critical for SOS1 function. Why these particular residues are important for SOS1 function awaits future investigation. In any case, our data on the sos1 mutant lesions provide a wealth of information that will be invaluable for detailed structure-function analysis in the future.
SOS1 gene expression is up-regulated by NaCl stress. This up-regulation is consistent with the role of SOS1 in Na+ tolerance. It has been known that NaCl stress also up-regulates the expression of genes encoding plasma membrane H+-ATPases (30). Increased H+-ATPase expression would provide a greater proton motive force that is necessary for elevated Na+/H+ antiporter activity.
The SOS3 calcium sensor physically interacts with the SOS2 protein kinase (17). In the presence of calcium, SOS3 activates SOS2 kinase activity. The SOS3–SOS2 kinase complex represents a regulatory pathway that specifically controls Na+ and K+ homeostasis and plant salt tolerance. Results presented in this paper suggest that one output of this pathway is the up-regulation of SOS1 expression under NaCl stress. The sos3 mutation abolishes SOS1 up-regulation in both the root and shoot. In the sos2 mutant, SOS1 up-regulation in the shoot but not in root was disrupted. The fact that SOS1 expression is still up-regulated in the root of sos2 mutant indicates that there may be a functionally redundant root-specific SOS2-like kinase or kinases. The regulation of SOS1 gene expression by the SOS2/SOS3 pathway is consistent with previous genetic evidence suggesting that SOS1 functions in the same pathway as SOS2 and SOS3 (12, 13).
SOS1 is essential for the homeostasis of both Na+ and K+. Under NaCl stress, sos1 mutant plants accumulate less Na+ as well as less K+ (11, 31). SOS1 gene expression is concentrated in cells surrounding the xylem, suggesting that SOS1 may function in loading Na+ into the xylem for long-distance transport (unpublished data). A xylem-loading function of SOS1 would be consistent with sos1 mutant plants accumulating less Na+. Preferential expression of SOS1 at the symplast/xylem boundary would also help explain the K+ transport defect of sos1 mutant plants. It is well known that K+ and Na+ transport is closely linked at the xylem/symplast interface (32). The effect of SOS1 on K+ transport might be through its effect on H+ gradient across the cell membrane of stellar cells. For example, a K+-H+ symporter activity could be coupled with SOS1 via H+ cycling, and such a symporter may be required for high affinity K+ transport into the xylem. It is also possible that a K+/Na+ symporter is coupled with SOS1 via Na+ cycling.
Acknowledgments
We sincerely thank B. Stevenson and J. Liu for technical assistance and Ray A. Bressan, Robert T. Leonard, and Hans Bohnert for insightful discussions and suggestions. This work was supported by National Institutes of Health Grant R01GM59138.
Abbreviations
- BAC
bacterial artificial chromosome
- SSLP
simple sequence length polymorphism
- MS
Murashige and Skoog
- ABA
abscisic acid
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF256224).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120170197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120170197
References
- 1.Niu X, Bressan R A, Hasegawa P M, Pardo J M. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacoby B. In: Handbook of Plant and Crop Stress. Pessarakli M, editor. New York: Dekker; 1999. pp. 97–123. [Google Scholar]
- 3.Rubio F, Gassman W, Schroeder J I. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- 4.Schachtman D P, Kumar R, Schroeder J I, Marsh E L. Proc Natl Acad Sci USA. 1997;94:11079–11084. doi: 10.1073/pnas.94.20.11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amtmann A, Sanders D. Adv Bot Res. 1998;29:76–112. [Google Scholar]
- 6.Gaxiola R A, Rao R, Sherman A, Grisafi P, Alper S L, Fink G R. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apse M P, Aharon G S, Snedden W A, Blumwald E. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 8.Schachtman D P, Liu W. Trends Plant Sci. 1999;4:281–287. doi: 10.1016/s1360-1385(99)01428-4. [DOI] [PubMed] [Google Scholar]
- 9.Haro R, Garciadeblas B, Rodriguez-Navarro A. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 10.Jia Z, McCullough N, Martel R, Hemmingsen S, Young P G. EMBO J. 1992;11:1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S-J, Lei D, Zhu J-K. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Zhu J-K. Proc Natl Acad Sci USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J-K, Liu J, Xiong L. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo J M. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- 16.Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3703–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bechtold N, Ellis J, Pelletier G. C R Acad Sci (Paris) 1993;316:1194–1199. [Google Scholar]
- 19.Counillon L, Pouyseegur J. Biochim Biophys Acta. 1993;1172:343–345. doi: 10.1016/0167-4781(93)90228-6. [DOI] [PubMed] [Google Scholar]
- 20.Utsugi J, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Biochim Biophys Acta. 1998;1398:330–334. doi: 10.1016/s0167-4781(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 21.Hahnenberger K M, Jia Z, Young P G. Proc Natl Acad Sci USA. 1996;93:5031–5036. doi: 10.1073/pnas.93.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dibrov P, Smith J J, Young P G, Fliegel L. FEBS Lett. 1997;405:119–124. doi: 10.1016/s0014-5793(97)00169-5. [DOI] [PubMed] [Google Scholar]
- 23.Prior C, Potier S, Souciet J L, Sychrova H. FEBS Lett. 1996;387:89–93. doi: 10.1016/0014-5793(96)00470-x. [DOI] [PubMed] [Google Scholar]
- 24.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. J Biol Chem. 1989;264:20297–20302. [PubMed] [Google Scholar]
- 25.Nass R, Cunningham K W, Rao R. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 26.Nass R, Rao R. J Biol Chem. 1998;273:21054–21060. doi: 10.1074/jbc.273.33.21054. [DOI] [PubMed] [Google Scholar]
- 27.Numata M, Petrecca K, Lake N, Orlowski J. J Biol Chem. 1998;273:6951–6959. doi: 10.1074/jbc.273.12.6951. [DOI] [PubMed] [Google Scholar]
- 28.Orlowski J, Grinstein S. J Biol Chem. 1997;272:22373–22376. doi: 10.1074/jbc.272.36.22373. [DOI] [PubMed] [Google Scholar]
- 29.Silva N L, Haworth R S, Singh D, Fliegel L. Biochemistry. 1995;34:10412–10420. doi: 10.1021/bi00033a013. [DOI] [PubMed] [Google Scholar]
- 30.Niu X, Narasimhan M L, Salzman R A, Bressan R A, Hasegawa P M. Plant Physiol. 1993;103:713–718. doi: 10.1104/pp.103.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding L, Zhu J-K. Plant Physiol. 1997;113:795–799. doi: 10.1104/pp.113.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacan D, Durand M. Plant Physiol. 1996;110:705–711. doi: 10.1104/pp.110.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]