Abstract
1-O-β-acyl acetals serve as activated donors in group transfer reactions involved in plant natural product biosynthesis and hormone metabolism. However, the acyltransferases that mediate transacylation from 1-O-β-acyl acetals have not been identified. We report the identification of a cDNA encoding a 1-O-β-acylglucose-dependent acyltransferase functioning in glucose polyester biosynthesis by Lycopersicon pennellii. The acyltransferase cDNA encodes a serine carboxypeptidase-like protein, with a conserved Ser-His-Asp catalytic triad. Expression of the acyltransferase cDNA in Saccharomyces cerevisiae conferred the ability to disproportionate 1-O-β-acylglucose to diacylglucose. The disproportionation reaction is regiospecific, catalyzing the conversion of two equivalents of 1-O-β-acylglucose to 1,2-di-O-acylglucose and glucose. Diisopropyl fluorophosphate, a transition-state analog inhibitor of serine carboxypeptidases, inhibited acyltransferase activity and covalently labeled the purified acyltransferase, suggesting the involvement of an active serine in the mechanism of the transacylation. The acyltransferase exhibits no carboxypeptidase activity; conversely, the serine carboxypeptidases we have tested show no ability to transacylate using 1-O-acyl-β-glucoses. This acyltransferase may represent one member of a broader class of enzymes recruited from proteases that have adapted a common catalytic mechanism of catabolism and modified it to accommodate a wide range of group transfer reactions used in biosynthetic reactions of secondary metabolism. The abundance of serine carboxypeptidase-like proteins in plants suggests that this motif has been used widely for metabolic functions.
Activation and transacylation of many fatty acids and phenolic acids are known to proceed by means of a thioester-dependent mechanisms. However, an alternative mechanism of activation involving the UDPglucose-dependent transglucosylation of phenolic and fatty acids as 1-O-β-acyl acetals, a species exhibiting group transfer potentials (−35.7 kJ⋅mol−1) (1) similar to that of thioesters, is now recognized. Although the role of activated acetals such as caffeoyl β-glucose in transacylation reactions of quinic acid was first proposed by Kojima and Uritani (2) for chlorogenic acid biosynthesis, it is now known that other 1-O-β-acyl acetals are also used in a wide range of transacylation reactions. In Brassicaceae, 1-O-sinapoyl-β-glucose serves as an acyl donor for the formation of sinapoylcholine, sinapoylmalate, and disinapoylglucose (1, 3, 4). 1-O-galloyl-β-glucose (β-glucogallin) acts as a donor in biosynthesis of gallotannins (5). Hydroxycinnamoylglucose and its hydroxylated derivative caffeoyl glucose are intermediates for the formation of chlorogenic acid (3-O-caffeoylquinic acid) in Ipomoea (1, 6, 7) in an alternative route to the caffeoyl-CoA-dependent biosynthesis of this compound (8). Chlorogenic acid, itself a high-energy acyl donor similar to 1-O-acyl-β-glucose, serves as the caffeoyl donor for the transacylation of isochlorogenic acid (3,5-di-O-caffeoylquinic acid), caffeoylglucaric acid, and caffeoylgalactonic acid (9–11). For the synthesis of a number of feruloylated betacyanins, 1-O-feruloyl-β-glucose is the feruloyl donor (12). In the conjugation of the plant growth hormone indoleacetic acid, a UDPglucose-dependent reaction activates indoleacetic acid to form 1-O-indolylacetyl-β-glucose. Indolylacetyl-myo-inositol is synthesized by transfer of the indolylacetyl moiety from indolylacetylglucose to inositol (13–15). It is evident that 1-O-acylglucoses and similar high-energy molecules occupy a prominent position in the secondary metabolism of higher plants. Despite the importance of indoleacetic acid and phenolic metabolism in plant biochemistry, the mechanistic basis of these acyl transfer reactions has largely gone unexplored.
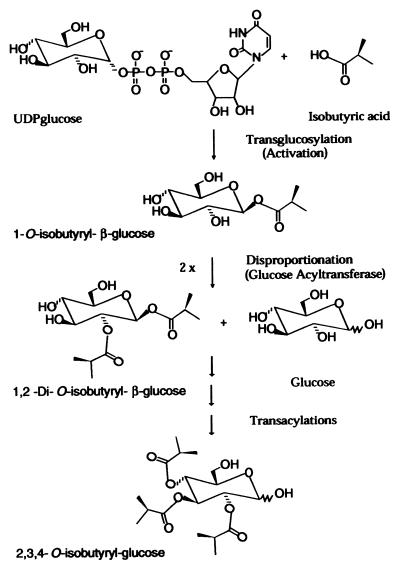
Previously we identified a UDPglucose-dependent mechanism of fatty acid activation and 1-O-acyl-β-glucose-mediated transacylations in the synthesis of glucose polyesters (16–19). Glucose polyesters, which are secreted in glandular trichome exudates of the wild tomato species Lycopersicon pennellii contribute to insect resistance (20–26). In L. pennellii glucose polyesters (2,3,4-tri-O-acylglucose), the fatty acid substituents range in length from 4:0 to 12:0. Fatty acid activation in acylglucose biosynthesis is catalyzed by chain length-specific UDPglucose:fatty acid glucosyltransferases to form high-energy 1-O-acyl-β-glucoses, which serve as acyl donors for the synthesis of glucose polyesters (16, 17). Enzymatic disproportionation of two equivalents of 1-O-acyl-β-glucose results in the formation of 1,2-diacyl-β-glucoses (Fig. 1). This reaction is catalyzed by a glucose acyltransferase that was recently purified to homogeneity (19). In this paper, we report the cloning and heterologous expression of this glucose acyltransferase and its relation to the serine carboxypeptidase gene family, and we suggest a mechanism for the evolution of acyl transfer by activated 1-O-β-acyl acetals.
Figure 1.
Acylglucose biosynthetic pathway. Fatty acids are activated by means of UDPglucose-dependent transglucosylation to form the high-energy acyl donor molecule, 1-O-acyl-β-glucose. Two equivalents of the activated 1-O-acyl-β-glucose intermediate then undergo enzyme-catalyzed disproportionation to form 1,2-substituted diacylglucose and glucose. In this figure, isobutyrate represents the acyl group; however, branched and straight short (C4–C5) to medium (C10–C12) chain fatty acids are used in acylglucose biosynthesis and are activated by chain length-specific UDPglucose:fatty acid transglucosylases.
Materials and Methods
Cloning of the Acyltransferase cDNA.
Reverse transcription–PCR (RT-PCR) was used to clone an internal fragment of the cDNA. RNA was extracted from L. pennellii as reported (27). Poly(A)+ mRNA was obtained by using PolyATtract mRNA isolation kit (Promega). Oligo(dT)15 was used for the first-strand cDNA synthesis. Superscript II RNase H− reverse transcriptase from GIBCO/BRL was used; reaction conditions were as described by the manufacturer. Primers were designed according to the peptide sequences from the purified 34-kDa subunit of the acyltransferase (19). Two primers, pt50 and re24, were selected based on their low level of degeneracy, and tomato codon usage tables (ftp://ftp.bio.indiana.edu) were used to further decrease the degeneracy of the primers. The sequences of the primers are as follows:
pt50: GA(A/G)CA(C/T)TT(C/T)AT(C/T)GT(T/G)GA(A/G)AC and
re24: A(A/G)ICCCAT(A/G)TG(A/G)TT(A/T)GC(A/G)TA.
PCR conditions were as reported (28). The major band from the PCR was purified from the agarose gel by using a Bio 101 Geneclean kit and ligated to the PCRII plasmid vector (Invitrogen) by the AT cloning method. Clones were sequenced, and an acyltransferase clone, pAGTF, was identified by the presence of peptide sequences of the purified enzyme.
5′ Rapid amplification of the cDNA ends (RACE) and 3′RACE (29, 30) were used to obtain a full-length acyltransferase cDNA. Poly(A)+ mRNA was obtained as in RT-PCR. GIBCO/BRL 5′RACE and 3′RACE kits were used. All of the gene specific primers (GSP) used below were designed from pAGTF. Forward primers (FOR) and reverse primers (REV) are described below:
GSP REV1: ATCTGTTAGAGCATTGCCTT,
GSP REV2: GCCTGTCACCAACTTCAATA,
GSP FOR1: GTTGGAGAAGAGGAAAAAGT, and
GSP FOR2: TACAATCTGAGAGAGACCCA.
The PCR products were ligated to pAMP1 vector (GIBCO/BRL). The 5′RACE and 3′RACE procedures were as described by the manufacturer, except for the following modification: enrichment of templates for the gene of interest was added to the normal 3′RACE procedure because another gene in the cDNA pool with higher abundance possessed sequences similar to the adapter primer provided by the kit. Different clones from separate PCRs were used to confirm the sequence of the gene (because of low fidelity of Taq DNA polymerase).
Acyltransferase Expression in Saccharomyces cerevisiae.
5′- and 3′-end sequences obtained from the 5′RACE and 3′RACE were used to amplify a full-length cDNA of the glucose acyltransferase. BamHI and EcoRI restriction sites were added at the 5′- and 3′-ends, respectively.
5′ primer: AAGGATCCAATGGCGCGGGTCACACTGTT and
3′ primer: AGAATTCTTAACGTCAACCTCGAATGA.
Procedures were the same as described for RT-PCR. PCR products were purified and cut by BamHI and EcoRI and ligated to pYES2 (Invitrogen) prepared with the same restriction enzymes. Ligation was performed at 14°C overnight, and the products were used to transform XL-1 Blue cells. Clones were sequenced, and a clone with the acyltransferase cDNA, pYAGT2, was identified.
KT1115 (MATa leu2-3 leu2-112 ura-52) (31) was transformed with pYAGT2. KT1115 was a gift from Ralph Dewey (North Carolina State University, Raleigh, NC). Uracil dropout medium [0.13% (wt/vol) amino acids without uracil, 0.17% (wt/vol) yeast nitrogen base without amino acids and ammonium sulfate, 0.5% (wt/vol) (NH4)2SO4, and 2% (wt/vol) glucose] was used as selective medium. Baffled flasks were used to grow yeast cultures for expression. Transformed yeast cells were inoculated in uracil dropout medium with 5% (wt/vol) raffinose instead of 2% (wt/vol) glucose. After the culture density reached 0.4–0.5 OD600, galactose was added into the medium to a final concentration of 2% (wt/vol) to induce transcription of the pYES2 insert. Cells were sampled at different time points after galactose addition.
Functional expression of the acyltransferase was assayed by using permeabilized cells. Ten milliliters of induced culture was centrifuged, and the pellet was washed with 10 ml of ice-cold H2O, and then with 10 ml of ice-cold freeze-thaw buffer [FT buffer: 100 mM Hepes⋅NaOH, pH 7.5/20% (vol/vol) glycerol/0.1% (vol/vol) Triton X-100]. The pellet was resuspended in 50 μl of ice-cold FT buffer and frozen at −80°C overnight or longer (32). Samples were thawed at 30°C, and 50 μl of the cell suspension was removed for enzymatic assays and GC-MS analyses. Immunoblot analyses were performed as reported (33) with the following modification. Tris-buffered saline (50 mM Tris⋅HCl, pH 7.5/1 mM MgCl2/10 mM NaCl) was used for all incubations; and both primary and secondary antibodies were used at 1:1000 dilution. The primary antibody was generated against recombinant L. pennellii glucose acyltransferase expressed in Escherichia coli (unpublished work).
Disproportionation Activity Assay and Acyltransferase Characterization.
Disproportionation activity was detected by measuring the formation of diacylglucose from 1-O-[1-14C]isobutyryl-β-glucose. Reactions were performed in a 15-μl solution containing 50 mM Hepes⋅NaOH at pH 7.5, 1 mM 1-O-[1-14C]isobutyryl-β-glucose (105 cpm), and 5000-fold purified acyltransferase from L. pennellii LA1376 leaves (19) or variable amounts of commercially available serine carboxypeptidases. 1-O-isobutyryl-β-d-glucose and 1-O-[1-14C]isobutyryl-β-glucose were synthesized as previously described (17). The reaction was carried out at 37°C for 30 min. Five microliters of the reaction solution was loaded on a TLC plate and developed with chloroform/methanol/water (75:22:3). Autoradiography was then performed on the TLC plates (16, 17).
1H-NMR (500 MHz) analysis of the acyltransferase reaction was performed in a 1 mM solution of synthetic 1-O-isobutyryl-β-glucose (17) in 2H2O at 23°C. The reaction was initiated by the addition of 50 μg of L. pennellii glucose acyltransferase (19). A total of 120 transients per spectrum were collected for 1 h. This substrate, kindly provided by Gurdev Ghangas (Cornell University), contains small amounts of non-anomerically substituted isobutyrylglucose as an unavoidable consequence of nonenzymatic acyl migration during storage.
For GC-MS analysis of disproportionation products, the reaction was performed with 50 μl of permeabilized yeast cells and 25 μl of 12 mM 1-O-isobutyryl-β-glucose at room temperature for 3 h. After the reaction, the cells were pelleted at 12,000 × g for 1 min, the supernatant was removed, and 500 μl of MeOH was added to the supernatant. The sample was dried under N2 and acetylated with 60 μl of pyridine and 30 μl of acetic anhydride overnight. The acetylated sample was dried under N2 at 40°C, and resuspended in 200 μl of methylene chloride. One microliter was injected into a DB5 column (25 m × 0.22 mm) in a Hewlett–Packard 5890 gas chromatograph with an HP 5970 mass selective detector. The GC program was 80°C for 1 min, followed by an increase to 200°C at 12°C per min, to 260°C at 5°C per min, and was held at 260°C for another 15 min.
Inhibition of disproportionation by diisopropyl fluorophosphate (DFP) and benzylsuccinic acid was studied. DFP or d,l-benzylsuccinic acid was dissolved in isopropyl alcohol at various concentrations and preincubated with approximately 0.5 μg of 1200-fold purified acyltransferase (19) from L. pennellii LA1376 leaves at 37°C for 30 min. Routine disproportionation activity assay was then performed after preincubation with each inhibitor.
[14C]DFP was used to covalently label glucose acyltransferase. Ten micrograms of partially purified acyltransferase as used in the inhibition study was incubated with 0.65 mM [1,3-14C]DFP (10 μCi) (NEN) in 100 μl of H2O at 37°C for 30 min. Sodium deoxycholate (100 μl, pH 8.8) was then added to the mixture to a final concentration of 150 μg/ml. The sample was incubated on ice for 30 min. Two hundred microliters of 14% trichloroacetic acid was added to the sample and incubated for 30 min. The sample was centrifuged at 12,000 × g for 30 min, and the pellet was washed with 300 μl of 80% acetone. The pellet was resuspended by boiling in 25 μl of loading buffer and loaded onto an SDS/10% polyacrylamide gel for electrophoresis. Autoradiography was performed on the dried gel.
Results
cDNA Encoding Glucose Acyltransferase.
RT-PCR was used to amplify an internal fragment of the glucose acyltransferase cDNA (GenBank accession no. AF248647). Two peptide sequences derived from the 34-kDa subunit of the glucose acyltransferase were used as primers for RT-PCR to amplify a fragment of 632 bp possessing an ORF of 210 amino acids. Six of the seven peptides sequenced from the 34-kDa acyltransferase subunit (19) were present in the deduced amino acid sequence of this cDNA fragment.
The entire 1548-bp sequence of the cDNA was obtained by 5′RACE and 3′RACE (29, 30) using sequences from the 632-bp cDNA fragment. The sequence begins with an ATG codon and has a 1392-bp ORF, encoding a 464-aa polypeptide with a predicted molecular mass of 53 kDa. The deduced protein sequence contains all of the seven polypeptide sequences from the 34-kDa subunit (Fig. 2). Because all of the seven polypeptide sequences were derived from tryptic digest, the deduced amino acid before each peptide in the cDNA sequence should be either R or K. This is true for all peptides except PT50 (Fig. 2). Hence, PT50 represents the mature N terminus of the 34-kDa protein. Therefore, the deduced sequence indicates that the enzyme is synthesized as a preprotein bearing a hydrophobic N-terminal 18-aa signal peptide; cleavage of the signal peptide at this site is also predicted (http://www.cbs.dtu.dk/services/SignalP/). The N-terminal sequence of the 24-kDa subunit is also present in the deduced amino acid sequence of the cDNA. Thus, the 34-kDa and 24-kDa subunits of the acyltransferase are encoded by the same gene and result from posttranslational processing of the primary translation product. Four possible N-glycosylation sites (NXT/S) are present in the deduced amino acid sequence (three on the 34-kDa subunit, one on the 24-kDa subunit); glycosylation of the enzyme is indicated by its affinity for Con A during purification (19). The site for posttranslational proteolytic processing should be between PT60 and N24 (Fig. 2). Thus, the deduced molecular mass for the large subunit of the acyltransferase is between 30 kDa and 33 kDa. The value for the small subunit is 20 kDa. Glycosylation may contribute to the differences between the molecular masses calculated from the gene and the estimated molecular mass from the purified protein.
Figure 2.
Deduced amino acid sequence of the glucose acyltransferase (AT) and its alignment with barley serine carboxypeptidase I (BI). The numbers on the right are the residue numbers for the last amino acid on each line. Straight lines above the AT sequence indicate peptide sequences obtained from the 34-kDa and 24-kDa subunits. PT sequences are tryptic peptide sequences from the 34-kDa subunit. N24 is the N terminus of the 24-kDa subunit. ●, The R or K N-terminal to tryptic cleavage sites. Between AT and BI, colons are used to link the identical sequences, and dots are used to link the similar sequences. *, The active catalytic Ser-158, His-446, and Asp-393 sites in BI. @, The sites corresponding to those involved in the binding of substrate C-terminal carboxylate group in yeast serine carboxypeptidase.
Acyltransferase Expression in S. cerevisiae.
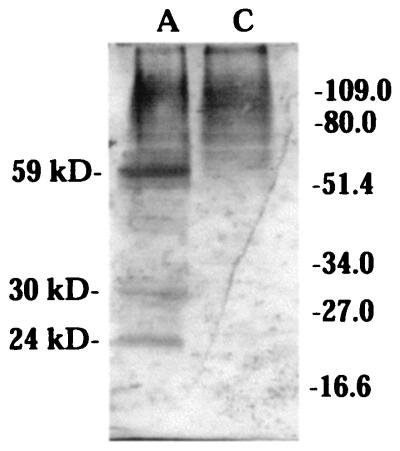
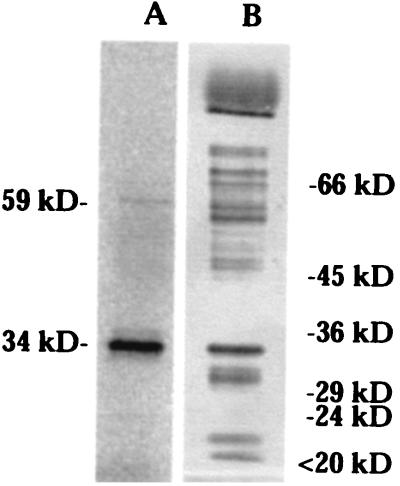
The full-length acyltransferase cDNA was cloned into the BamHI and EcoRI sites of pYES2 (Invitrogen) to form pYAGT2, and transferred to S. cerevisiae KT1115 (ura3). Raffinose was substituted for glucose as the carbon source to avoid the inhibition effect of GAL1 by glucose. Immunoblotting of the yeast cell extract using acyltransferase antisera revealed 59-kDa, 30-kDa, and 24-kDa immunoreactive bands (Fig. 3), indicating the unprocessed and proteolytically processed forms of the protein.
Figure 3.
L. pennellii acyltransferase expression in yeast. (A) Immunoblot of yeast cells expressing unprocessed (59 kDa) and processed (30 kDa and 24 kDa) forms of the acyltransferase. (C) Immunoblot of control yeast cells harboring vector only.
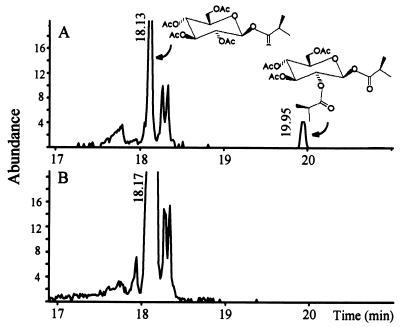
Triton X-100 permeabilized cells (32) were used to study enzyme activity in situ. S. cerevisiae expressing the acyltransferase cDNA has the ability to synthesize diacylglucose when permeabilized and provided 1-O-acyl-β-glucose. Although total ion chromatograms are extremely complex because of the diversity of yeast carbohydrate metabolism, extracted ion chromatograms selected for display of components fragmenting to the isobutyryl ion, m/z 71, provide a facile display of isobutyrylglucose-related metabolism (Fig. 4). The tetraacetate derivatives of the substrate, 1-O-isobutyryl-β-glucose eluted at 18.1 min, and the triacetate derivatives of the product, diisobutyrylglucose, eluted at 19.95 min. Yeast cells expressing the L. pennellii glucose acyltransferase disproportionate 1-O-isobutyryl-β-glucose to form diisobutyrylglucose (Fig. 4A). Vector-only control cells do not disproportionate 1-O-isobutyryl-β-glucose (Fig. 4B). Hence, it is clear that this cDNA encodes an acyltransferase that catalyzes disproportionation reactions characteristic of glucose polyester synthesis in L. pennellii glandular trichomes.
Figure 4.
GC-MS analysis of 1-O-isobutyryl-β-glucose disproportionation in yeast cells. Permeabilized cells were provided with 1-O-isobutyryl-β-glucose for 3 h at room temperature, and the supernatant was peracetylated. (A) m/z 71 extracted ion chromatograph from cells expressing the acyltransferase. (B) m/z 71 extracted ion chromatograph from reaction performed with control cells harboring vector only. The 18.1-min peak in A and B is the tetraacetate derivative of the substrate 1-O-isobutyryl-β-glucose. The 19.95-min peak in A is the triacetate derivative of diisobutyrylglucose.
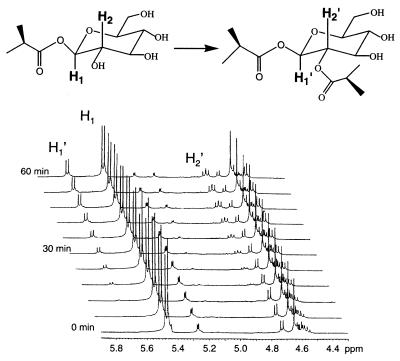
In addition, the disproportionation reaction catalyzed by this enzyme is regiospecific. Both the yeast-expressed (Fig. 4) and native enzyme catalyze the formation of a single diacylglucose product. In a 1H-NMR time-course study (Fig. 5), the reaction of 1-O-isobutyryl-β-glucose with the native acyltransferase is characterized initially by the appearance of an anomeric proton signal (δ 5.7 ppm, d, J = 8.4 Hz) downfield shifted from the anomeric proton of the starting material (δ 5.45, d, J = 8.4 Hz) by 0.25 ppm, and a new dd (δ 4.8, J = 8.4 Hz) downfield shifted from the clustered signals derived from H-2, -3, and -4. The appearance of these signals is consistent only with the acylation of the H-2 hydroxyl by isobutyrate, leading to formation of 1,2-di-O-isobutyryl-β-glucose and deshielding of the proton at H-2. Concomitant with the transfer of the isobutyryl moiety from one equivalent of 1-O-isobutyryl-β-glucose, one equivalent of β-glucose is liberated, leading to the appearance of the anomeric proton of α-glucose (5.15 ppm, J = 4 Hz) as the β-glucose anomerizes. After 1 h, the newly formed 1,2-di-O-isobutyryl-β-glucose accumulates sufficiently to act as an acyl donor, consistent with previous observations (17). Acyl transfer from 1,2-di-O-isobutyryl-β-glucose leads to the formation of α- and β-2-O-isobutyrylglucoses and regenerates 1-O-isobutyryl-β-glucose when β-glucose is the acceptor; consequently, the spectra become difficult to interpret after this time (not shown). However, it is clear that the initial reaction catalyzed by this acyltransferase shows a strong preference for acyl transfer to the 2-hydroxyl of 1-O-isobutyryl-β-glucose.
Figure 5.
1H-NMR of the disproportionation reaction catalyzed by L. pennellii glucose acyltransferase. The zero-time spectrum was acquired immediately after addition of acyltransferase to the NMR tube. Acylation at H-2 is indicated by the time-dependent appearance of a downfield-shifted anomeric proton (H1′), and appearance of the downfield-shifted H-2′ (in the nonacylated substrate, the signal for H-2 is present in the complex cluster of signals around δ4.6).
Glucose Acyltransferase Is a Serine Carboxypeptidase-Like Protein.
The deduced acyltransferase protein sequence shows sequence similarities to serine carboxypeptidases, such as barley carboxypeptidase I, II, and III, wheat serine carboxypeptidase II, and yeast carboxypeptidase Y when blastp is performed (http://www.ncbi.nlm.nih.gov). Amino acid similarity and identity reache 76% and 35%, respectively, when the acyltransferase is compared with barley carboxypeptidase I (Fig. 2 and ref. 34).
Serine carboxypeptidases possess a characteristic Ser-His-Asp catalytic triad. The Ser residue is unusually reactive because of its interaction with the His imidazole nitrogen. In the catalytic mechanism of serine carboxypeptidases, this Ser undergoes acylation as a consequence of its nucleophilic attack on the carbonyl carbon of the peptide substrate, forming a tetrahedral acyl-enzyme intermediate. This intermediate is then decomposed by water, liberating the bound peptide and regenerating the reactive Ser. The Ser-His-Asp catalytic triad is also responsible for the activities of endopeptidases, such as trypsin and subtilisin, as a result of convergent molecular evolution. Additionally, some lipases possess Ser-His-Asp or similar Ser-His-Glu catalytic triad centers (35, 36).
The sequences around the Ser, His, and Asp catalytic triad residues are conserved between the glucose acyltransferase and barley serine carboxypeptidases I (Fig. 2). The heterotetrameric composition of the acyltransferase is also similar to that of barley serine carboxypeptidase I (37, 38). This suggests that the acyltransferase is a serine carboxypeptidase-like enzyme that utilizes the Ser-His-Asp catalytic motif to catalyze acyl transfer and exchange reactions. In the glucose acyltransferase, the putative active Ser is present on the 34-kDa subunit, and the active His and Asp are on the 24-kDa subunit.
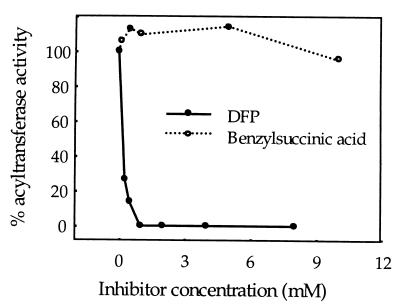
DFP is a transition state inhibitor of serine carboxypeptidases. DFP reacts irreversibly with the active Ser residue, forming a covalent DFP-enzyme complex. If transacylation catalyzed by the glucose acyltransferase employs an active serine-based catalytic mechanism, DFP should inhibit the disproportionation activity and covalently modify the active serine in the acyltransferase (Ser-174 in the 34-kDa subunit). Disproportionation of 1-O-acyl-β-glucose is inhibited 50% by 170 μM DFP and is completely inhibited by 1 mM DFP (Fig. 6). [14C]DFP specifically labeled the 34-kDa subunit in a 1200-fold purified acyltransferase preparation (Fig. 7). Therefore, an active Ser residue appears to play a critical function in catalysis of acyl transfer from 1-O-acyl-β-glucose. A minor 59-kDa DFP-labeled band is likely to represent the unprocessed form of the acyltransferase. This suggests that the unprocessed enzyme also adopts a conformation sufficient to activate Ser by a proximal His.
Figure 6.
Effects of DFP and benzylsuccinic acid treatment on disproportionation activity of glucose acyltransferase. DFP and benzylsuccinic acid were preincubated with the acyltransferase, and disproportionation reactions were performed with 1-O-[1-14C]isobutyryl-β-glucose.
Figure 7.
DFP labeling of the glucose acyltransferase. (A) Autoradiograph of acyltransferase labeled with [1,3-14C]DFP. The enzyme was purified 1,200-fold from L. pennellii leaves. The 34-kDa band is the large subunit of the acyltransferase in which the putative active serine is located. The minor band of 59 kDa is probably the unprocessed acyltransferase. (B) Silver stain of the same protein preparation.
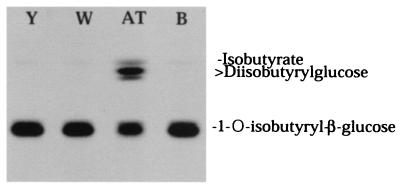
Benzylsuccinic acid is a competitive inhibitor of serine carboxypeptidases, such as yeast carboxypeptidase Y, barley serine carboxypeptidases, and wheat serine carboxypeptidase II (39–41), and is widely used as an affinity support for purification of plant serine carboxypeptidases. Although benzylsuccinic acid is a potent inhibitor of wheat serine carboxypeptidase II, it has no effect on disproportionation activity up to 10 mM (Fig. 6), suggesting that charge or conformational differences at active sites exist between the glucose acyltransferase and general serine carboxypeptidases. Similarly, the glucose acyltransferase does not exhibit activity toward model substrates reported for a wound-induced tomato serine carboxypeptidase (not shown) (42). Furthermore, wheat and yeast carboxypeptidases are incapable of disproportionating 1-O-acyl-β-glucose even when assayed at very high concentrations of this substrate (Fig. 8).
Figure 8.
TLC autoradiograph of reactions of yeast serine carboxypeptidase (Y), wheat serine carboxypeptidase II (W), and L. pennellii glucose acyltransferase (AT) with 1-O-[14C]isobutyryl-β-glucose. B, The reaction blank without enzyme. Variable amounts of Y and W have been used without any indication of disproportionation ability.
Discussion
Heterologous expression of the L. pennellii glucose acyltransferase cDNA results in disproportionation of 1-O-β-acylglucoses by yeast cells. The reaction catalyzed by the L. pennellii acyltransferase is regiospecific. The deduced sequence of the cDNA shows high sequence similarity to serine carboxypeptidases, and is highly conserved around the active site Ser, His, and Asp residues characteristic of serine carboxypeptidases. Inhibition of disproportionation activity by DFP, a transition state inhibitor of active Ser-based enzymes, and covalent DFP labeling of the enzyme demonstrate the involvement of an active serine residue in the transacylation process. Hence, the glucose acyltransferase utilizes a serine carboxypeptidase-like catalytic triad to accomplish the synthesis of glucose polyesters. By analogy to serine proteases, the mechanism of acyl transfer is likely to involve nucleophilic attack of the active Ser hydroxyl on the ester carbonyl carbon of 1-O-β-acylglucose, forming a tetrahedral enzyme-bound acyl intermediate and releasing free glucose. This transition state would then be subject to attack by the 2-hydroxyl of a second 1-O-β-acylglucose, resulting in formation of 1,2-di-O-β-acylglucose, and regenerating the active Ser. Attack of the acyl-enzyme intermediate by β-glucose or water would result in the anomeric acyl exchange and esterase activities, respectively, also reported to be catalyzed by preparations of this enzyme (16, 17, 19).
Despite the similarity between glucose acyltransferase and serine carboxypeptidases, no carboxypeptidase activity could be detected with a range of substrates used by many serine carboxypeptidases, including a wound-induced carboxypeptidase reported from tomato leaves (42). Furthermore, yeast and wheat serine carboxypeptidases do not possess the ability to disproportionate 1-O-β-glucoses. Therefore, although closely related to and apparently derived from serine carboxypeptidases, this glucose acyltransferase does not appear to retain carboxypeptidase function.
Difference in residues involved in substrate binding sites may reveal the reasons for the lack of carboxypeptidase activity from the acyltransferase. Three-dimensional structure (43) and site-directed mutagenesis studies of yeast carboxypeptidase Y (44) indicate that the side chains of Trp-49, Asn-51, Glu-65, and Glu-145 are involved in the recognition of the C-terminal carboxylate of peptide substrates by forming a hydrogen bond network. Mutations at these sites, especially on Asn-51 and Glu-145, dramatically decrease the kcat/Km value for carboxypeptidase activity and may even eliminate the ability of the enzyme to interact with the carboxylate of the peptide substrate. Serine carboxypeptidases share conserved sequence around the sites of Asn-51 and Glu-145 analogous to those in carboxypeptidase Y (44); these residues are not conserved in this glucose acyltransferase or in the deduced sequences of three other serine carboxypeptidase-like acyltransferases isolated from Solanum berthaualtii (GenBank accession nos. AF006078, AF006079, and AF006080). In general, sequence comparison of serine-carboxypeptidase-like acyltransferases with bona fide proteinases of the same class does not immediately reveal characteristics that can be used to ascertain the functional activity of their products.
Serine carboxypeptidases are primarily known for catabolic or processing functions, e.g., protein turnover or modification of precursor proteins. However, recruitment of serine carboxypeptidase motifs for other functions—primarily hydrolytic—have been reported. For example, a cyanogenic hydroxynitrile lyase from sorghum shares significant sequence similarity to serine carboxypeptidase and is inactivated by DFP (45, 46), and fungal cutinase also employs the catalytic triad of serine proteinases (47). In addition, an acyl-CoA: alcohol acyltransferase involved in the production of ethyl hexanoate is inhibited by DFP, but no sequence is available for this enzyme (48).
The L. pennellii glucose acyltransferase is an example of recruitment of a common mechanism of catabolism to a synthetic plant secondary metabolic pathway. We predict that similar adaptations have also occurred in enzymes involved in biosynthetic pathways that use 1-O-acylglucoses or similar high-energy acyl donors in transacylation reactions. For example, the formation of 1,2-di-sinapoylglucose (3) is analogous to the L. pennellii glucose acyltransferase's disproportionation reaction leading to synthesis of 1,2-di-acylglucose, and the sinapoyltransferase has an apparent Mr of 55,000, near the value for the unprocessed glucose acyltransferase (3). Similarly, the mechanistically analogous sinapoyltransferase involved in the synthesis of sinapoyl malate has a Mr around 52,000 (2). The transacylation involved in the formation of indolylacetyl-myo-inositol is similar to the L. pennellii acyl exchange reaction in its utilization of 1-O-acyl-β-glucose as the acyl donor, a cyclic hexahydric alcohol as the acyl acceptor, and an acyltransferase of Mr 59,000, similar to that of the unprocessed L. pennellii glucose acyltransferase (15). Analogous adaptations may also have occurred in the evolution of the 1-O-β-galloylglucose-using galloyltransferases in 1,2,3,4,6-pentagalloylglucose biosynthesis, and the acyltransferases involved in the β-1-O-acyl acetal-dependent formation of chlorogenic acid, isochlorogenic acid, acylated betacyanins, caffeoylglucaric acid, and caffeoylgalactaric acid (4, 9–12). The multiplicity and diversity of serine carboxypeptidase-like proteins in plants supports the proposition that this family may have acquired a broad repertoire of metabolic functions. Currently, the Arabidopsis genome lists 38 serine carboxypeptidase-like genes, suggesting the possibility that more than 60 of these genes will be known when the genome sequence is complete. Functional analysis of these genes will reveal how widely recruitment of serine-based proteases has occurred in the evolution of acyltransferases involved in indoleacetic acid, phenolic, and other biosynthetic activities.
Abbreviations
- DFP
diisopropyl fluorophosphate
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF248647, AF006078, AF006079, and AF00608).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110154197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110154197
References
- 1.Mock H, Strack D. Phytochemistry. 1993;32:575–579. [Google Scholar]
- 2.Kojima H, Uritani I. Plant Cell Physiol. 1972;18:1075–1084. [Google Scholar]
- 3.Grawe W, Bachhuber P, Mock H, Strack D. Planta. 1992;187:236–241. doi: 10.1007/BF00201945. [DOI] [PubMed] [Google Scholar]
- 4.Dahlbender B, Strack D. Phytochemistry. 1986;25:1043–1046. [Google Scholar]
- 5.Gross G G. In: Phenolic Metabolism in Plants. Stafford H A, Ibrahim R K, editors. New York: Plenum; 1992. pp. 297–324. [Google Scholar]
- 6.Villegas R J A, Kojima M. J Biol Chem. 1986;261:8729–8733. [PubMed] [Google Scholar]
- 7.Tanaka M, Kojima M. Arch Biochem Biophys. 1991;284:151–157. doi: 10.1016/0003-9861(91)90277-p. [DOI] [PubMed] [Google Scholar]
- 8.Zenk M H. In: Biochemistry of Plant Phenolics. Swain T, Harborne J B, Van Sumere C F, editors. Vol. 12. New York: Plenum; 1977. pp. 139–176. [Google Scholar]
- 9.Villegas R J A, Shimokawa T, Okuyama H, Kojima M. Phytochemistry. 1987;26:1577–1581. [Google Scholar]
- 10.Strack D, Gross W, Wray V, Grotjahn L. Plant Physiol. 1987;83:475–478. doi: 10.1104/pp.83.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strack D, Gross W. Plant Physiol. 1990;92:41–47. doi: 10.1104/pp.92.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokern M, Heuer S, Strack D. Bot Acta. 1992;105:146–151. [Google Scholar]
- 13.Szerszen J B, Szczyglowski K, Bandurski R S. Science. 1994;265:1699–1701. doi: 10.1126/science.8085154. [DOI] [PubMed] [Google Scholar]
- 14.Kowalczyk S, Bandurski R S. Biochem J. 1991;279:509–514. doi: 10.1042/bj2790509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kesy J, Bandurski R S. Plant Physiol. 1990;94:1598–1604. doi: 10.1104/pp.94.4.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghangas G S, Steffens J C. Proc Natl Acad Sci USA. 1993;90:9911–9915. doi: 10.1073/pnas.90.21.9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghangas G S, Steffens J C. Arch Biochem Biophys. 1995;316:370–377. doi: 10.1006/abbi.1995.1049. [DOI] [PubMed] [Google Scholar]
- 18.Kuai J P, Ghangas G S, Steffens J C. Plant Physiol. 1997;115:1581–1587. doi: 10.1104/pp.115.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li A X, Eannetta N, Gangus G S, Steffens J C. Plant Physiol. 1999;121:453–460. doi: 10.1104/pp.121.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burke B A, Goldsby G, Mudd J B. Phytochemistry. 1987;26:2567–2571. [Google Scholar]
- 21.Goffreda J C, Mutschler M A, Tingey W M. Entomol Exp Appl. 1988;48:101–108. [Google Scholar]
- 22.Goffreda J C, Mutschler M A, Ave D A, Tingey W M, Steffens J C. J Chem Ecol. 1989;15:2135–2148. doi: 10.1007/BF01207444. [DOI] [PubMed] [Google Scholar]
- 23.Hawthorne D J, Shapiro J A, Tingey W M, Mutschler M A. Entomol Exp Appl. 1992;65:65–73. [Google Scholar]
- 24.Rodriguez A E, Tingey W M, Mutschler M A. J Econ Entomol. 1993;86:34–39. [Google Scholar]
- 25.Juvik J A, Shapiro J A, Young T E, Mutschler M A. J Econ Entomol. 1994;87:482–492. [Google Scholar]
- 26.Liedl B E, Lawson D M, White K K, Shapiro J A, Cohen D E, Carson W G, Trumble J T, Mutschler M A. J Econ Entomol. 1995;88:742–748. [Google Scholar]
- 27.Callahan A, Morgens P, Walton E. Hortic Sci. 1989;24:356–358. [Google Scholar]
- 28.Lee C C, Caskey C T. In: PCR Protocols: A Guide to Methods and Applications. Innis M A E A, editor. San Diego: Academic; 1990. pp. 46–53. [Google Scholar]
- 29.Frohman M A. In: PCR Protocols: A Guide to Methods and Applications. Innis M A E A, editor. San Diego: Academic; 1990. pp. 28–38. [Google Scholar]
- 30.Loh E. Methods. 1991;2:11–19. [Google Scholar]
- 31.Gietz D, Jean A S, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miozzari G F, Niederberger P, Hutter R. Anal Biochem. 1978;90:220–233. doi: 10.1016/0003-2697(78)90026-x. [DOI] [PubMed] [Google Scholar]
- 33.Ausubel F, Brent R, Kingston R, Moore D, Smith J, Seidman J, Struhl K. Current Protocols in Molecular Biology. Brooklyn, NY: Greene; 1987. [Google Scholar]
- 34.Myers E, Miller W. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 35.Schrag J D, Li Y, Wu S, Cygler M. Nature (London) 1991;351:761–764. doi: 10.1038/351761a0. [DOI] [PubMed] [Google Scholar]
- 36.Schrag J D, Winkler F K, Cygler M. J Biol Chem. 1992;267:4300–4303. [PubMed] [Google Scholar]
- 37.Breddam K, Sorensen S B, Ottesen M. Carlsburg Res Commun. 1983;48:217–230. [Google Scholar]
- 38.Liao D I, Breddam K, Sweet R M, Bullock T, Remington S J. Biochemistry. 1992;31:9796–9812. doi: 10.1021/bi00155a037. [DOI] [PubMed] [Google Scholar]
- 39.Johansen J T, Breddam K, Ottesen M. Carlsburg Res Commun. 1976;41:1–14. [Google Scholar]
- 40.Ray L E. Carlsburg Res Commun. 1976;41:169–182. [Google Scholar]
- 41.Bullock T L, Branchaud B, Remington S J. Biochemistry. 1994;33:11127–11134. doi: 10.1021/bi00203a009. [DOI] [PubMed] [Google Scholar]
- 42.Walker-Simmons M, Ryan C A. Phytochemistry. 1980;19:43–47. [Google Scholar]
- 43.Endrizzi J A, Breddam K, Remington S J. Biochemistry. 1994;33:11106–11120. doi: 10.1021/bi00203a007. [DOI] [PubMed] [Google Scholar]
- 44.Mortensen U H, Remington S J, Breddam K. Biochemistry. 1994;33:508–517. doi: 10.1021/bi00168a016. [DOI] [PubMed] [Google Scholar]
- 45.Wajant H, Mundry K. Plant Sci. 1993;89:127–133. [Google Scholar]
- 46.Wajant H, Mundry K, Pfizenmaier K. Plant Mol Biol. 1994;26:735–746. doi: 10.1007/BF00013758. [DOI] [PubMed] [Google Scholar]
- 47.Koller W, Kolattakudy P E. Biochemistry. 1982;21:3083–3090. doi: 10.1021/bi00256a008. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi H, Hasuo T, Amachi T, Akita O, Hara S, Yoshizawa K. Agric Biol Chem. 1989;53:1551–1556. [Google Scholar]