Abstract
The principal pathological features of Alzheimer's disease (AD) are extracellular amyloid plaques and intracellular neurofibrillary tangles, the latter composed of the microtubule-binding protein tau assembled into paired helical and straight filaments. Recent studies suggest that these pathological entities may be functionally linked, although the mechanisms by which amyloid deposition promotes pathological tau filament assembly are poorly understood. Here, we report that tau is proteolyzed by multiple caspases at a highly conserved aspartate residue (Asp421) in its C terminus in vitro and in neurons treated with amyloid-β (Aβ) (1–42) peptide. Tau is rapidly cleaved at Asp421 in Aβ-treated neurons (within 2 h), and its proteolysis appears to precede the nuclear events of apoptosis. We also demonstrate that caspase cleavage of tau generates a truncated protein that lacks its C-terminal 20 amino acids and assembles more rapidly and more extensively into tau filaments in vitro than wild-type tau. Using a monoclonal antibody that specifically recognizes tau truncated at Asp421, we show that tau is proteolytically cleaved at this site in the fibrillar pathologies of AD brain. Taken together, our results suggest a novel mechanism linking amyloid deposition and neurofibrillary tangles in AD: Aβ peptides promote pathological tau filament assembly in neurons by triggering caspase cleavage of tau and generating a proteolytic product with enhanced polymerization kinetics.
Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by accelerated neuronal cell death leading to dementia (1). Its hallmark pathologic features are extracellular amyloid plaques and intraneuronal fibrillar structures, the latter including neurofibrillary tangles (NFTs), neuropil threads, and dystrophic neurites invading amyloid plaques (2). Amyloid plaques are formed by the extracellular deposition of proteolytic fragments of the amyloid precursor protein (APP) termed amyloid-β (Aβ) (1, 3), whereas the fibrillar pathologies are composed of the microtubule-associated protein tau assembled into polymeric filaments (paired helical and straight filaments) (2). The pathogenic role of amyloid deposition in AD is underscored by the evidence that each of the disease-causing mutations in familial AD results in enhanced production of amyloidogenic Aβ peptides; these peptides are sufficient to induce apoptosis in cultured neurons (1, 3). Furthermore, the recent observation that tau mutations cause hereditary frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), a class of diseases characterized by NFT-like deposition of polymeric tau and dementia without amyloid plaques, emphasizes the critical role that tau plays in neurodegenerative events (4–6). Although amyloid plaques and NFTs have been largely regarded as independent neuropathologic entities, recent work suggests they may be functionally linked: mutation of APP that results in amyloid deposition or direct intracranial injection of Aβ peptide increases NFT formation in transgenic mice expressing an FTDP-17-causing tau mutant (7, 8). However, the molecular mechanism(s) by which the extracellular accumulation of Aβ peptides promotes the intracellular assembly of pathologic tau filaments is poorly understood.
In AD, the tau present in NFTs is aberrantly phosphorylated and often proteolytically truncated at its C terminus (9, 10). One such proteolytic event is cleavage of tau at Glu391 by a yet-to-be-identified protease; tau truncated at this site is present in NFTs in brains of AD patients (10). Such post-translational modifications are thought to impair tau's ability to bind/stabilize microtubules, and they also drive tau filament assembly in vitro (11–13). Indeed, we have recently demonstrated that the C terminus of tau inhibits filament assembly, and that removal of as few as 12 amino acid residues from its C terminus accelerates tau filament assembly in vitro (12).
Because members of the caspase family of cysteine proteases are activated in apoptotic neurons in AD and play a critical role in Aβ-induced neuronal apoptosis (14–16), we postulated that caspases might contribute to the proteolytic truncation of tau observed in the neurofibrillar pathologies of AD. In support of this hypothesis is the recent observation that tau is cleaved by caspase-3 in vitro and in neurons subjected to hypokalemia or staurosporine (17–19), although the functional consequences of its proteolytic cleavage and its potential contribution to AD have not been delineated. In this report, we demonstrate that tau is cleaved by multiple caspases at Asp421 in its C terminus, and that the resulting N-terminal caspase cleavage product (amino acids 1–421) assembles more rapidly into filaments than WT tau. In addition, we show that tau is specifically cleaved at Asp421 in neurons treated with amyloidogenic Aβ peptide and in the characteristic fibrillar pathologies in AD. Hence, our findings suggest a previously unrecognized link between amyloid and NFTs, whereby Aβ exposure triggers caspase cleavage of tau, which in turn promotes the assembly of tau into pathological filaments.
Materials and Methods
Caspase Cleavage Reactions in Vitro. 35S-labeled WT (four-repeat) human tau protein was incubated with buffer control, 2.5 or 25 ng of caspase-1, -2, -3, -6, -7, or -8 for 1 h at 37°C, as described (20, 21); the cleavage products were detected by autoradiography of SDS-polyacrylamide gels. 35S-labeled WT or mutant D421E tau proteins were synthesized from the corresponding cDNAs in vitro by using the TNT T7 Quick Coupled Transcription/Translation System (Promega). A cDNA-encoding mutant D421E tau was made from the WT human tau cDNA by using the QuikChange (Stratagene) site-directed mutagenesis kit with the following oligonucleotide primers: 5′-AGCATCGACATGGTAGAATCGCCCCAGCTCGCC-3′ and 5′-GGCGAGCTGGGGCGATTCTACCATGTCGATGCT-3′. The mutation was verified by DNA sequencing.
TauC3 mAb Production. A mouse mAb was generated against a peptide corresponding to the C terminus of tau truncated at Asp421. Specifically, the peptide CSSTGSIDMVD, which corresponds to tau residues 412–421 with a Cys added to the N terminus, was synthesized by Cell Essentials (Boston), and this peptide was coupled through the cysteine to maleimide-activated keyhole limpet hemocyanin (Pierce). The mice were immunized eight times over a period of 12 mo with 100 μg of conjugated peptide administered s.c. In two of the last three immunizations, an additional immunization (100 μg of conjugated peptide) was also administered by i.p. injection. In the penultimate immunization, however, 200 μg of recombinant 1–421 truncated tau was administered s.c. along with an i.p. injection of 100 μg of conjugated peptide. Four days after the final immunization, the mice were killed and their splenocytes fused to SP2/o myeloma cells as described (22). Two weeks later, positive clones were selected on the basis of their ability to bind to recombinant truncated tau (amino acids 1–421) but not to full-length tau. One cell line (TauC3) was obtained, subcloned four times, adapted to reduced serum medium, and placed in a bioreactor. Thirty-two milligrams of purified IgG1 antibody was obtained and stored in Hepes saline buffer (pH 7.4) containing 50% glycerol at –80°C prior to use.
Expression of Recombinant WT or Truncated Tau Proteins in Escherichia coli. Full-length human tau protein and the various truncation mutants coupled to a 6×-His tag were expressed in E. coli and purified by Ni-NTA agarose chromatography followed by size exclusion chromatography as described (23).
Treatment of Cortical Neurons with Fibrillar Aβ. Primary cortical neurons were prepared from the cortex of embryonic day 18 rats, plated onto poly-l-lysine coated six-well plates, with or without glass coverslips, and grown in Neurobasal media (Invitrogen) containing B27 supplements (Invitrogen) and 2 mM l-glutamine, as described (24, 25). After 4 days in culture, cortical neurons were treated for 0–24 h with 10 μM Aβ (1–42) (obtained from Mary Jo Ladu, Evanston Northwestern Healthcare Research Institute) that had been induced to form fibrils by preincubation in culture medium (designated “fibrillar” Aβ). Specifically, 0.45 mg of Aβ peptide was dissolved in 20 μl of DMSO and diluted to a 100-μM stock solution in medium, which was then incubated with gentle shaking at room temperature for 4 days. Fibrillar Aβ was then diluted to 10 μM and applied to neuron cultures (0.2% DMSO was added to control cultures). For the caspase inhibitor experiments, cortical neurons were preincubated in the presence or absence of 50 μM zVAD-fmk (ICN) for 1 h and then treated for 3 h with 10 μM fibrillar Aβ.
Immunoblotting. After treatment, cortical neurons were washed with PBS and extracted for 3 min in microtubule stabilization buffer (130 mM Hepes/4.2 mM MgCl2/10 mM EGTA, pH 6.9) and 0.2% Triton X-100. The cells were then rinsed in microtubule stabilization buffer without Triton X-100 and scraped in RIPA lysis buffer (50 mM Tris·HCl/150 mM NaCl/1% NP-40/0.5% deoxycholate/0.1% SDS/5 mM EDTA, pH 7.5). Lysates were then centrifuged at ∼17,000 × g for 15 min. The protein concentration of the supernatants was determined by standard Lowry assay. Immunoblotting was performed as detailed (21) by using the following mAbs: Tau1 (100 ng/ml) (22), Tau5 (100 ng/ml) (26), TauC3 (1 μg/ml), or poly (ADP-ribose) polymerase (BD PharMingen; 1:1,000 dilution).
Immunofluorescence and Confocal Microscopy. Cortical neurons were plated on coverslips as described above. After 4 days in culture, neurons were incubated with 20 μM fibrillar Aβ for 8 h. Neurons were then washed with PBS, incubated for 30 sec in microtubule stabilization buffer, and fixed for 10 min in 4% paraformaldehyde at room temperature. Coverslips were preincubated with 1% nonfat dry milk in PBS containing 1% BSA and 1.5% Triton X-100 for 1 h at room temperature, prior to incubation in blocking buffer containing the primary Abs: TauC3 (1 μg/ml) or 5H1 β-tubulin mAb [IgM subtype (27), 1:200 dilution] overnight at 4°C. After washing with PBS, the secondary Ab was applied for 1 h at room temperature: FITC-conjugated anti-IgM antibody to detect 5H1 (1:100 dilution) or rhodamine-conjugated anti-IgG Ab to detect TauC3 (1:100). Laser-scanning confocal microscopy was performed as described (28) by using a Zeiss LSM 510 laser scanning system. Bandpass filters were chosen to minimize “bleed through” between channels. For apoptosis assays, primary cortical neurons were treated with 20 μM Aβ for 0–12 h. The percentage of TauC3-positive and -negative neurons with apoptotic nuclei (condensed chromatin or fragmented nucleus) was determined by staining with 10 μg/ml Hoescht 33258 (Sigma) as described (29). At least 100 TauC3-positive or -negative neurons were scored for apoptosis in four independent experiments with primary neurons prepared on 2 different days.
Tau Filament Assembly in Vitro. Recombinant WT or truncated tau proteins (4 μM) were incubated with 75 μM arachidonic acid (Cayman Chemical, Ann Arbor, MI) for 0–6 h to induce polymerization, and the kinetics of polymer assembly was measured by right-angle laser light scattering as described (30). The morphology of glutaraldehyde-fixed tau filaments was examined by negative-stain electron microscopy (JEOL JEM-1120 transmission electron microscope), as detailed (30).
Immunohistochemistry. Immunostaining was performed on 40-μm-thick frozen sections of human hippocampus from AD brain provided by the Neuropathology Core of the Northwestern University Cognitive Neurology and Alzheimer's Disease Center. Brightfield and laser-scanning confocal microscopy were performed as described (28). For brightfield microscopy, Tau5 was used at 20 ng/ml, and a tissue culture supernatant containing TauC3 was used at a dilution of 1:2,500. For confocal microscopy, TauC3 was applied at 400 ng/ml along with Thiazin red (Sigma) at 0.001%. In this case, the Ab was detected by a fluorescein-conjugated secondary Ab (1:100 goat anti-mouse, Kirkegaard & Perry Laboratories).
Results
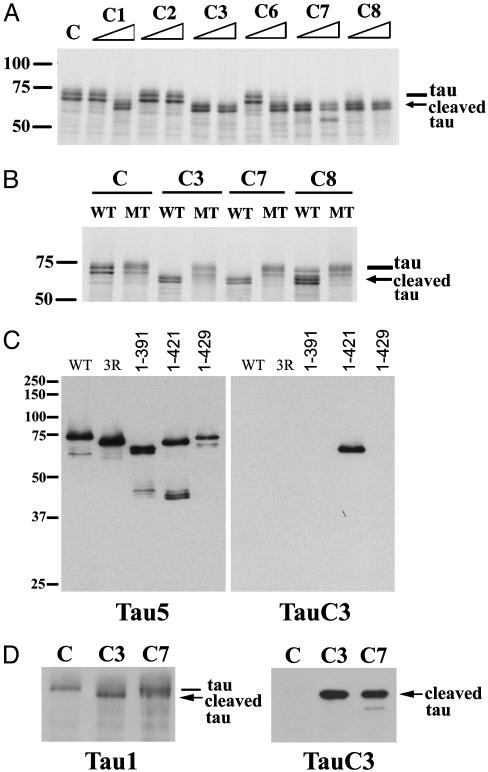
Human Tau Is Cleaved by Multiple Caspases at Asp421in Vitro. To determine whether family members other than caspase-3 proteolyze tau in vitro, we incubated 35S-labeled WT human tau with a panel of recombinant caspases. As shown in Fig. 1 A, 35S-labeled WT tau (which appears as a doublet, likely due to transcription initiation at an internal ATG site) was cleaved by caspases-1, -3, -6, -7, and -8 into a major proteolytic product ≈5 kDa smaller than WT tau (indicated by arrow, also a doublet). Caspases-3, -7, and -8 proteolyzed tau most efficiently, with cleavage of WT tau occurring at the lower of the two enzyme concentrations tested. On the basis of the observed size of the proteolytic product and a search of the amino acid sequence of tau for caspase cleavage motifs, we postulated that tau was cleaved by caspases at Asp421 in its C terminus (DMVD421-S422), the same cleavage site previously described for caspase-3 (18, 19). Hence, we made a mutant tau construct in which Asp421 was converted to a Glu residue (D421E), thereby creating a site resistant to caspase cleavage. As shown in Fig. 1B, 35S-labeled WT tau, but not mutant (MT) D421E tau, was proteolyzed by caspases-3, -7, and -8. These results demonstrate unequivocally that Asp421 is the principal site cleaved by each of these caspases in vitro.
Fig. 1.
Human tau is cleaved by multiple caspases at Asp421 in vitro. (A) 35S-labeled WT human tau (doublet) was incubated with buffer control (C) or 2.5 or 25 ng of caspases-1, -2, -3, -6, -7, or -8 (C1–C8) for 1 h. The major caspase cleavage product (doublet) is indicated by the arrow. (B) 35S-labeled WT or mutant (MT) D421E tau was incubated with control buffer or 2.5 ng of caspases-3, -7, or -8 for 1 h. In both A and B, the cleavage products were resolved by SDS/PAGE and visualized by autoradiography. (C) TauC3 mAb specifically recognizes tau that is truncated at Asp421. Recombinant human WT tau (containing four microtubule-binding repeats), three-repeat tau (produced by alternative splicing, labeled 3R), or truncated tau proteins encoding amino acids 1–391, 1–421, or 1–429 was immunoblotted with Tau5 (Left) or TauC3 (Right) mAbs. (D) Recombinant human WT tau was incubated with control buffer (C) or 25 ng of caspase-3 or -7 (C3 or C7) for 1 h, and the reaction products were immunoblotted with Tau1 (Left) or TauC3 (Right) mAbs. The molecular mass (kDa) of markers is shown at the left of A–C.
TauC3 mAb Specifically Recognizes Tau Truncated at Asp421. To examine the functional consequences of caspase cleavage of tau at Asp421, we generated a mAb (designated TauC3) that specifically recognizes tau truncated at this site. As shown in Fig. 1C Right, recombinant tau truncated at Asp421 (amino acids 1–421, designated “truncated” tau for brevity) was detected by immunoblotting with TauC3, whereas WT tau (containing four microtubule-binding repeats), tau containing three microtubule-binding repeats (produced by alternative splicing, designated 3R), or tau proteins truncated at amino acid residues Glu391 or Ala429 were not recognized by TauC3. All of these proteins were detected by Tau5, which reacts with residues 210–230 in the “proline-rich” domain (Fig. 1C Left) (23, 26). To further confirm the specificity of TauC3 for caspase-cleaved tau, we incubated WT tau protein with buffer control or 25 ng of caspase-3 or -7 for 1 h and analyzed the reaction products by immunoblotting with TauC3. TauC3 specifically recognized tau cleaved by caspase-3 or -7 but not uncleaved tau incubated with buffer control (Fig. 1D Right), whereas both uncleaved tau and tau proteolyzed by caspase-3 or -7 were detected by Tau1, which recognizes an epitope N-terminal to the caspase cleavage site (dephosphorylated residues 189–207) (22, 23). These findings underscore the exquisite specificity of TauC3 in recognizing tau proteolyzed by caspases at Asp421.
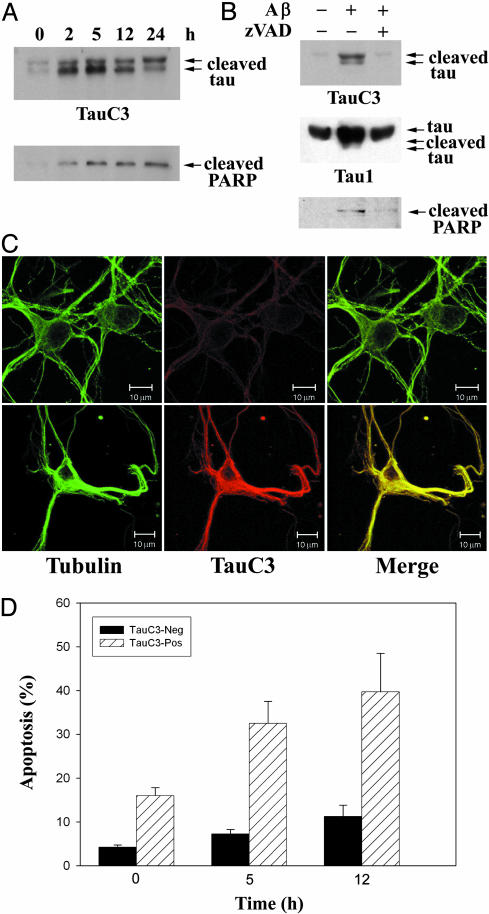
Tau Is Proteolyzed at Asp421 in Neurons Undergoing Aβ-Induced Apoptosis. Because treatment of neurons with amyloidogenic Aβ peptides activates caspases (16), we postulated that tau might be cleaved at Asp421 during the induction of Aβ-induced neuronal apoptosis. To this end, we incubated primary cortical neurons with 10 μM fibrillar Aβ (1–42) peptide for 0–24 h. As shown in Fig. 2 A Upper), TauC3 detected truncated tau (seen as a prominent doublet likely reflecting cleavage of the two major tau isoforms) in cortical neurons as early as 2 h after incubation with Aβ. A very faint doublet was also observed in untreated neurons (designated time 0) that were incubated for 24 h in the absence of Aβ (in 0.2% DMSO vehicle); this small amount of cleaved tau in untreated neurons reflects the few neurons undergoing apoptosis in the absence of Aβ treatment. The time course of Aβ-induced tau cleavage was similar to that of poly(ADP-ribose) polymerase (PARP) (Fig. 2 A Lower), a substrate of multiple caspases including caspase-3 and -7 (31, 32), suggesting that the Aβ-induced proteolysis of tau might be mediated by caspases. TauC3 also detected truncated tau in hippocampal neurons treated with Aβ (1–40) but not in untreated hippocampal neurons (data not shown). To determine unambiguously the role of caspases in this proteolytic event, we preincubated cortical neurons in the presence and absence of 50 μM zVAD-fmk, a broad-spectrum caspase inhibitor, and then treated them with 10 μM fibrillar Aβ for an additional 3 h. As shown in Fig. 2B, zVAD-fmk potently inhibited Aβ-induced cleavage of tau and PARP (with Tau1, truncated tau partly comigrated with full-length tau). A readily detectable amount of Triton X-100-insoluble truncated tau appeared to colocalize with microtubules in Aβ-treated neurons (Fig. 2C). Importantly, a significant subset of TauC3-positive neurons treated with Aβ had condensed or fragmented nuclei indicative of apoptosis, although most TauC3-positive neurons had intact nuclei (Fig. 2D). In contrast, Aβ-treated neurons that were Tau C3-negative rarely had apoptotic nuclei (Fig. 2D). These results suggest that tau proteolysis likely precedes the apoptotic destruction of the nucleus. Taken together, these findings demonstrate that tau is selectively and rapidly cleaved at Asp421 by caspases in cortical neurons during the induction of apoptosis by Aβ.
Fig. 2.
Treatment of cortical neurons with fibrillar Aβ induces tau proteolysis at Asp421. (A) Primary cultures of cortical neurons prepared from embryonic day-18 rats were treated with 10 μM fibrillar Aβ (1–42) for 0–24 h, and lysates were immunoblotted with TauC3 (Upper) or PARP (Lower) mAbs. (B) Cortical neurons were preincubated in the absence or presence of 50 μM zVAD-fmk for 1 h and then treated for an additional 3 h with 10 μM fibrillar Aβ (1–42) prior to immunoblot analysis. (C) Laser-scanning confocal images of untreated control neurons (Upper) or a neuron treated with fibrillar Aβ (Lower) analyzed by indirect immunofluorescence with tubulin or TauC3 mAbs; the merged images are shown (Right). (Bar=10 μm.) (D) Tau proteolysis at Asp421 is an early event in Aβ-induced neuronal apoptosis. Primary cortical neurons were treated with 20 μM Aβ for 0–12 h, and TauC3-negative (black bars) or TauC3-positive neurons (hatched bars) were scored for apoptosis by nuclear morphology (mean ± SEM).
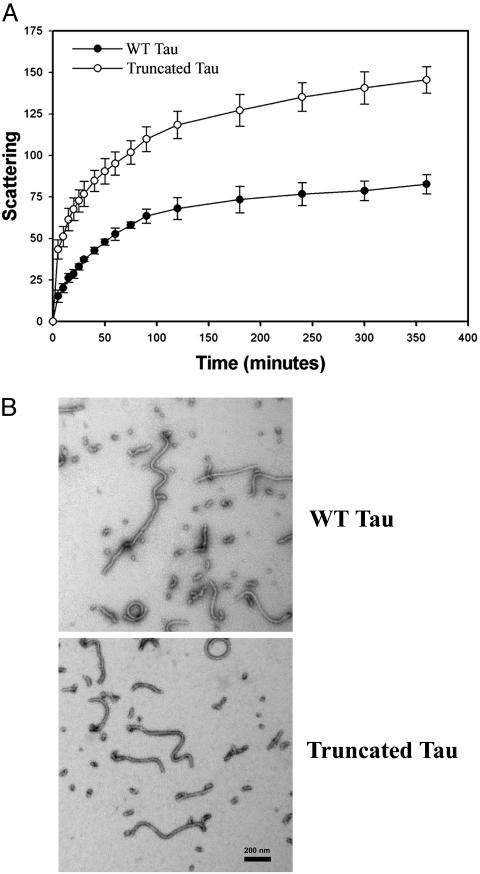
Truncation of Tau at Asp421 Increases the Rate and Extent of Tau Filament Assembly in Vitro. Because we have previously demonstrated that the C terminus of tau inhibits its assembly into polymeric filaments in vitro (12), we hypothesized that caspase cleavage of tau would promote tau filament assembly. To test this hypothesis, we compared the kinetics of filament assembly in vitro of WT and truncated tau proteins by laser light scattering. As shown in Fig. 3A, truncated tau assembled into filaments more rapidly than WT tau after stimulation of tau polymerization with arachidonic acid. Moreover, because the scattering intensity is proportional to the total mass of polymeric filaments (30), truncated tau formed approximately twice the filament mass as did WT tau. Importantly, the tau filaments (≈15 nm in diameter) formed by truncated tau (Fig. 3B Lower) are morphologically similar to those formed by WT tau (Fig. 3B Upper) and resemble the unpaired straight filaments present in NFTs. These findings suggest that truncated tau might promote the formation of NFTs and other neurofibrillar pathologies in AD via its enhanced propensity to assemble into filaments and/or by a diminished tendency to disassemble from these polymeric filaments.
Fig. 3.
Truncation of tau at Asp421 increases the rate and extent of tau filament assembly in vitro.(A) Kinetic analysis of the assembly of recombinant WT or truncated (1–421) tau proteins into tau filaments in vitro. Tau polymerization was induced by the addition of 75 μM arachidonic acid to 4 μM recombinant WT or truncated tau proteins and measured by right-angle laser light scattering. Scattering intensities were measured on a scale of 0–256 and are proportional to the total mass of polymerized tau filaments (30). The data are presented as the mean ± SEM of three independent experiments. (B) Representative electron micrographs of tau filaments assembled in vitro by recombinant WT tau (Upper) or truncated tau (Lower) proteins. (Bar = 200 nm.)
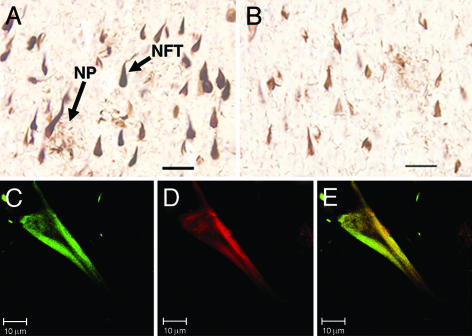
TauC3 Labels the Neurofibrillar Pathologies of AD. When used to probe tissue sections taken from AD brains, TauC3 detected NFTs and dystrophic neurites forming neuritic plaques in the CA1 layer of the hippocampus (Fig. 4A). The pattern of immunostaining with TauC3 was similar to that observed with Tau5 in adjacent sections (Fig. 4B), indicating that at least a subset of those neurons exhibiting pathological aggregates of tau contain tau truncated at the caspase cleavage site. Indeed, TauC3 immunoreactivity (Fig. 4C) frequently colocalized with Thiazin red staining (Fig. 4D) in NFTs examined by confocal microscopy (merged image in Fig. 4E). The latter reagent is a reliable marker of β-pleated sheet conformation in the neurofibrillar pathology of AD (33), implying that caspase-truncated tau often resides within the polymeric filaments.
Fig. 4.
The characteristic neurofibrillar pathologies of AD contain tau truncated at Asp421. The CA1 hippocampal region from a patient with AD was analyzed by immunohistochemistry by using TauC3 (A) and Tau5 (B) mAbs. NFT and dystrophic neurites forming neuritic plaques (NP), two hallmark pathologic features of AD, are indicated. (Bars = 50 μm.) In C–E, laser-scanning confocal images of a single NFT from a patient with AD were stained with TauC3 (C) or Thiazin red (D); the latter is a marker for compact filaments (33). The merged image is shown in E. (Bar = 10 μm.)
Discussion
We have demonstrated that proteolytic cleavage of tau at Asp421 by multiple caspases generates a truncated tau protein (amino acids 1–421) with a “gain-of-function” phenotype, i.e., enhanced filament assembly in vitro. Importantly, the DMVD421-S422 caspase cleavage motif in tau has been strictly conserved from mouse to humans but is not present in other members of the tau superfamily, including MAP2, which otherwise has a high degree of homology to tau in its C terminus (34). Caspase proteolysis of tau at Asp421 removes 20 amino acids from its C terminus, a domain we have previously demonstrated to inhibit tau filament assembly in vitro (12). We have postulated that this inhibitory effect is due to tau's C terminal tail folding back on the microtubule-binding repeat region and disrupting the latter region's polymerization-promoting effects (12). Together with our observation that this specific proteolytic event occurs in cortical neurons treated with Aβ and in the hallmark neurofibrillar pathologies of AD, these findings suggest a mechanism linking extracellular amyloid plaques and intracellular NFTs: Aβ-induced caspase activation and the subsequent proteolysis of tau at Asp421 that promotes pathological filament assembly and/or inhibits disassembly, thereby stabilizing filaments. Indeed, the recent demonstration that tau-deficient neurons are resistant to Aβ-induced neurotoxicity suggests that tau may be a critical downstream target for neurodegenerative processes initiated by amyloid (35).
We have also shown that detergent-insoluble truncated tau appears to colocalize, at least to some extent, with microtubules, an observation consistent with the previously reported binding of C-terminal deletion mutants of tau to microtubules in vitro, albeit with varying isoform-dependent affinities (36, 37). Together with our observation that tau truncation promotes filament assembly, these latter findings suggest that the assembly of truncated tau into pathological filaments may be initiated on microtubules, as has been previously suggested for WT tau in vitro (38). Alternatively, our results do not exclude the possibility that a cytosolic pool of truncated tau may initiate polymerization. Clearly, the regulation of tau assembly in diseased neurons in vivo is likely to be quite complex. Indeed, because Aβ treatment also activates the cdk5/p25 complex and other kinases that phosphorylate tau at multiple sites in its C terminus, including Ser422 at the caspase cleavage site (7, 35, 39), it will be important in future studies to examine whether phosphorylation of tau alters its sensitivity to caspase cleavage and to determine the relative contribution of these proteolytic and phosphorylation events in promoting tau filament assembly.
In addition, our results suggest that caspase-cleaved tau appears to reside in a highly stable detergent-insoluble fraction in AD brain. Although TauC3 readily detects caspase-cleaved tau in NFTs by immunostaining, we have been unable to detect truncated tau by immunoblotting SDS-soluble lysates of AD brain or sarkosyl-insoluble paired helical filament preparations (data not shown). These findings are reminiscent of those seen with the MN423 mAb, which detects tau truncated at Glu391 (40). Similar to TauC3, the MN423 mAb does not immunoblot tau truncated at Glu391 in AD brain lysates, but it does recognize NFTs by immunostaining (33, 40). Hence, these two truncation-specific antibodies appear to detect an extremely detergent-insoluble subset of tau polymers in NFTs. It remains to be determined whether tau cleavage at Asp421 necessarily precedes truncation at Glu391 in AD brain or whether these distinct proteolytic events are functionally unrelated.
It was previously shown that tau is cleaved by caspase-3 at Asp421 in vitro and in neurons induced to undergo apoptosis by treatment with hypokalemia or the protein kinase inhibitor staurosporine (17–19). The current findings directly implicate this proteolytic event in the pathogenesis of AD, link amyloid exposure to tau cleavage, and demonstrate the functional consequences of tau truncation in promoting its pathological assembly into filaments. Indeed, our observation that tau is specifically cleaved at Asp421 in the neurofibrillar pathologies of AD provides unequivocal evidence that caspases are activated in degenerating neurons in this disease. Of note, the percentage of tangles stained with the TauC3 antibody appears to vary with the brain region assayed and the stage of the disease in a way that suggests that caspase cleavage of tau may be a relatively early event in AD (F.G.-S., R.W.B., A.L.G., V.L.C., and L.I.B., unpublished results). In support of this notion is our finding that tau is rapidly cleaved (within 2 h) in Aβ-treated neurons prior to the apoptotic disassembly of the nucleus. Taken together, the results presented here argue strongly for a pathogenic role of caspase-cleaved tau in AD.
Acknowledgments
We are indebted to members of the Cryns and Binder laboratories for their critical reading of this manuscript, to Dr. R. Talanian for providing recombinant caspases, and to Dr. A. Ferreira for help with the cortical neuron cultures. This work was supported in part by a grant from the Alzheimer's Association (to V.L.C.), by National Institutes of Health Grants NS31957 (to V.L.C.) and AG09466 (to L.I.B.), by a fellowship (Programa de Mejoramiento de la Calidad y la Equidad de la Educación Superior AUS0006, Chile; to A.Z.), and by an institutional research grant to Northwestern University from the Howard Hughes Medical Institute (to V.L.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AD, Alzheimer's disease; NFTs, neurofibrillary tangles; Aβ, amyloid-β.
References
- 1.Yuan, J. & Yankner, B. A. (2000) Nature 407, 802–809. [DOI] [PubMed] [Google Scholar]
- 2.Trojanowski, J. Q., Schmidt, M. L., Shin, R. W., Bramblett, G. T., Rao, D. & Lee, V. M. (1993) Brain Pathol. 3, 45–54. [DOI] [PubMed] [Google Scholar]
- 3.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 4.Hutton, M., Lendon, C. L., Rizzu, P., Baker, M., Froelich, S., Houlden, H., Pickering-Brown, S., Chakraverty, S., Isaacs, A., Grover, A., et al. (1998) Nature 393, 702–705. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini, M. G., Murrell, J. R., Goedert, M., Farlow, M. R., Klug, A. & Ghetti, B. (1998) Proc. Natl. Acad. Sci. USA 95, 7737–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, L. N., Poorkaj, P., Wszolek, Z., Geschwind, D. H., Nasreddine, Z. S., Miller, B., Li, D., Payami, H., Awert, F., Markopoulou, K., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 13103–13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotz, J., Chen, F., van Dorpe, J. & Nitsch, R. M. (2001) Science 293, 1491–1495. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, J., Dickson, D. W., Lin, W. L., Chisholm, L., Corral, A., Jones, G., Yen, S. H., Sahara, N., Skipper, L., Yager, D., et al. (2001) Science 293, 1487–1491. [DOI] [PubMed] [Google Scholar]
- 9.Grundke-Iqbal, I., Iqbal, K., Tung, Y. C., Quinlan, M., Wisniewski, H. M. & Binder, L. I. (1986) Proc. Natl. Acad. Sci. USA 83, 4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak, M., Kabat, J. & Wischik, C. M. (1993) EMBO J. 12, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso, A. C., Zaidi, T., Grundke-Iqbal, I. & Iqbal, K. (1994) Proc. Natl. Acad. Sci. USA 91, 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraha, A., Ghoshal, N., Gamblin, T. C., Cryns, V., Berry, R. W., Kuret, J. & Binder, L. I. (2000) J. Cell Sci. 113, 3737–3745. [DOI] [PubMed] [Google Scholar]
- 13.Alonso, A., Zaidi, T., Novak, M., Grundke-Iqbal, I. & Iqbal, K. (2001) Proc. Natl. Acad. Sci. USA 98, 6923–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gervais, F. G., Xu, D., Robertson, G. S., Vaillancourt, J. P., Zhu, Y., Huang, J., LeBlanc, A., Smith, D., Rigby, M., Shearman, M. S., et al. (1999) Cell 97, 395–406. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa, T., Zhu, H., Morishima, N., Li, E., Xu, J., Yankner, B. A. & Yuan, J. (2000) Nature 403, 98–103. [DOI] [PubMed] [Google Scholar]
- 16.Troy, C. M., Rabacchi, S. A., Friedman, W. J., Frappier, T. F., Brown, K. & Shelanski, M. L. (2000) J. Neurosci. 20, 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canu, N., Dus, L., Barbato, C., Ciotti, M. T., Brancolini, C., Rinaldi, A. M., Novak, M., Cattaneo, A., Bradbury, A. & Calissano, P. (1998) J. Neurosci. 18, 7061–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasulo, L., Ugolini, G., Visintin, M., Bradbury, A., Brancolini, C., Verzillo, V., Novak, M. & Cattaneo, A. (2000) J. Neurochem. 75, 624–633. [DOI] [PubMed] [Google Scholar]
- 19.Chung, C. W., Song, Y. H., Kim, I. K., Yoon, W. J., Ryu, B. R., Jo, D. G., Woo, H. N., Kwon, Y. K., Kim, H. H., Gwag, B. J., et al. (2001) Neurobiol. Dis. 8, 162–172. [DOI] [PubMed] [Google Scholar]
- 20.Cryns, V. L., Byun, Y., Rana, A., Mellor, H., Lustig, K. D., Ghanem, L., Parker, P. J., Kirschner, M. W. & Yuan, J. (1997) J. Biol. Chem. 272, 29449–29453. [DOI] [PubMed] [Google Scholar]
- 21.Cryns, V. L., Bergeron, L., Zhu, H., Li, H. & Yuan, J. (1996) J. Biol. Chem. 271, 31277–31282. [DOI] [PubMed] [Google Scholar]
- 22.Binder, L. I., Frankfurter, A. & Rebhun, L. I. (1985) J. Cell Biol. 101, 1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmel, G., Mager, E. M., Binder, L. I. & Kuret, J. (1996) J. Biol. Chem. 271, 32789–32795. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez, A., Toro, R., Cáceres, A. & Maccioni, R. B. (1999) FEBS Lett. 459, 421–426. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez, A., Muñoz, J. P. & Maccioni, R. B. (2001) Exp. Cell Res. 64, 266–274. [DOI] [PubMed] [Google Scholar]
- 26.LoPresti, P., Szuchet, S., Papasozomenos, S. C., Zinkowski, R. P. & Binder, L. I. (1995) Proc. Natl. Acad. Sci. USA 92, 10369–10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thurston, V. C., Pena, P., Pestell, R. & Binder, L. I. (1997) Cell Motil. Cytoskeleton 38, 100–110. [DOI] [PubMed] [Google Scholar]
- 28.Ghoshal, N., Garcia-Sierra, F., Fu, Y., Beckett, L. A., Mufson, E. J., Kuret, J., Berry, R. W. & Binder, L. I. (2001) J. Neurochem. 77, 1372–1385. [DOI] [PubMed] [Google Scholar]
- 29.Kamradt, M. C., Chen F., Sam S. & Cryns, V. L. (2002) J. Biol. Chem. 277, 38731–38736. [DOI] [PubMed] [Google Scholar]
- 30.Gamblin, T. C., King, M. E., Dawson, H., Vitek, M. P., Kuret, J., Berry, R. W. & Binder, L. I. (2000) Biochemistry 39, 6136–6144. [DOI] [PubMed] [Google Scholar]
- 31.Lazebnik, Y. A., Kaufmann, S. H., Desnoyers, S., Poirier, G. G. & Earnshaw, W. C. (1994) Nature 371, 346–347. [DOI] [PubMed] [Google Scholar]
- 32.Slee, E. A., Adrain, C. & Martin, S. J. (2001) J. Biol. Chem. 276, 7320–7326. [DOI] [PubMed] [Google Scholar]
- 33.Mena, R., Edwards, P. C., Perez-Olvera, O. & Wischik, C. M. (1995) Acta Neuropathol. 89, 50–56. [DOI] [PubMed] [Google Scholar]
- 34.Dammerman, M., Yen, S. H. & Shafit-Zagardo, B. (1989) J. Neurosci. Res. 24, 487–495. [DOI] [PubMed] [Google Scholar]
- 35.Rapoport, M., Dawson, H. N., Binder, L. I., Vitek, M. P. & Ferreira, A. (2002) Proc. Natl. Acad. Sci. USA 99, 6364–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goode, B. L. & Feinstein, S. C. (1994) J. Cell Biol. 124, 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goode, B. L., Chau, M., Denis, P. E. & Feinstein, S. C. (2000) J. Biol. Chem. 275, 38182–38189. [DOI] [PubMed] [Google Scholar]
- 38.Ackmann, M., Wiech, H. & Mandelkow, E. (2000) J. Biol. Chem. 275, 30335–30343. [DOI] [PubMed] [Google Scholar]
- 39.Patrick, G. N., Zukerberg, L., Nikolic, M., de la Monte, S., Dikkes, P. & Tsai, L. H. (1999) Nature 402, 615–622. [DOI] [PubMed] [Google Scholar]
- 40.Harrington, C. R., Mukaetova-Ladinska, E. B., Hills, R., Edwards, P. C., Montejo de Garcini, E., Novak, M. & Wischik, C. M. (1991) Proc. Natl. Acad. Sci. USA 88, 5842–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]